Back to Journals » Drug Design, Development and Therapy » Volume 15
Study on the Inhibitory Effect of Curcumin on GBM and Its Potential Mechanism
Authors Su X, Chen S , Lu H, Li H , Qin C
Received 14 February 2021
Accepted for publication 3 June 2021
Published 28 June 2021 Volume 2021:15 Pages 2769—2781
DOI https://doi.org/10.2147/DDDT.S306602
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Tin Wui Wong
Xiaotao Su,1 Shaohua Chen,2 Hongyu Lu,1 Haoyu Li,3 Chao Qin1
1Department of Neurology, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, People’s Republic of China; 2Department of Urology, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, People’s Republic of China; 3Department of Ophthalmology, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, People’s Republic of China
Correspondence: Haoyu Li
Department of Ophthalmology, The First Affiliated Hospital of Guangxi Medical University, 6 Shuangyong Road, Nanning, 530021, People’s Republic of China
Email [email protected]
Chao Qin
Department of Neurology, The First Affiliated Hospital of Guangxi Medical University, 6 Shuangyong Road, Nanning, 530021, People’s Republic of China
Email [email protected]
Background: Glioblastoma (GBM) is the most prevalent malignant tumor of the central nervous system (CNS). However, current GBM treatments are ineffective, signifying the great importance of exploring new therapeutic targets. Curcumin has been found to be a natural compound with an anticancer potential. However, its targets and mechanisms in GBM are still unclear.
Methods: Differentially expressed genes (DEGs) were screened from the GBM dataset in the GEO database and intersected with the target genes of curcumin to select potential target genes. Subsequently, survival analysis was performed with the GEPIA database to confirm the effect of target genes on the prognosis of GBM, and functional enrichment analysis was performed using the DAVID database. In vitro, CCK-8 assay was used to screen the appropriate concentration of curcumin; scratch and transwell invasion assays were used to evaluate the effect of curcumin on the migration and invasion abilities of GBM cells. Furthermore, RT-qPCR and Western blotting were used to detect changes in target genes and flow cytometry was used to assess the apoptosis level.
Results: A total of 16 target genes of curcumin and GBM were obtained, among which ENO1, MMP2, and PRKD2 significantly affected the prognosis (P < 0.05). We further selected ENO1 for functional enrichment analysis and found that it was enriched in the glycolytic pathway. Meanwhile, in vitro experiments showed that curcumin could inhibit the migration and invasion of U251 cells and promote apoptosis (P < 0.05).
Conclusion: ENO1 could be a possible target for curcumin in the suppression of GBM cells.
Keywords: glioblastoma, curcumin, ENO1, migration, apoptosis, computational biology
Introduction
Glioblastoma (GBM) is the most prevalent primary tumor of the central nervous system (CNS) and accounts for 60% to 70% of primary brain tumors.1 GBM is more common in the elderly and its incidence increases with age.2 Currently, the treatment of GBM mainly includes maximal surgical resection, supplemented by postoperative radiotherapy, and chemotherapy. However, due to the highly invasive nature of GBM, it is difficult to remove the tumor tissue completely. Most of the patients relapse just a few months after treatment and their relative survival rates remain low, with a median survival time of only 14–15 months and a 5-year survival rate of less than 10%.3,4 The efficacy of chemotherapy drugs such as temozolomide (TMZ) is often limited by drug resistance while it simultaneously increases adverse effects in patients. Therefore, the treatment of GBM still faces great challenges and the search for potential targets for GBM treatment remains necessary.
Curcumin, a traditional Chinese medicine, is a bright yellow and hydrophobic polyphenol extracted from the rhizome of the herb Curcuma longa.5 Recent studies have elucidated that curcumin has anti-cancer effects in lung, rectal, and breast cancer,6–8 similar effects also exhibited in the GBM. In a recent study, curcumin was found to downregulate p-AKT/mTOR, thereby reducing invasiveness and increasing the apoptosis level of GBM.9 However, the target of curcumin inhibition in GBM is still controversial.10,11 The identification of specific targets of curcumin for GBM treatment will help deepen the understanding of its pharmacological mechanisms and, more significantly, provide a theoretical basis for the rational use of curcumin in clinical practice.
