Back to Journals » International Journal of Women's Health » Volume 15
Study on the Relationship Between PTPRO Methylation in Plasma and Efficacy Neoadjuvant Chemotherapy in Patients with Early Breast Cancer
Authors Liu XW, Hong MJ, Qu YY
Received 28 June 2023
Accepted for publication 7 October 2023
Published 2 November 2023 Volume 2023:15 Pages 1673—1680
DOI https://doi.org/10.2147/IJWH.S428038
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Elie Al-Chaer
Xiang-Wei Liu,1 Mei-Juan Hong,2 Yan-Yu Qu3
1Department of Breast Surgery, The First People’s Hospital of Foshan, Foshan, 528000, People’s Republic of China; 2Ultrasound Diagnosis and Treatment Center, The First People’s Hospital of Foshan, Foshan, 528000, People’s Republic of China; 3Departmentof Pathology, The Second People’s Hospital of Foshan, Foshan, 528000, People’s Republic of China
Correspondence: Yan-Yu Qu, Email [email protected]
Objective: This study aimed to explore the correlation between PTPRO methylation in plasma and the efficacy of neoadjuvant chemotherapy (NAC) for early breast cancer (BC).
Methods: Eighty-two patients with early BC undergoing NAC were included. PTPRO methylation status in plasma before and after NAC was detected using methylation-specific PCR and the relationship between PTPRO methylation and NAC efficacy was analyzed.
Results: The rate of pathologic complete response (pCR) was only 25.0% (12/48) in patients with positive PTPRO methylation result before NAC, but 61 0.8% (21/34) in pre-NAC methylation-negative patients (OR = 0.24, 95% CI: 0.09– 0.65, P = 0.005). In addition, the pCR rate was 12.1% (4/33) in patients with positive PTPRO methylation results both before and after NAC, but 53.3% (8/15) in patients with pre-NAC positive methylation and post-NAC negative methylation results (OR = 0.12, 95% CI: 0.03– 0.52, P = 0.004).
Conclusion: Plasma PTPRO methylation is a potential biomarker for predicting the efficacy of NAC in early BC.
Keywords: breast cancer, neoadjuvant chemotherapy, PTPRO, methylation, liquid biopsy
Introduction
Breast cancer (BC) is the most prevalent malignant cancer among Chinese women, seriously threatening women’s health and lives.1 Chemotherapy is an important treatment for BC. Neoadjuvant chemotherapy (NAC) is a systemic therapy with cytotoxic agents administered prior to surgical treatment, mainly for patients with stage II and III BC and inflammatory BC.2 The maximum lesions diameter and clinical stage of breast cancer patients decreased significantly after NAC than before, while the apparent diffusion coefficient increased considerably.3 Meanwhile, NAC could monitor the sensitivity of patients to chemotherapy regimens, thus converting inoperable BC into operable BC and giving non-breast-conserving patients the chance of breast-conserving. NAC can also. A good prognosis is suggested if patients achieve a pathologic complete response (pCR) after completing NAC.4 However, NAC is not effective for all BC patients, with pCR rates typically ranging from 5% to 38%.5 A significant proportion of patients are insensitive to NAC and even experience tumor progression during NAC. Consequently, the timing of surgery is delayed, and the treatment opportunity is missed.6 Therefore, the prediction of whether NAC is effective for BC patients is highly meaningful for the precise treatment of BC.
Epigenetics is a heritable molecular mechanism that regulates gene expression without altering the actual sequence of DNA.7 DNA methylation, histone modification, chromatin remodeling, and RNA-mediated targeting regulate many biological processes that are critical to cancer development.8 A recent review by Sher et al provides a comprehensive overview of the important role of multiple epigenetic disorders in the progression and survival of BC.9 A clinical study by Paydar et al showed that compared with benign breast tumors, malignant breast tumor samples displayed aberrant patterns of histones marks with hypomethylation of H4K20 and hypoacetylation of H3K18 and promoter methylation, and there was a negative significant correlation between H3K9ac levels and tumor size, indicating gene promoter hypermethylation along with histone modification may play an important role in the progression and prognosis of breast cancer.10 In addition, several clinical studies have shown the role of DNA methylation in early diagnosis of breast cancer,11 chemotherapy response,12 immune infiltration and survival.13 In conclusion, epigenetic modification plays an important role in the progression and prognosis of breast cancer.
