Back to Journals » OncoTargets and Therapy » Volume 14
SQLE Mediates Metabolic Reprogramming to Promote LN Metastasis in Castration-Resistant Prostate Cancer
Authors Xu Z, Huang L, Dai T, Pei X, Xia L, Zeng G, Ye M, Liu K, Zeng F, Han W, Jiang S
Received 26 April 2021
Accepted for publication 13 July 2021
Published 24 July 2021 Volume 2021:14 Pages 4285—4295
DOI https://doi.org/10.2147/OTT.S315813
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jianmin Xu
Zhenzhou Xu,* Liang Huang,* Tao Dai,* Xiaming Pei, Longzheng Xia, Gongqian Zeng, Mingji Ye, Kan Liu, Fuhua Zeng, Weiqing Han, Shusuan Jiang
Department of Urology, The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Hunan Cancer Hospital, Changsha, 410013, Hunan, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhenzhou Xu; Shusuan Jiang
Department of Urology, The Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Hunan Cancer Hospital, 283# Tongzipo Road, Yuelu District, Changsha, Hunan, 4100013, People’s Republic of China
Tel +86 18874728621
; +86 18608408523
Fax +86 73189762270
Email [email protected]; [email protected]
Background: Almost all metastatic hormone-sensitive prostate cancers (mHSPC) will develop into metastatic castration-resistant prostate cancer (mCRPC) after androgen deprivation therapy (ADT). The expression level of squalene monooxygenase (SQLE) is increased in CRPC cells and regulates cholesterol metabolism. This study verified the biological function and mechanisms of SQLE in CRPC.
Methods: The expression of SQLE in human prostate cancer cells was overexpressed or silenced and its efficacy on cell survival was determined by the MTS test. Energy metabolism phenotype test was evaluated by XF real-time ATP rate assay, XF cell mitochondrial stress test, XF glycolysis stress test and XF mito fuel flex test. Cell migration and invasion were evaluated by colony formation assays and transwell assays; the expression of mRNA and protein was assessed by RT-qPCR and Western blot, respectively. Moreover, BALB/c nude mice model was performed to evaluate the lymph node metastasis.
Results: In our study, we found that the expression level of SQLE was significantly increased in bicalutamide-resistant-C4-2B cells compared to LNCaP cells. SQLE knockdown partly restored the sensitivity of drug-resistant cells to bicalutamide and reduced lymph node metastasis by inhibiting fatty acid oxidation in mitochondria. We also found that terbinafine, the specific inhibitor of SQLE, can enhance the sensitivity of prostate cancer cells to bicalutamide.
Conclusion: Our study revealed that SQLE is involved in the progression of castration resistance in CRPC through mediating metabolic reprogramming, presenting SQLE as a new target for the treatment of mCRPC.
Keywords: SQLE, bicalutamide, CRPC, ADT, lymph node metastasis
Background
Prostate cancer (PC) is a leading cause of male death worldwide, and it is the most frequently diagnosed cancer among men aged 65–74 years old.1 Due to the popularization of PSA screening, the incidence of PC has substantially increased in the past few decades. The prognosis of PC varies greatly, being highly dependent on some factors, such as stage of diagnosis, race, and age.2 Following these prognostic risk stratifications, the management options for local prostate cancer include active surveillance, watchful waiting, hormonal therapy, radical prostatectomy, and radical radiotherapy.3 However, for advanced castration-resistant prostate cancer (CRPC), the prognosis is not good due to the lack of effective therapeutic targets. Therefore, there is an urgent need to discover new and effective therapeutic targets in mCRPC.