In conclusion, our current study used bioinformatics techniques to analyze the targeting of curcumin, based on which and a combination of in vitro experiments we were the first to discover that curcumin targets ENO1 and may exert anti-tumor effects by downregulating it. This finding provides a theoretical basis for further understanding of the anti-tumor mechanism of curcumin and, more importantly, a novel avenue for future clinical treatment of GBM. Secondly, HIF-1α, the transcription factor of glycolysis-related genes, was found to be downregulated after curcumin treatment in our study, suggesting that glycolysis-related metabolic abnormalities may be the mechanism of GBM progression, which also provides clues to dissect the dynamic changes of tumor microenvironment in GBM.
Materials and Methods
Analysis of Potential Target Genes for Curcumin and GBM
Curcumin’s molecular formula was downloaded from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP, http://lsp.nwu.edu.cn/tcmsp.php) and analyzed with the Druggable Pharmacophore Models of the PharmMapper Server (http://www.lilab-ecust.cn/pharmmapper/) to obtain its potential molecular targets.
The Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/gds) database was used to retrieve microarray data of tumor tissues and non-tumor tissues of GBM patients (GSE4290 and GSE19728). The differentially expressed genes (DEGs) were analyzed by R.4.0.2.
A Venn plot (http://bioinformatics.psb.ugent.be/webtools/Venn/) was used to acquire the target genes, visualized by the overlap of the DEGs and the potential molecular targets of curcumin, which were considered to be the target genes of curcumin in the treatment of GBM.
Survival Analysis of Potential Target Genes
The screened potential target genes were categorized into high expression and low expression groups based on the median of their transcriptional level, retrieved from the clinical information on GBM patients in the Cancer Genome Atlas (TCGA, https://Cancergenome.nih.gov/) database. Survival analysis was performed with the gene expression profiling interactive analysis (GEPIA, http://gepia.cancer-pku.cn/index.html) database. In addition, the Chinese Glioma Genome Atlas (CGGA: http://www.cgga.org.cn/) database was further used to verify the results.
Functional Enrichment Analysis of Target Genes
In order to further explore the functions of curcumin in GBM, the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses were performed using the Database for Annotation, Visualization, and Integrated Discovery (DAVID: https://david.ncifcrf.gov/tools.jsp). Additionally, the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING: v.11.0: https://string-db.org/) database and Cytoscape (v.3.6.1) software were used to establish and visualize a protein–protein interaction (PPI) network, which evaluated the physical and functional relationships that exist between the proteins of these target genes. Meanwhile, the function of target genes was also analyzed by GeneMANIA (http://genemania.org/), which is an online analysis tool that can predict gene function based on existing genomic and proteomics data. Finally, gene set enrichment analysis (GSEA) was conducted to find the enriched pathway of target genes. The criteria for defining a statistically significant enriched gene set were P < 0.05 and false discovery rate (FDR) < 0.025.
Reagents
Curcumin with a purity of more than 98% was obtained from Sigma (Sigma-Aldrich, St. Louis, MO, USA) and dissolved with dimethyl sulfoxide (DMSO, Solarbio Life Sciences, Beijing, China).
Cell Culture and Treatments
The U251 cells, used as the GBM cells, were obtained from Procell Life Science & Technology (Wuhan, China) and cultured in Dulbecco’s modified Eagle medium (DMEM, GIBCO, NY, USA), which was supplemented by 1% penicillin/streptomycin (GIBCO, NY, USA) and 10% fetal bovine serum (FBS, GIBCO, NY, USA), in a humidified incubator set at 37°C, 5% CO2, and 95% air atmosphere. All the following experiments were repeated in triplicate, if not otherwise stated.