DNA methylation is an epigenetic mechanism, involving the transfer of methyl groups to cytosine bases by DNA methyltransferases.14 DNA methylation is a reversible process, occurring mainly in dinucleotide-rich CpG islands.14 Methylation in the promoter region of tumor-suppressor genes can lead to silencing of anti-oncogenes.15 Methylated circulating tumor DNA (met-ctDNA) can be detected in 20% to 47% of early BC,16 with a significantly higher positive rate than that of traditional tumor markers (eg CEA, CA15-3). Met-ctDNA has been used to predict the response of BC to chemotherapy and has shown good specificity and sensitivity.17
Protein Tyrosine Phosphatase Receptor-type O (PTPRO) belongs to the R3 receptor-type protein tyrosine phosphatase family, which functions as an anti-oncogene in a variety of tumors.18,19 PTPRO regulates multiple kinases and pathways (AKT/mTOR/SREBP1/ACC1 pathway, MAPK/PPARα/ACOX1 pathway, TLR4/NF-κB pathway, etc) and has been implied to play a regulatory role in immune cell infiltration in various cancers.20–22 Moreover, studies have shown that PTPRO gene promoter methylation and functional inhibition are frequently present in BC.23 A study by Huang et al24 demonstrated 74.3% of BC tissues had PTPRO methylation; when PTPRO methylation was observed in cancer tissues, methylated PTPRO was detectable in the plasma of 62.1% of patients. Similarly, Li et al25 revealed 54.1% of BC tissues had PTPRO methylation and the overall survival rate was reduced in patients with higher methylation levels; when PTPRO methylation was present in cancer tissues, 54.7% of patients exhibited PTPRO methylation in plasma. At the cellular level, PTPRO phosphatase activity shortens the half-life of epidermal growth factor receptor 2 (HER2) by increasing intracellular degradation of HER2.26 These studies suggest that PTPRO methylation in plasma is a promising BC biomarker.
Based on the above research background, this study is intended to examine PTPRO methylation in the plasma from BC patients before and after NAC and analyze its relationship with NAC efficacy. We aimed to explore whether PTPRO methylation in plasma could be used as a potential biomarker.
Study Subjects and Methods
Study Subjects
Eighty-two patients with early BC admitted to The First People’s Hospital of Foshan between September 2016 and June 2018 were selected. The study was approved by the Ethics Committee of the First People’s Hospital of Foshan and conducted in accordance with the approval guidelines (L-2023-5).
Inclusion and Exclusion Criteria
Inclusion criteria were as follows: (1) women aged >18 years; (2) a diagnosis of early intensive BC confirmed by histopathologic examination; (3) women who gave informed consent to NAC; (4) women who gave informed consent and volunteered to participate in this study.
Exclusion criteria were as follows: (1) severe cardiac, hepatic, or renal insufficiency or a combination of other high-risk diseases; (2) history of neurological disorders, psychiatric disorders, and recent use of psychiatric medications; (3) coagulation disorders or other contraindications to surgery; (4) incomplete clinical data.
Treatment Methods
Chemotherapy Regimens
All patients received four cycles of anthracyclines (liposomal epirubicin or liposomal doxorubicin), followed by four cycles of taxanes (docetaxel or paclitaxel or albumin-bound paclitaxel). Chemotherapy was given every three weeks and recorded as a cycle. Patients received a total of eight chemotherapy cycles. For patients with HER2-positive BC, trastuzumab (Herceptin) targeted therapy was received in conjunction with taxanes-based chemotherapy.
Evaluation of Chemotherapy Efficacy
Postoperative specimens from post-NAC patients were collected for pathological examination to clarify whether pCR was achieved. The pCR was defined as the absence of invasive BC at the primary site and a negative result of regional lymph nodes.
Extraction of Circulating DNA
Before NAC and before surgery, 5 mL of venous blood from each patient was collected in anticoagulant tubes and mixed. Then the tubes were placed on ice, followed by centrifugation (4 °C, 1000 × g, 15 min). The supernatant plasma was collected, divided, and stored at −80 °C or on ice until use. Circulating DNA in plasma was extracted according to the instructions of the Circulating DNA Nucleic Acid Extraction Kit (AmoyDx, Xiamen, China).