The proliferation and progression of PC cells are both strongly dependent on androgens.4 Androgen deprivation therapy (ADT) is the initial recommended treatment for metastatic hormone-sensitive prostate cancers (mHSPC). Although mHSPC patients initially exhibit a good response to ADT treatment, they eventually develop resistance to ADT and progress to mCRPC.3 As a member of the nonsteroidal anti-androgen, bicalutamide is often used in conjunction with androgen deprivation to block AR (androgen receptor) activity and tumor growth in mHSPC patients. However, with the increase of cell endogenous androgens, simply blocking AR receptors cannot achieve the desired effect. Therefore, the androgen biosynthesis inhibitor abiraterone, which inhibits 17 α-hydroxylase/C17,20-lyase (CYP17) is widely used in patients with CRPC.5 Nevertheless, a quarter of CRPC patients are not sensitive to abiraterone. The drug resistance mechanism of CRPC remains unclear. Therefore, revealing the drug resistance mechanism of CRPC is essential for the treatment of patients with CRPC.
Aberrant cholesterol metabolism is increasingly appreciated as being essential for cancer initiation and progression.6 Squalene monooxygenase (also known as squalene epoxidase, SQLE), encoded by the human SQLE gene and located on human chromosome 8q24.1, is one of key rate-limiting enzymes in downstream cholesterol synthesis.7 It catalyzes the conversion of squalene to 2, 3-oxidosqualene and is the rate-limiting enzyme in sterol biosynthesis, which is essential for prostate function.8 Abnormal SQLE expression has been reported in some tumors, including breast, pancreatic, liver, and prostate cancers, and lung SCC (squamous cell carcinoma).8 Previous studies showed the expression of SQLE in breast cancer indicates a poor outcome.9 SQLE loss results in accumulation of the upstream metabolite squalene, which is normally undetectable.10 In ALK+ anaplastic large cell lymphoma, squalene alters the cellular lipid profile and protects cancer cells from ferroptotic cell death, providing a growth advantage under conditions of oxidative stress and in tumor xenografts.10
Cholesterol is essential for rapid cancer growth and has been specifically linked to PC progression to CRPC.11 Its levels are elevated in patient serum and bone metastasis post-androgen deprivation therapy (ADT), and hypercholesterolemia correlates with increased PC-specific mortality.12 In a recent study, researchers identified high mRNA expression of SQLE as strongly associated with high Gleason grade and as a strong predictor of lethal prostate cancer, independent from standard clinicopathologic factors.7 However, the regulating mechanism of SQLE in CRPC is not clear, prompting our interest in its exploration.
The purpose of this study was to assess the mechanism of SQLE in CRPC using cellular and animal models. To our knowledge, this is the first report of the relationship between SQLE and fatty acid metabolic reprogram in CRPC. We examined the potential of SQLE knockdown to overcome bicalutamide resistance and inhibit lymph node metastasis and evaluated its contribution to mitochondrial oxidative phosphorylation (OXPHOS) and lipid biosynthesis in CRPC.
Materials and Methods
Cell Culture and Treatment
Human PCa cells lines LNCaP, PC3, DU145C4, C4, C4-2 and C4-2B were obtained from the American Type Culture Collection (Rockville, MD, USA). All cells were cultured in RPMI-1640 (Gibco-Life Technologies) containing 1% penicillin/streptomycin (Beyotime Biotechnology, Shanghai, China) and 10% fetal bovine serum (Gibco-Life Technologies). LNCaP, androgen-independent prostate cancer cells (PC3, DU145), and bicalutamide-resistant prostate cancer cells (C4, C4-2, and C4-2B) were established in our laboratory. For experiments involving bicalutamide exposure, actively growing cells were continuously incubated with the indicated dose of bicalutamide in 10% FBS at 37°C for 48 h prior to analysis.
Establishment of Stable SQLE-Overexpression/Interfering Cell Lines
To generate SQLE stable overexpression/knockdown PC cells, LNCaP cells transfected with LV-control and LV-SQLE, and C4-2B cells transfected with scramble and sh-SQLE using Lipofectamine 3000 (Invitrogen, Waltham City, Massachusetts, USA), plasmids followed by 1000 μg/mL G418 screening were pool collected, and positive clones were selected and identified for stable overexpression or abrogated SQLE levels. The target sequences of scramble and sh-SQLE were 5′-GCGGAGCGGCT CCCAGTTCG-3′ and 5′-TCCAATAAAGTTTCTGTATCAT −3′, respectively.