Cell Viability Assay
A Cell Counting Kit-8 (CCK-8) assay (Solarbio Life Sciences, Beijing, China) followed by spectrophotometric absorbance measurement at 450nm was performed to estimate cell proliferation at 24 h. The GBM cells were cultured at a density of 5×103 cells/well on 96-well plastic culture plates. The U251 cells were treated with 10, 20, 30, 40, 50, and 100 μM curcumin for 24 h. For each concentration gradient, a blank control group using a normal medium without curcumin and DMSO and a vehicle group with the addition of DMSO matching that is contained in the gradient curcumin solution were also prepared. All experiments complied with the manufacturer’s instructions.
Scratch Wound Assay
As described previously,12 cells in the six-well plastic culture plates in the state of 100% confluence were scratched with an aseptic pipette tip, which resulted in a cell-free gap on the gel. Following the rinsing of the cells with serum-free DMEM, the widths at 0 h were recorded with a photo of the gaps. Then, an individual cell sample was incubated with serum-free DMEM as negative control. Curcumin at a concentration of 20 and 30 μM was applied to the other groups, respectively. The migration status at the gaps was recorded with photos at 24 and 48 h.
Transwell Invasion Assay
To evaluate the invasive ability of the cells, a transwell invasion assay with Boyden chambers (BD Bio-sciences, Franklin Lakes, NJ, USA) of 8-mm pore size membranes coated by Matrigel was utilized. The GBM cells were first transferred to the upper chamber, which contained serum-free media, while the lower chamber contained media supplemented with 10% FBS. Following a few hours of incubation, cells that had permeated the membrane were stained with both methanol and 0.1% crystal violet. Then, a microscope was used to observe and count the invaded cells in random fields of each well at 100× magnification.
Flow Cytometry
Cell apoptosis was examined with the Annexin PE Annexin V apoptosis detection kit I (BD Bio-sciences, Franklin Lakes, NJ, USA). The mixture of culture media and cells was centrifuged to remove the supernatant, followed by washing it three times with PBS. The experimental procedures complied with the manufacturer’s instructions.
Real-Time Quantitative PCR
TRIzol reagent (TaKaRa Biotechnology, Dalian, China) was used to extract total RNA and the cDNA was generated with PrimeScript RT Reagent Kit (TaKaRa Biotechnology, Dalian, China). To conduct real-time quantitative PCR (RT-qPCR) amplification with an ABI 7500 Real-Time PCR system (Applied Biosystems, CA, USA) and SYBR Premix Ex TaqII (TaKaRa Biotechnology, Dalian, China), cDNA from the cells was utilized as template. In independent experiments, RT-qPCR was conducted to quantitatively determine the transcriptional levels of Bcl-2, Bax, Caspase-3, enolase 1 (ENO1), hypoxia-inducible factor 1-alpha (HIF-1α), and β-actin in cell samples with the gene-specific primers shown in Table 1. The expression levels of target genes were normalized to that of β-actin, and the results were stated as fold change in cycle threshold (Ct) value relative to the control group as assessed with the 2−ΔΔCt method. Only data with a Ct value < 35 was included in the analysis.
 |
Table 1 Primer Sequences of Quantitative Real-Time Polymerase Chain Reaction |
Western Blotting
The primary antibodies β-actin, Bax, Bcl-2, Caspase-3, ENO1, and HIF-1α were purchased from Cell Signaling Technology Inc. (MA, USA). Protein extracts were kept at −80 °C prior to utilization. The same amounts of denatured protein were processed with 10% SDS-PAGE. The resultant protein bands were placed on polyvinylidene difluoride (PVDF) membranes (Millipore, MA, USA). The LI-COR automatic chemiluminescence image analysis system was used for visualization. The Odyssey Fc Imaging System was used for the quantification of Western blotting signals. Figure 1 shows the workflow of bioinformatics analysis and experimental validation.
 |
Figure 1 The workflow of bioinformatics analysis and experimental validation. |
Statistics
All the calculation steps were conducted with GraphPad Prism v8.2.0 (GraphPad Software, San Diego, USA). The significance of different culture conditions was assessed using one- and two-way ANOVAs. The P-values of multiple comparisons were presented in the figures as asterisk symbols (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001). The criterion for defining statistical difference was set as P < 0.05.