Bisulfite Modification of DNA and Reference Selection
Bisulfite modification of plasma DNA was performed using EZ DNA Methylation-Gold TM Kit (Zymo Research, Irvine, CA, USA). The modified DNA was stored at −20 °C. PTPRO-methylated MCF-7 human cancer cell line and PTPRO-unmethylated NE-2 immortalized normal cell line were used as positive and negative controls, respectively.18
Methylation-Specific PCR
Before NAC, the methylation-specific PCR (MSP) was used to detect PTPRO methylation status in the plasma of all patients. For patients with a positive result of PTPRO methylation in pre-NAC plasma, MSP was carried out again after NAC. For patients with a negative result of PTPRO methylation in pre-NAC plasma, PTPRO methylation status in post-NAC plasma was not tested again. Primers synthesized by Invitrogen (Shanghai, China) were added, and the primer sequences were referred to a published literature.27 A 20 μL MSP reaction system was built as follows: 2 × premix 10 μL, forward and reverse primers (10 μmol/L) 0.4 μL each, DNA 2 μL, and ddH2O 7.2 μL. The reaction conditions were: pre-denaturation at 94 °C for 2 min; 95 °C for 45s, 60 °C for 45s, 72 °C for 45s, 35 cycles; extension at 72 °C for 10 min. PCR products were electrophoresed on 2% agarose gels with ethidium bromide. Images were acquired using a bio-rad UV imaging system (Bio-Rad, Hercules, CA, USA) and Image J software (National Institutes of Health, Bethesda, MD, USA) was used for grayscale value analysis and result calculation. Each sample was amplified simultaneously with methylated primers and unmethylated primers. If the gene promoter was aberrantly methylated, it is denoted by M; if it was not methylated, it is denoted by U. The presence of only the M band indicated complete methylation, and that of both M and U bands indicated partial methylation; both partial and complete methylation were considered as methylation-positive. The presence of only the U band indicated a methylation-negative state.
Statistical Analysis
SPSS 21.0 software (SPSS, Chicago, IL, USA) was used for statistical analysis, and P < 0.05 was considered statistically significant. Measurement data were expressed as means ± standard deviation (SD) and count data were expressed as n (%). Univariate analysis was performed first. The t-test was used for comparison between two groups for measurement data, and χ2 test was used for the count data. Factors with P < 0.05 in univariate analysis were further included in logistic regression analysis, and the odds ratio (OR) was calculated.
Results
General Clinical Data of Patients with Early Breast Cancer
A total of 82 patients with early BC were included in this study, aged from 34 to 65 years (average age: 47.9 ± 7.8 years). Preoperative clinical tumor stages were cT1 – 4 in 7, 59, 6, and 10 patients, respectively; preoperative clinical lymph node stages were cN0 – 3 in 19, 53, 8 and 2 cases, respectively. In terms of pathological classification, 79 patients had invasive ductal carcinoma, and 3 patients had other pathological types. Pathological histology was grade II in 64 cases and grade III in 18 cases. There were 34 estrogen receptor (ER)-negative patients and 48 ER-positive patients; there were 27 progesterone receptor (PR)-negative patients and 55 PR-positive patients; there were 45 HER2-negative patients and 37 HER2-positive patients. Twenty-eight (34.1%) BC patients with Ki67 ≤ 30% and 54 (65.9%) with Ki67 > 30%. Before NAC, plasma PTPRO methylation was negative in 34 (41.5%) patients, and it was positive in 48 (58.5%) patients (Table 1).
 |
Table 1 Univariate Analysis of Clinical Data and NAC Efficacy in Patients with Breast Cancer |
Univariate Analysis of NAC Efficacy in Patients with Early Breast Cancer
We evaluated the efficacy of NAC in BC patients using pathological response. Seventy-eight BC patients underwent mastectomy after NAC, and the other four patients underwent breast-conserving surgery; all patients received axillary lymph node dissection. pCR was achieved in 33 (40.2%), but not in the other 49 (59.8%) patients. Univariate analysis found that PTPRO methylation status before NAC (P = 0.001) and HER2 status (P = 0.01) were correlated factors for post-NAC pCR in patients with early BC. However, age, cT, cN, pathological type, histological grade, ER, PR, Ki67 were not significantly correlated with post-NAC pCR (Table 1).