MTS Assay
The MTS assay was used to detect the sensitivity of PC cells to bicalutamide. Cells (3×103 cells/well) were seeded into 96-well plates and cultured overnight. Then, cells were treated with various concentrations of bicalutamide for 24 h. Next, 20 μL MTS was added in 200 μL medium per well, and absorbance was measured at 490 nm after the cells were incubated for 4 h. The experiment was performed in triplicate.
XF Real-Time ATP Rate Assay
Total ATP production was measured according to instructions provided by Seahorse Bioscience (Agilent, US). Cells were exposed to these compounds as follows: oligomycin (1μM) and a mixture of rotenone (1μM) and antimycin A (1μM), which can report multiple parameters, including glycolytic ATP, mitochondrial respiratory ATP rate, and total ATP production rates. Data were analyzed using XF Wave Software (Seahorse Bioscience, Agilent, US). All data were normalized to protein content.
XF Cell Mitochondrial Stress Test
Mitochondrial stress was measured according to instructions provided by Seahorse Bioscience (Agilent, US). Briefly, cells were metabolically perturbed by the addition of three compounds in succession, and OCR (O2 consumption rate) was measured prior to and after addition of each compound as follows: oligomycin (1μM); carbonyl cyanide-4-(trifluoromethoxy) phenylhydrazone (FCCP) (0.6μM); and a mixture of rotenone (1μM) and antimycin A (1μM). Data were analyzed using XF Wave software. Cellular protein was measured using a BCA assay kit (Pierce, Thermo Fisher, US) following standard procedures. All data were normalized to protein content.
XF Glycolysis Stress Test
Cells were exposed to three compounds, with measurements after each addition as follows: glucose (10 mM), oligomycin (1 μM), and 2-deoxy-D-glucose (2-DG, 100mM). Data were analyzed with XF Wave software. Protein was quantified as described above. All data were normalized to protein content.
XF Mito Fuel Flex Test
A mitochondrial fuel flex test was performed to measure the dependency, capacity, and flexibility of cells to oxidize three critical mitochondrial fuels: glucose, glutamine, and LCFAs. Cells were exposed to BPTES (3 μM), etomoxir (4 μM), or UK5099 (2 μM) in succession, and OCR was measured prior to and after the addition of each compound. Data were assessed with XF Wave software. Protein was quantified as described, and all data were normalized to protein content.
Clone Formation Assay
Stably SQLE-overexpression/interfering cells (300 cells/well) were added to six-well plates and incubated for 14 days. Then, the cells were fixed with 4% paraformaldehyde and stained with crystal violet. Each treatment group was replicated in three wells, and the colony formation experiment was repeated three times.
Transwell Migration and Invasion Assays
A transwell chamber (8μm, 24-well format; Corning, USA), with, or without a diluted Matrigel (BD Biosciences, New Jersey, USA) coating, was used to assess the migration and invasion of cultured cells. Briefly, 5×105/100 μL of cells (migration assays) and 1×106/100 μL of cells (invasion assays) were seeded in serum-free RPMI-1640 to the top chamber. The cells in the top chamber that had not migrated through the filter were wiped off with a cotton swab, while those that had migrated to the bottom surface were fixed in pre-cold methanol for 20 min and stained with 0.5% crystal violet, then counted under a microscope. Migration and invasion rates are expressed as the ratio of the treated group value to the control group value. The experiment was performed in triplicate.
Quantitative Real-Time PCR (qRT-PCR)
Total RNA of tissue was extracted using an RNA extraction kit, according to the manufacturer’s protocol. The cDNA was reversing transcribed using a Takara PrimeScript RT-PCR Kit (Takara, Shiga, Japan), according to the manufacturer’s instructions. qRT-PCR was performed using a TB Green Fast qPCR Mix (Takara, Shiga, Japan) in the Roche Light Cycler 96 instrument, and the instrument software (Life science, Roche, Mannheim, Germany) was used to determine the relative expression levels of target genes. Primers were as follows: SQLE: L-5ʹ- CTCCAAGTTCAGGAAAAGCCTGG-3ʹ, R-5ʹ- GAGAACTGGACTCGGGTTAGCT-3ʹ; ACTIN: L-5ʹ-gaaggtgaaggtcggagtc-3ʹ, R-5ʹ-gaagatggtgatgggatttc-3ʹ. ACTIN was used as the reference and normalization control. The average of three independent analyses for each gene was calculated. The experiment was performed in triplicate.