Results
Curcumin and GBM Share Common Targets Shown by Bioinformatics Analysis
We obtained a total of 158 potential target genes for curcumin based on the matching of TCMSP and PharmMapper services. Two GEO datasets (GSE4290 and GSE19728) and a total of 86 GBM patients and 27 controls were analyzed in this study. In the end, 16 target genes were acquired (Figure 2), including ENO1, matrix metalloproteinase 2 (MMP2), dihydropyrimidine dehydrogenase (DPYD), glutamate ionotropic receptor N-methyl-D-aspartate type subunit 2A (GRIN2A), protein kinase D2 (PRKD2), tripartite motif containing 5 (TRIM5), transgelin 2 (TAGLN2), zinc finger protein 36, C3H1 Type-Like 2 (ZFP36L2), glycogen phosphorylase L (PYGL), glutamate decarboxylase 1 (GAD1), guanylate-binding protein 1 (GBP1), potassium voltage-gated channel subfamily J member 3 (KCNJ3), tumor protein P53 (TP53), lactate dehydrogenase D (LDHD), phosphodiesterase 2A (PDE2A), and homeostatic iron regulator (HFE).
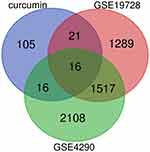 |
Figure 2 Venn plot of overlapping genes in the GEO database of DEGs and target genes of curcumin. |
Survival Analysis of 16 Target Genes Using Data from TCGA
Based on the overall survival (OS), the significance of these target genes in the prognosis of GBM was estimated (Figure 3). The results showed that ENO1, MMP2, and PRKD2 were related to the prognosis of GBM (P < 0.05).
 |
Figure 3 Survival analysis of 16 target genes. |
Functional Enrichment Analysis
To further evaluate the functions of these target genes, we performed GO and KEGG analysis of gene enrichment. GO analysis included biological process (BP), cellular component (CC), and molecular function (MF). As shown in Table 2, the target genes were primarily enriched in protein binding (GO:0005515), plasma membrane (GO:0005886), cytosol (GO:0005829) process, and the beta-alanine metabolism (hsa00410) pathway. As can be seen in the table, PDE2A, PYGL, PDYD, GAD1, and ENO1 were involved in most of the processes. Meanwhile, the PPI network constructed by the STRING database showed the interaction of 16 target genes (Figure 4). Among the three genes with statistically significant effects on GBM prognosis, ENO1 had the highest number of connecting nodes.
 |
Table 2 Go and KEGG Pathway Analyzed by DAVID |
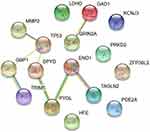 |
Figure 4 PPI network constructed by the STRING database. Abbreviations: PPI, protein–protein interaction; STRING, the Search Tool for the Retrieval of Interacting Genes/Proteins. |
Overall, we focused on ENO1 while performing GSEA and GeneMANIA (Figure 5). GSEA results indicated that high expression of ENO1 promoted the gluconeogenesis pathway, the fructose and mannose metabolism, and the pentose phosphate pathway. Meanwhile, GeneMANIA showed the networks of ENO1.
 |
Figure 5 The functional enrichment analysis of ENO1 performed by (A) GSEA and (B) GeneMANIA. Abbreviation: GSEA, gene set enrichment analysis. |
Expression and Survival Analysis of ENO1 Using Data from CGGA
Moreover, mRNA-seq_693 (693 gliomas and 249 GBM patients included), mRNA-seq_325 (325 gliomas and 139 GBM patients included), mRNA-array_301 (301 gliomas and 124 GBM patients included) datasets of CGGA database were used to supplement the verification of ENO1 expression differences and survival analysis (Figure S1). It can be seen that the expression of ENO1 showed a trend of increasing with the increase in the tumor grade of glioma (P <0.05). The analysis results obtained from the mRNA-seq_325 dataset showed that the high expression of ENO1 was related to the poor prognosis of primary GBM.