Logistic Regression Analysis of NAC Efficacy in Patients with Early Breast Cancer
Further logistic regression analysis was performed (Table 2). The pCR rate was only 25.0% (12/48) in early BC patients with methylated PTPRO (methylation-positive) before NAC, while the pCR rate was 61.8% (21/34) in those with unmethylated PTPRO (methylation-negative) before NAC (OR = 0.24, 95% CI: 0.09–0.65, P = 0.005). The pCR rate was 56.8% (21/37) in HER2-positive patients and 26.7% (12/45) in HER-negative patients (OR = 2.95, 95% CI: 1.11–7.87, P = 0.03). The results suggested that plasma PTPRO methylation status before NAC and HER2 status were relevant factors for post-NAC pCR in BC patients.
 |
Table 2 Logistic Regression Analysis of NAC Efficacy in Breast Cancer Patients |
Univariate Analysis of NAC Efficacy in Early Breast Cancer Patients with Positive Pre-NAC PTPRO Methylation
After NAC treatment, we evaluated the factors affecting the efficacy of NAC in BC patients with PTPRO methylation-positive results in plasma (Table 3). Age, cT, cN, pathological type, histological grade, ER, PR, HER2 and Ki67 were not significantly associated with post-NAC pCR. Only PTPRO methylation status after NAC was a correlate of pCR (OR = 0.12, 95% CI: 0.03–0.52, P = 0.004). We found that after NAC treatment, PTPRO methylation status in plasma changed from positive to negative in 15 of 48 (31.3%) patients with early BC, and the pCR rate reached 53.3% (8/15). Plasma PTPRO methylation status remained positive in the other 33 (68.7%) patients and their post-NAC pCR rate was only 12.1% (4/33).
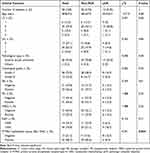 |
Table 3 Univariate Analysis of NAC Efficacy in Early Breast Cancer Patients with Positive Pre-NAC PTPRO Methylation |
Discussion
NAC is increasingly used in treating BC and has gradually become a new mode in BC treatment.28 However, not all BC patients can benefit from NAC, and the pCR rate of early BC patients after NAC treatment in this study was only 40.2% (33/82). Therefore, it is necessary to identify patients who are not sensitive to NAC in advance to ensure more precise treatment. Liquid biopsy refers to the extraction of patient’s body fluid (such as blood) by non-invasive means and analysis of tumor-derived biomarkers (such as met-ctDNA, ctDNA). Liquid biopsy is applied to early diagnosis, prognosis evaluation, and monitoring of treatment effects. Liquid biopsy has the advantages of non-invasiveness, repeatability, timeliness, which can realize the standardized process of diagnosis.29 The use of liquid biopsy for predicting the efficacy of NAC in BC is also increasingly studied.30
The present study showed that the pre-NAC positive rate of plasma PTPRO methylation in BC patients was 58.5%. This positive rate is basically equal to the positive rate of 54.1%25 – 62.2%24 reported in previous research, but higher than the positive rate of 17.0% – 46.2% for traditional tumor markers (CEA, CA125, CA153).30 Pre-NAC plasma PTPRO methylation status was an independent influencing factor of pCR (OR = 0.24, 95% CI: 0.09–0.65, P = 0.005). The pCR rate was only 25.0% in patients with a positive result of pre-NAC PTPRO methylation, which was significantly lower than the pCR rate of 61.8% in methylation-negative patients. This suggests that plasma PTPRO methylation is a promising predictor of NAC efficacy in BC.
This study showed that changes in plasma PTPRO methylation status before and after NAC were also significantly associated with NAC efficacy (OR = 0.12, 95% CI: 0.03–0.52, P = 0.004). The pCR rate was only 12.1% in patients with positive PTPRO methylation results before and after NAC, but 53.3% in patients with pre-NAC methylation-positive and post-NAC methylation-negative results. This result has some implications for the selection of surgical modality for patients after NAC. Patients with positive PTPRO methylation of the PTPRO gene both before and after NAC have a low pCR rate and thus breast-conserving surgery should be carefully selected.