Western Blot
The levels of targeting proteins were determined by Western blot assays. A total of 30 μg of cell lysates were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto a PVDF membrane. Membranes were incubated overnight at 4°C with primary antibodies, including SQLE (1:1000) and ACTIN monoclonal antibody (1:5000). The bound antibodies were detected by horseradish peroxidase-conjugated second antibodies for 2 h at room temperature and visualized using Pierce ECL Western Blotting Substrate (Thermo Scientific, Massachusetts, USA). The relative levels of individual proteins to control ACTIN were analyzed by ImagJ2 software (Madison, WI, USA).
Mice and the Footpad Implantation Model
All male NOD/SCID nude mice were acquired from Hunan SJA Laboratory Animal Co., Ltd., Changsha, Hunan, China. All mice were fed with ad libitum access to a standard diet and water in a pathogen-free environment in The Institutional Animal Care and Use Committee of Affiliated Cancer Hospital, Xiangya School of Medicine, Central South University. Before all surgical and sampling procedures, the mice were anesthetized by intraperitoneal injection of a combination of anesthetics (80 mg/kg of ketamine and 12 mg/kg of xylazine). For the cell footpad implantation model, 5×105 LNCaP, C4-2B, and C4-2B_shSQLE cells were implanted subcutaneously into the footpad region of the hind limb of 8-week-old NOD/SCID nude mice. Primary tumor volume was monitored and calculated according to the formula 0.5*A*B2 (A is the largest diameter of a tumor and B is its perpendicular diameter). Popliteal LNs (pLNs), as mapped by pathology colleagues, were dissected for further experiments and analyses.
Statistical Analysis
Results are expressed as means ± SEM. Statistical analysis was performed using ANOVA followed by a two-sample t-test or one-way ANOVA Dunnett test (P<0.05 was considered statistically significant).
Results
SQLE is Related to Bicalutamide Resistance in Prostate Cancer Cells
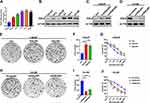
To systematically investigate the expression of SQLE in prostate cancer cells, we first analyzed the mRNA and protein expression of SQLE in androgen-dependent prostate cancer cells (LNCaP), androgen-independent prostate cancer cells (PC3, DU145), and castration-resistant prostate cancer cells (C4, C4-2, and C4-2B). The results showed that the mRNA and protein levels of SQLE were highly elevated in androgen-independent cells compared to androgen-dependent cells (P<0.05) (Figure 1A and B). To determine the impact of SQLE expression on cell growth, we constructed LNCaP stable cell lines ectopically expressing SQLE and C4-2B stable cell lines silencing SQLE (Figure 1C and D). Next, we observed that overexpressed SQLE significantly promoted the colony formation of the cells, whereas silencing of SQLE expression strongly suppressed colony formation, as observed in colony formation assays (P <0.05) (Figure 1E, F, H and I). To assess the effects of SQLE on the resistance of prostate cancer cells to bicalutamide, cells were exposed to bicalutamide before the survival fraction was analyzed for cell proliferation using CCK-8 assay. The results demonstrated that overexpression of SQLE increased the survival capacity of LNCaP cells after bicalutamide at 0, 0.1, 1, 10, 100, and 1000 μM (Figure 1G). In contrast, silencing of SQLE by sh-SQLE contributed to the sensitivity of C4-2B cells to bicalutamide (Figure 1J). These results indicate that SQLE is highly expressed in castration-resistant prostate cancer cells and significantly promotes the resistance of prostate cancer cells to bicalutamide.