Effects of Curcumin on the Activity of GBM Cells
CCK-8 was used to determine the effect of curcumin on GBM cell activity to evaluate its anti-proliferative ability (Figure 6). The results showed that there was no significant inhibitory effect of DMSO on U251 cells in the vehicle group compared to the control group at these concentration gradients (P > 0.05). In contrast to the vehicle group, the inhibitory effect of curcumin on the proliferation of U251 cells was dose-dependent. Among them, curcumin of 50 and 100 μM had statistically different inhibitory effects on U251 cells than the vehicle group (P < 0.05). As the concentration of curcumin increased, the cell morphology became smaller and the adherence decreased. Curcumin began to show toxicity to U251 cells when its concentration reached 40 μM. In order to improve measurement of the effects on GBM cell invasion, migration, and apoptosis, we finally selected the non-toxic concentrations (20 and 30μM) in subsequent experiments to exclude the influence of poor cell status on the results.
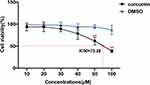 |
Figure 6 Effects of curcumin on the activity of U251 cells, which were treated with curcumin at corresponding concentrations. **P < 0.01. |
Curcumin Inhibited Migration and Invasion of GBM Cells
A wound-healing assay was conducted to preliminary assess the effects of curcumin on migration. As shown in Figure 7, the migration distance of U251 cells treated with 20 and 30 μM of curcumin was significantly shorter compared to that of the negative control (NC) group after 24 and 48 h of incubation (P < 0.05). Meanwhile, the results of the transwell assay showed that U251 cells treated with 20 and 30 μM curcumin had reduced migration and invasion capacities compared to the control group (Figure 8).
 |
Figure 7 The effect of curcumin on the migration of U251 cells, which were treated with curcumin at corresponding concentrations. **P < 0.01. Abbreviation: NC, negative control. |
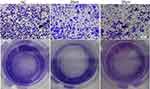 |
Figure 8 The effect of curcumin on the invasion capacity of U251 cells, which were treated with curcumin at corresponding concentrations. Abbreviation: NC, negative control. |
Curcumin Increased Apoptosis in U251 Cells
The apoptosis level in the 20 and 30 μM curcumin-treated group was higher compared to that of the control group according to the outcomes of the flow cytometry assay (P < 0.05; Figure 9).
The effect of curcumin on apoptosis of U251 cells was further confirmed by RT-qPCR analysis and Western blotting (Figures 10 and 11). The expression of Bax and cleaved Caspase-3 was increased and Bcl-2 expression was decreased in U251 cells after treatment with 20 and 30 μM curcumin, indicating that curcumin enhanced the apoptosis of GBM cells (P < 0.05).
 |
Figure 10 The effect of curcumin on the expression of HIF-1α and ENO1 and apoptosis level in U251 cells confirmed by RT‐qPCR. *P < 0.05 and **P < 0.01. Abbreviation: NC, negative control. |
 |
Figure 11 The effect of curcumin on the expression of HIF-1α and ENO1 and apoptosis level in U251 cells confirmed by Western blotting. *P < 0.05 and **P < 0.01. Abbreviation: NC, negative control. |
Changes in HIF-1α and ENO1 Expression Levels After Curcumin Treatment
RT-qPCR analysis and Western blotting were used to detect the changes of HIF-1α and ENO1 at the mRNA expression level and protein expression level after curcumin treatment, respectively (Figures 10 and 11). After 20 and 30 μM curcumin-treated, both HIF-1α and ENO1 expression in U251 cells showed a tendency to decrease (P < 0.05).