This study also has some limitations. The number of cases was small and there may have been bias. We failed to detect the pre-NAC methylation status of PTPRO gene in the primary tumor specimens obtained by needle biopsy and did not study the concordance between the methylation of PTPRO gene in the primary tumor and blood. Additionally, the effect of plasma PTPRO methylation on the long-term survival of patients has not been observed because of short follow-up time. In future studies, we will continue to expand the sample size and observe the effect of PTPRO methylation on patient survival.
Conclusion
In conclusion, plasma PTPRO methylation status is a valid predictor of NAC efficacy in BC. The pCR rate is low in patients with positive methylation results before NAC, especially in patients with positive methylation results both before and after NAC. This study provides a new option for NAC efficacy prediction and precision medicine of BC. It is expected that the predictive efficiency can be further improved by combining other predictors in future studies.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Ethics Approval and Consent to Participate
The study was approved by the Ethics Committee of the First People’s Hospital of Foshan and conducted in accordance with the approval guidelines (L-2023-5). All patients provided written informed consent prior to enrollment in the study. My study complies with the Declaration of Helsinki.
Disclosure
The authors declare that they have no competing interests.
References
1. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
2. Derks MGM, van de Velde CJH. Neoadjuvant chemotherapy in breast cancer: more than just downsizing. Lancet Oncol. 2018;19(1):2–3. doi:10.1016/S1470-2045(17)30914-2
3. Liu Y, Wu M, Tan W, Gong J, Ma J. Efficacy evaluation of neoadjuvant chemotherapy in breast cancer by MRI. Contrast Media Mol Imaging. 2022;2022:4542288. doi:10.1155/2022/4542288
4. Fayanju OM, Ren Y, Thomas SM, et al. The clinical significance of breast-only and node-only Pathologic Complete Response (pCR) After Neoadjuvant Chemotherapy (NACT): a Review of 20,000 breast cancer patients in the National Cancer Data Base (NCDB). Ann Surg. 2018;268(4):591–601. doi:10.1097/SLA.0000000000002953
5. von Minckwitz G, Untch M, Nuesch E, et al. Impact of treatment characteristics on response of different breast cancer phenotypes: pooled analysis of the German neo-adjuvant chemotherapy trials. Breast Cancer Res Treat. 2011;125(1):145–156. doi:10.1007/s10549-010-1228-x
6. Caudle AS, Gonzalez-Angulo AM, Hunt KK, et al. Predictors of tumor progression during neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28(11):1821–1828. doi:10.1200/JCO.2009.25.3286
7. Holliday R. Epigenetics: an overview. Dev Genet. 1994;15(6):453–457. doi:10.1002/dvg.1020150602
8. Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi:10.1016/j.cell.2012.06.013
9. Sher G, Salman NA, Khan AQ, et al. Epigenetic and breast cancer therapy: promising diagnostic and therapeutic applications. Semin Cancer Biol. 2022;83:152–165. doi:10.1016/j.semcancer.2020.08.009
10. Paydar P, Asadikaram G, Nejad HZ, et al. Epigenetic modulation of BRCA-1 and MGMT genes, and histones H4 and H3 are associated with breast tumors. J Cell Biochem. 2019;120(8):13726–13736. doi:10.1002/jcb.28645
11. Widschwendter M, Evans I, Jones A, et al. Methylation patterns in serum DNA for early identification of disseminated breast cancer. Genome Med. 2017;9(1):115. doi:10.1186/s13073-017-0499-9
12. Hartmann O, Spyratos F, Harbeck N, et al. DNA methylation markers predict outcome in node-positive, estrogen receptor-positive breast cancer with adjuvant anthracycline-based chemotherapy. Clin Cancer Res. 2009;15(1):315–323. doi:10.1158/1078-0432.CCR-08-0166