The Mitochondrial OXPHOS Produced by Fatty Acid is the Essential Energy for Castration-Resistant Prostate Cancer Cells
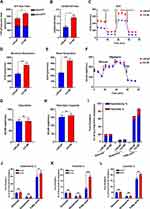
Metabolic reprogramming is an important basis for the development of many tumors, including prostate cancer. To evaluate the metabolic patterns in prostate cancer, we performed an energy metabolism phenotype test in C4-2B cells. As shown in Figure 2A, the ATP production of glycolysis in LNCaP and C4-2B cells was no different. However, the ATP production of OXPHOS (oxidative phosphorylation) in C4-2B cells was significantly increased compared to LNCaP cells. In C4-2B and LNCaP cells, no difference was observed in ECAR ratio, glycolysis value, and capacity (Figure 2F–H), indicating unchanged glycolysis levels. However, the signs of OXPHOS, such as the oxidative respiration in the mitochondrial oxidative stress test, the energy metabolism OCR/ECAR ratio, and the mitochondrial basal respiration, increased in C4-2B cells (Figure 2B, C and E). These findings suggest that OXPHOS is the main source of energy in castration-resistant prostate cancer cells. The mitochondrial maximum respiration value significantly increased in C4-2B cells, indicating that the cells are more resistant to stress (Figure 2D). The majority of energy produced by mitochondria is mainly derived from three fuels: glucose, glutamine, and fatty acids.13 As the mitochondrial substrate analysis tests showed, fatty acid synthesis obviously increased in prostate cancer cells, especially in C4-2B cells, compared to the other two fuel sources (glucose and glutamine) (Figure 2I). The results were similar to the previous study that indicated that a significant hallmark of PC is abnormal lipid metabolism, as first observed by Medes and colleagues in 1953.14 In C4-2B cells, the dependence, flexibility, and capacity of fatty acids in mitochondria were significantly higher than in LNCaP cells (P<0.05) (Figure 2J–L). Altogether, these results mean that fatty acid oxidation in mitochondria is essential to castration-resistant prostate cancer cells.
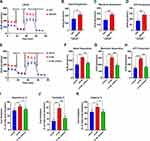
SQLE Supports Elevated Mitochondrial OXPHOS of Fatty Acids in Castration-Resistant Prostate Cancer Cells
To determine the relationship between SQLE expression and the OXPHOS of castration-resistant prostate cancer cells, the results of mitochondrial stress test exhibited that ectopically expressing SQLE significantly increased the OXPHOS level in LNCaP cells (Figure 3A). The basal and maximal respiration were also higher in the SQLE-overexpressing LNCaP cell line (P<0.05) (Figure 3B and C), with higher ATP was higher than in the control group (P<0.05) (Figure 3D). These results indicated that high expression of SQLE contributed to enhance OXPHOS in prostate cancer cells. Conversely, SQLE-deficient C4-2B cell lines showed a lower OXPHOS level than the control group (Figure 3E–H). As shown in Figure 3I–K, the dependency, flexibility, and capacity of fatty acid oxidation were higher in C4-2B cells than in SQLE-deficient C4-2B cells (P<0.05). In summary, SQLE mediated the mitochondrial OXPHOS level of fatty acids in prostate cancer cells.
Bicalutamide Resistance Evokes an Increase in SQLE Levels That Facilitates Prostate Cancer Metastasis
To determine the functional significance of SQLE on invasion and metastasis in PCa, we perturbed the expression of SQLE in PCa cell lines and studied the effect of this regulation on cell invasion and migration. We found that the invasion and migration ability of C4-2B cells with high SQLE expression was significantly stronger than that of LNCaP cells with low SQLE expression (P<0.05). However, blockage of SQLE obviously attenuated the invasion and migration of C4-2B (P<0.05) (Figure 4A–C). In the plantar popliteal lymph node (LN) metastasis model, the LN metastasis area induced by LNCaP cells with low SQLE expression was smaller than that of C4-2B cells with high SQLE expression (P<0.05). However, the knockdown of SQLE significantly decreased the LN metastasis area of C4-2B cells (P<0.05) (Figure 4D and E). The tumor weights of the three groups were not different, respectively (Figure 4F). Therefore, the results indicate that SQLE could promote the LN metastasis of castration-resistant prostate cancer in vivo and in vitro.