Discussion
Curcumin has received increasing attention in recent years due to its antitumor effects,6–8 however its mechanisms of action are still unclear. We first used bioinformatics to determine curcumin’s possible targets of action in GBM and then performed survival analysis on these target genes to verify their clinical value. With this approach, our study finally narrowed down the number of potential targets to 16, with ENO1, MMP2, and PRKD2 as the potential target genes for curcumin in affecting the prognosis of GBM. Finally, combined with the results of the PPI network analysis, we chose to perform preliminary in vitro experiments to validate ENO1 as a target.
Enhanced glycolysis is one of the metabolic characteristics of GBM.13 Enolase is a key glycolytic enzyme, and ENO1 is its major isoform expressed in GBM.14 Several studies have confirmed that ENO1 is highly expressed in different types of cancers including glioma, liver, and breast cancer, and positively correlated with tumor progression and venous invasion.15–19 HIF-1α is one of the regulators of oxygen homeostasis. When cells undergo hypoxia, HIF-1α can act as a major transcription factor that activates the encoded glycolytic enzymes, including ENO1. Previous studies have shown that HIF-1α causes the upregulation of ENO1 in hypoxic cancer tissue and brain cells.13,15,18–20 Using bioinformatics technology, we found that ENO1 was upregulated in GBM and it promoted the gluconeogenesis pathway, the fructose and mannose metabolism pathway, and the pentose phosphate pathway, which is consistent with the results of previous studies. Our in vitro experiments demonstrated that the proliferation, invasion, as well as migration ability of GBM cells decreased after curcumin treatment. In addition, the expression of ENO1 as well as HIF-1α was inhibited. Therefore, we hypothesized that ENO1 may be a target of curcumin. Curcumin can inhibit the expression of HIF-1α and ENO1, which could cause inhibition of the glycolytic processes in tumor cells leading to an impaired energy supply to GBM, which in turn affects tumor proliferation, invasion, and migration.
Although we did not overexpress or knockdown ENO1 to verify this mechanism of curcumin in our experiments, Sanzey et al used intracranial GBM xenografts and found that knockdown of seven glycolytic genes, including ENO1, resulted in a dramatic increase in mouse survival.21 Chen et al demonstrated with both in vitro and in vivo experiments that downregulation of ENO1 could lead to a decrease in cell proliferation and migration abilities.22 Furthermore, Song et al found that knockdown of ENO1 inhibited the PI3K/Akt pathway, resulting in the suppression of glioma cell growth, migration, and invasive progression.19 These findings all support our hypothesis.
Moreover, ENO1 has a complex relationship with apoptosis. Overexpression of ENO1 in pulmonary artery smooth muscle cells induces an anti-apoptotic phenotype through the AMPK-Akt pathway.23 In pancreatic cancer cells, modulation of ENO1 expression can affect the activation of p38MAPK and thus cause apoptosis of the cancer cell.24 In cardiomyocytes, ENO1 promotes apoptosis through AMPK dephosphorylation.25 We observed that the apoptosis of glioma cells increased after curcumin treatment (Figures 9–11). Based on the results of our study, we hypothesized that curcumin may be involved in the promotion of apoptosis by inhibiting the expression of ENO1, however the exact mechanism remains to be further investigated. Nevertheless, it is worth noting that the ways in which curcumin promotes apoptosis in glioma cells may vary. We found that curcumin promoted both apoptosis in GBM cells as well as expression of Caspase-3 and Bax, in addition to decreasing the expression of Bcl-2 (Figures 10 and 11). These findings suggested that curcumin could induce inherent apoptotic signaling in GBM cells and promote apoptosis.
In summary, after investigating the potential efficacy of curcumin in inhibiting GBM in vitro and its potential targets, we obtained the following results: firstly, curcumin can inhibit migration and invasion of GBM cells; secondly, curcumin could promote GBM cell apoptosis; and thirdly, ENO1 may be the target of curcumin’s effect on GBM. In vitro experiments have demonstrated that curcumin can inhibit expression of ENO1 in GBM cells. Although some scattered studies have touched upon the potential mechanisms, systematic studies are still pending. We believe that our results provide some direction for future studies.