13. Li D, Zhao W, Zhang X, et al. NEFM DNA methylation correlates with immune infiltration and survival in breast cancer. Clin Epigenetics. 2021;13(1):112. doi:10.1186/s13148-021-01096-4
14. Subramaniam D, Thombre R, Dhar A, Anant S. DNA methyltransferases: a novel target for prevention and therapy. Front Oncol. 2014;4:80. doi:10.3389/fonc.2014.00080
15. Gyorffy B, Bottai G, Fleischer T, et al. Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. Int J Cancer. 2016;138(1):87–97. doi:10.1002/ijc.29684
16. Kloten V, Becker B, Winner K, et al. Promoter hypermethylation of the tumor-suppressor genes ITIH5, DKK3, and RASSF1A as novel biomarkers for blood-based breast cancer screening. Breast Cancer Res. 2013;15(1):R4. doi:10.1186/bcr3375
17. Takahashi H, Kagara N, Tanei T, et al. Correlation of methylated circulating tumor DNA with response to neoadjuvant chemotherapy in breast cancer patients. Clin Breast Cancer. 2017;17(1):61–69 e3. doi:10.1016/j.clbc.2016.06.006
18. Gan J, Zhang H. PTPRO predicts patient prognosis and correlates with immune infiltrates in human clear cell renal cell carcinoma. Transl Cancer Res. 2020;9(8):4800–4810. doi:10.21037/tcr-19-2808
19. Hou X, Du J, Fang H. PTPRO is a therapeutic target and correlated with immune infiltrates in pancreatic cancer. J Cancer. 2021;12(24):7445–7453. doi:10.7150/jca.64661
20. Xu D, Wang X, Yan S, et al. Interaction of PTPRO and TLR4 signaling in hepatocellular carcinoma. Tumour Biol. 2014;35(10):10267–10273. doi:10.1007/s13277-014-2302-5
21. Dai W, Xiang W, Han L, et al. PTPRO represses colorectal cancer tumorigenesis and progression by reprogramming fatty acid metabolism. Cancer Commun (Lond). 2022;42(9):848–867. doi:10.1002/cac2.12341
22. Dong H, Xie C, Yao Z, et al. PTPRO-related CD8+ T-cell signatures predict prognosis and immunotherapy response in patients with breast cancer. Front Immunol. 2022;13:947841. doi:10.3389/fimmu.2022.947841
23. Dong H, Ma L, Gan J, et al. PTPRO represses ERBB2-driven breast oncogenesis by dephosphorylation and endosomal internalization of ERBB2. Oncogene. 2017;36(3):410–422. doi:10.1038/onc.2016.213
24. Huang YT, Li FF, Ke C, et al. PTPRO promoter methylation is predictive of poorer outcome for HER2-positive breast cancer: indication for personalized therapy. J Transl Med. 2013;11:245. doi:10.1186/1479-5876-11-245
25. Li SY, Li R, Chen YL, et al. Aberrant PTPRO methylation in tumor tissues as a potential biomarker that predicts clinical outcomes in breast cancer patients. BMC Genet. 2014;15:67. doi:10.1186/1471-2156-15-67
26. Yu M, Lin G, Arshadi N, et al. Expression profiling during mammary epithelial cell three-dimensional morphogenesis identifies PTPRO as a novel regulator of morphogenesis and ErbB2-mediated transformation. Mol Cell Biol. 2012;32(19):3913–3924. doi:10.1128/MCB.00068-12
27. Motiwala T, Kutay H, Ghoshal K, et al. Protein tyrosine phosphatase receptor-type O (PTPRO) exhibits characteristics of a candidate tumor suppressor in human lung cancer. Proc Natl Acad Sci U S A. 2004;101(38):13844–13849. doi:10.1073/pnas.0405451101
28. Cai GX, Cai ZJ, Chen QJ, et al. Current status and development of chemotherapy of breast cancer: the main topics and agreements of China South Breast Cancer Symposium. Chin J Gen Surg. 2019;28(11):13.
29. Chinese Society of Laboratory Medicine, Laboratories. NCfC. Expert consensus on the application of liquid biopsy in clinical tumor diagnosis and treatment and medical laboratory practice. Chin J Lab Med. 2018;41(10):10.
30. Chen SH. Clinical value of combined detection of CA15-3, CA125, and CEA in breast cancer. Lab Med. 2013;28(3):3.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