Inhibition of SQLE Activity Elevates Sensitivity of Androgen-Independent Prostate Cancer to Bicalutamide
We further investigated whether knockdown of SQLE could enhance the bicalutamide sensitivity of androgen-independent prostate cancer cells (PC3 and DU145). The results showed that blockage of SQLE decreased the survival capacity of PC3 and DU145 cells after bicalutamide at 0, 0.1, 1, 10, 100, and 1000 μM (P<0.05) (Figure 5A and B). Subsequently, we used the inhibitor terbinafine (a SQLE inhibitor) to endogenously block the expression of SQLE and analyze whether terbinafine can enhance the sensitivity of bicalutamide. As shown in Figure 5C and D, the survival capacity of PC3 and DU145 cells treated with 5 μM and 10 μM terbinafine significantly decreased after bicalutamide at 0, 0.1, 1, 10, 100, and 1000 μM (P<0.05). These results suggest that targeting SQLE enhanced the bicalutamide sensitivity of PC3 and DU145 cells.
Discussion
Prostate cancer is strongly dependent on androgens (ARs). However, the response to ADT treatment is not the same in all patients with prostate cancer. This is due to the heterogeneity of prostate cancer and increased ectopic androgen synthesis.15 Drugs that block androgen synthesis in tumor cells and enhance the blocking efficiency of AR receptors, such as abiraterone and enzalutamide, are widely used in clinical practice. The inadequacy of blocking androgens, such as AR mutations, AR splices, and other androgen synthesis pathways, remains a major clinical problem.16,17 In this study, we demonstrated that cholesterol metabolism regulated by SQLE was an essential pathway to androgen biosynthesis even after androgen deprivation therapy in prostate cancer, implying that blocking SQLE would be a potential treatment in mCRPC.
Fatty acid and cholesterol metabolism are dysregulated in most patients with advanced PC.18,19 Previous studies have shown a positive association between serum cholesterol levels and high-grade prostate cancer.6,20 Prostate cancer can take in exogenous cholesterol through SR-B1 and synthesize endogenous cholesterol from acetyl-CoA.21 The accumulated cholesterol ester can produce free cholesterol by hormone-sensitive hydrolase (HSL) in the absence of an androgen environment, and eventually synthesize androgens to maintain the proliferation of prostate cancer cells. As a cholesterol-lowering therapy, HMG-CoA reductase inhibitors (statin lipid-lowering drugs) have reduced the risk of developing high-grade and aggressive prostate cancer.22 Cholesterol can affect the signal transduction of important signal pathways, such as the EFGR and AKT pathways, by regulating the dynamic changes of lipid rafts to maintain prostate cancer cell survival and growth.23
SQLE, a key enzyme of cholesterol biosynthesis, acted as an oncogene and played an important role in many tumors.24,25 SQLE mRNA expression has been significantly correlated with high-risk ER+ breast cancers.26 Alteration of SQLE expression modulates cell proliferation and migration by regulating ERK signaling.8 The oncogenic effect of SQLE in NPC (nasopharyngeal carcinoma) mainly results from cholesteryl ester accumulation and the activation of PI3K/AKT pathway.25 In prostate cancer, the expression of SQLE is not elevated in prostate cancers in general compared to normal prostatic tissue, however, if the SQLE expression in tumor tissue is elevated by 10%, its odds of lethal cancer increases by 40%. Thus, SQLE can distinguishes tumors at high risk of metastasis.27 However, higher SQLE expression is tightly associated with increased histologic markers of angiogenesis and is prognostic for lethal prostate cancer at diagnosis.27