Conclusion
In conclusion, the present study initially revealed that curcumin has an inhibitory effect on U251 cells as shown by bioinformatics analysis and in vitro experiments. ENO1 could be a potential target gene of curcumin and its anti-cancer mechanisms may be related to glycolysis and apoptosis pathways. Therefore, curcumin could be further evaluated as a promising therapeutic option that can improve GBM treatment.
Data Sharing Statement
The raw data related to the results of this research are included in the article.
Ethical Statement
Our study did not require an ethical board approval because it did not contain human or animal trials.
Author Contributions
HLi and CQ designed and guided the entire experiments. XS, SC and HLu performed experiments and analyzed the data. XS and SC took the lead in writing the manuscript. HLi and CQ revised the manuscript. CQ was responsible for funding acquisition. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval for the version to be published; and agree to be accountable for all aspects of the work.
Funding
This work was supported by The National Natural Science Foundation of China (No. 81860222) and the Science and Technology Plan Project of Guangxi (No. AB16380204).
Disclosure
The authors declare that there is no potential conflict of interest of this research.
References
1. McNeill KA. Epidemiology of brain tumors. Neurol Clin. 2016;34(4):981–998. doi:10.1016/j.ncl.2016.06.014
2. Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15 Suppl 2(Suppl2):ii1–ii56. doi:10.1093/neuonc/not151
3. Sasmita AO, Wong YP, Ling APK. Biomarkers and therapeutic advances in glioblastoma multiforme. Asia Pac J Clin Oncol. 2018;14(1):40–51. doi:10.1111/ajco.12756
4. Sabo B. Primary malignant brain tumours, psychosocial distress and the intimate partner experience: what do we know? Can J Neurosci Nurs. 2014;36(3):9–15.
5. Zlotogorski A, Dayan A, Dayan D, Chaushu G, Salo T, Vered M. Nutraceuticals as new treatment approaches for oral cancer--I: curcumin. Oral Oncol. 2013;49(3):187–191. doi:10.1016/j.oraloncology.2012.09.015
6. Zhu JY, Yang X, Chen Y, et al. Curcumin suppresses lung cancer stem cells via inhibiting Wnt/β-catenin and sonic hedgehog pathways. Phytother Res. 2017;31(4):680–688. doi:10.1002/ptr.5791
7. McFadden RM, Larmonier CB, Shehab KW, et al. The role of curcumin in modulating colonic microbiota during colitis and colon cancer prevention. Inflamm Bowel Dis. 2015;21(11):2483–2494. doi:10.1097/MIB.0000000000000522
8. Mittal L, Aryal UK, Camarillo IG, Raman V, Sundararajan R. Effective electrochemotherapy with curcumin in MDA-MB-231-human, triple negative breast cancer cells: a global proteomics study. Bioelectrochemistry. 2020;131:107350. doi:10.1016/j.bioelechem.2019.107350
9. Wang Z, Liu F, Liao W, et al. Curcumin suppresses glioblastoma cell proliferation by p-AKT/mTOR pathway and increases the PTEN expression. Arch Biochem Biophys. 2020;689:108412. doi:10.1016/j.abb.2020.108412
10. Zhao J, Zhu J, Lv X, et al. Curcumin potentiates the potent antitumor activity of ACNU against glioblastoma by suppressing the PI3K/AKT and NF-κB/COX-2 signaling pathways. Onco Targets Ther. 2017;10:5471–5482. doi:10.2147/OTT.S149708
11. Hesari A, Rezaei M, Rezaei M, et al. Effect of curcumin on glioblastoma cells. J Cell Physiol. 2019;234(7):10281–10288. doi:10.1002/jcp.27933
12. Ma C, Zhuang Z, Su Q, He J, Li H. Curcumin has anti-proliferative and pro-apoptotic effects on tongue cancer in vitro: a Study with Bioinformatics Analysis and in vitro experiments. Drug Des Devel Ther. 2020;14:509–518. doi:10.2147/DDDT.S237830
13. Gabriely G, Wheeler MA, Takenaka MC, Quintana FJ. Role of AHR and HIF-1α in glioblastoma metabolism. Trends Endocrinol Metab. 2017;28(6):428–436. doi:10.1016/j.tem.2017.02.009
14. Muller FL, Colla S, Aquilanti E, et al. Passenger deletions generate therapeutic vulnerabilities in cancer. Nature. 2012;488(7411):337–342. doi:10.1038/nature11331
15. Jiang BH, Agani F, Passaniti A, Semenza GL. V-SRC induces expression of hypoxia-inducible factor 1 (HIF-1) and transcription of genes encoding vascular endothelial growth factor and enolase 1: involvement of HIF-1 in tumor progression. Cancer Res. 1997;57(23):5328–5335.