Although the mechanism of androgen resistance is not fully clear, abnormal fatty acid metabolism has been shown to promote the resistance of CRPC cells to enzalutamide.28 Overexpression of carnitine palmitoyltransferase 1B (CPT1B), a rate-limiting step of fatty acid oxidation, is reported to significantly increase enzalutamide resistance via increasing AKT expression and phosphorylation in CRPC cells.28 This indicates that enhanced fatty acid oxidation could promote the drug resistance of CRPC cells. Our study has found bicalutamide resistance induced SQLE highly expression refer to increased mitochondrial OXPHOS, which supplied energy source to maintain proliferation and metastasis in CPRC cells. We demonstrated that SQLE was increased in androgen-independent PC cells (PC3 and DU145) and CRPC cells (C4, C4-2, and C4-2B). The overexpression of SQLE not only enhanced bicalutamide resistance in androgen-independent prostate cancer cells, but also increased the lymphatic metastasis ability of the cells in vivo. Moreover, the silencing of SQLE partly alleviated migration ability and resistance to bicalutamide, and weakened the mitochondrial OXPHOS in PC cells. More interestingly, targeting SQLE expressions by terbinafine enhanced the sensitivity to bicalutamide in PC cells. Thus, these results further illustrate that SQLE may play a crucial role in the acquisition of bicalutamide resistance in PC cells and might be a potential target against CRPC via regulating cholesterol metabolism.
This study was limited by the lack of data on SQLE expression in clinical CRPC patients. The relationship between SQLE expression and bicalutamide resistance has not been observed in actual clinical scenarios. In summary, our results showed that bicalutamide resistance enhanced SQLE expression and increases mitochondrial fatty acid oxidation in PC cells. Blockage of SQLE effectively suppressed bicalutamide resistance concomitant lymph node metastasis in prostate cancer cells. The findings emphasize that SQLE may be a potential biomarker and therapeutic target in CRPC patients.
Ethics and Consent Statement
This study was approved by the Ethics Committee of the Affiliated Cancer Hospital of Xiangya School of Medicine, Central South University, Hunan Cancer Hospital. The welfare and treatment of the laboratory animals followed the Chinese National Guidelines (GB/T 35892‐20181).
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This study was supported by grants from the Qihang Youth Fund of Hunan Cancer Hospital Scientific Research Climbing Project (Grant No. 2020QH003 and QH201907) and Natural Science Foundation for Youths of Jiangxi (Project No. 20202BABL216013).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Gupta SS, Martini A, Wagaskar VG, Tewari AK. Prostate cancer in a patient with giant prostatic hyperplasia: the robotic approach to a difficult problem. J Endourol Case Rep. 2019;5(3):99–101. doi:10.1089/cren.2019.0024
2. Liu Y, Zhou J, Liu C, et al. Comprehensive signature analysis of drug metabolism differences in the White, Black and Asian prostate cancer patients. Aging. 2021;13(12):16316–16340. doi:10.18632/aging.203158
3. Mai L, Li Y, Yang P, et al. Successful management of prostate cancer with bulky pelvic lymph node metastases after rapid development of castration-resistant prostate cancer: a case report with review of the literature. Transl Androl Urol. 2020;9(3):1428–1436. doi:10.21037/tau-20-725
4. Mills IG. Maintaining and reprogramming genomic androgen receptor activity in prostate cancer. Nat Rev Cancer. 2014;14(3):187–198. doi:10.1038/nrc3678