16. Trojanowicz B, Winkler A, Hammje K, et al. Retinoic acid-mediated down-regulation of ENO1/MBP-1 gene products caused decreased invasiveness of the follicular thyroid carcinoma cell lines. J Mol Endocrinol. 2009;42(3):249–260. doi:10.1677/JME-08-0118
17. Gao J, Zhao R, Xue Y, et al. Role of enolase-1 in response to hypoxia in breast cancer: exploring the mechanisms of action. Oncol Rep. 2013;29(4):1322–1332. doi:10.3892/or.2013.2269
18. Sharma NK, Sethy NK, Bhargava K. Comparative proteome analysis reveals differential regulation of glycolytic and antioxidant enzymes in cortex and hippocampus exposed to short-term hypobaric hypoxia. J Proteomics. 2013;79:277–298. doi:10.1016/j.jprot.2012.12.020
19. Song Y, Luo Q, Long H, et al. Alpha-enolase as a potential cancer prognostic marker promotes cell growth, migration, and invasion in glioma. Mol Cancer. 2014;13:65. doi:10.1186/1476-4598-13-65
20. Yeo EJ, Chun YS, Cho YS, et al. YC-1: a potential anticancer drug targeting hypoxia-inducible factor 1. J Natl Cancer Inst. 2003;95(7):516–525. doi:10.1093/jnci/95.7.516
21. Sanzey M, Abdul Rahim SA, Oudin A, et al. Comprehensive analysis of glycolytic enzymes as therapeutic targets in the treatment of glioblastoma. PLoS One. 2015;10(5):e0123544. doi:10.1371/journal.pone.0123544
22. Chen S, Zhang Y, Wang H, et al. WW domain-binding protein 2 acts as an oncogene by modulating the activity of the glycolytic enzyme ENO1 in glioma. Cell Death Dis. 2018;9(3):347. doi:10.1038/s41419-018-0376-5
23. Dai J, Zhou Q, Chen J, Rexius-Hall ML, Rehman J, Zhou G. Alpha-enolase regulates the malignant phenotype of pulmonary artery smooth muscle cells via the AMPK-Akt pathway. Nat Commun. 2018;9(1):3850. doi:10.1038/s41467-018-06376-x
24. Principe M, Borgoni S, Cascione M, et al. Alpha-enolase (ENO1) controls alpha v/beta 3 integrin expression and regulates pancreatic cancer adhesion, invasion, and metastasis. J Hematol Oncol. 2017;10(1):16. doi:10.1186/s13045-016-0385-8
25. Gao S, Li H, Feng XJ, et al. α-Enolase plays a catalytically independent role in doxorubicin-induced cardiomyocyte apoptosis and mitochondrial dysfunction. J Mol Cell Cardiol. 2015;79:92–103. doi:10.1016/j.yjmcc.2014.11.007
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.