5. Bjartell A, Lumen N, Maroto P, et al. Real-world safety and efficacy outcomes with abiraterone acetate plus prednisone or prednisolone as the first- or second-line treatment for metastatic castration-resistant prostate cancer: data from the prostate cancer registry. Target Oncol. 2021;16(3):357–367. doi:10.1007/s11523-021-00807-4
6. Gordon JA, Noble JW, Midha A, et al. Upregulation of scavenger receptor b1 is required for steroidogenic and nonsteroidogenic cholesterol metabolism in prostate cancer. Cancer Res. 2019;79(13):3320–3331. doi:10.1158/0008-5472.CAN-18-2529
7. Coates HW, Chua NK, Brown AJ. Consulting prostate cancer cohort data uncovers transcriptional control: regulation of the MARCH6 gene. Biochim Biophys Acta Mol Cell Biol Lipids. 2019;1864(11):1656–1668. doi:10.1016/j.bbalip.2019.08.006
8. Ge H, Zhao Y, Shi X, et al. Squalene epoxidase promotes the proliferation and metastasis of lung squamous cell carcinoma cells though extracellular signal-regulated kinase signaling. Thorac Cancer. 2019;10(3):428–436. doi:10.1111/1759-7714.12944
9. Brown DN, Caffa I, Cirmena G, et al. Squalene epoxidase is a bona fide oncogene by amplification with clinical relevance in breast cancer. Sci Rep. 2016;6(1):19435. doi:10.1038/srep19435
10. Garcia-Bermudez J, Baudrier L, Bayraktar EC, et al. Squalene accumulation in cholesterol auxotrophic lymphomas prevents oxidative cell death. Nature. 2019;567(7746):118–122. doi:10.1038/s41586-019-0945-5
11. Han W, Gao S, Barrett D, et al. Reactivation of androgen receptor-regulated lipid biosynthesis drives the progression of castration-resistant prostate cancer. Oncogene. 2018;37(6):710–721. doi:10.1038/onc.2017.385
12. Colli JL, Amling CL. High cholesterol levels are associated with reduced prostate cancer mortality rates during periods of high but not low statin use in the United States. Urol Oncol. 2009;27(2):170–173. doi:10.1016/j.urolonc.2007.11.029
13. Nacarelli T, Azar A, Altinok O, Orynbayeva Z, Sell C. Rapamycin increases oxidative metabolism and enhances metabolic flexibility in human cardiac fibroblasts. Geroscience. 2018;40(3):243–256. doi:10.1007/s11357-018-0030-2
14. Medes G, Thomas A, Weinhouse S. Metabolism of neoplastic tissue. IV. A study of lipid synthesis in neoplastic tissue slices in vitro. Cancer Res. 1953;13(1):27–29.
15. Barbalata CI, Tefas LR, Achim M, Tomuta I, Porfire AS. Statins in risk-reduction and treatment of cancer. World J Clin Oncol. 2020;11(8):573–588. doi:10.5306/wjco.v11.i8.573
16. Mitsiades N. A road map to comprehensive androgen receptor axis targeting for castration-resistant prostate cancer. Cancer Res. 2013;73(15):4599–4605. doi:10.1158/0008-5472.CAN-12-4414
17. Zong Y, Goldstein AS. Adaptation or selection–mechanisms of castration-resistant prostate cancer. Nat Rev Urol. 2013;10(2):90–98. doi:10.1038/nrurol.2012.237
18. Dlubek J, Rysz J, Jablonowski Z, Gluba-Brzozka A, Franczyk B. The correlation between lipid metabolism disorders and the prostate cancer. Curr Med Chem. 2020. doi:10.2174/0929867327666200806103744
19. Guo S, Ma B, Jiang X, Li X, Jia Y. Astragalus polysaccharides inhibits tumorigenesis and lipid metabolism through miR-138-5p/SIRT1/SREBP1 pathway in prostate cancer. Front Pharmacol. 2020;11:598. doi:10.3389/fphar.2020.00598
20. Jamnagerwalla J, Howard LE, Allott EH, et al. Serum cholesterol and risk of high-grade prostate cancer: results from the REDUCE study. Prostate Cancer Prostatic Dis. 2018;21(2):252–259. doi:10.1038/s41391-017-0030-9
21. Giunchi F, Fiorentino M, Loda M. The metabolic landscape of prostate cancer. Eur Urol Oncol. 2019;2(1):28–36. doi:10.1016/j.euo.2018.06.010
22. Skotland T, Ekroos K, Kauhanen D, et al. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur J Cancer. 2017;70:122–132. doi:10.1016/j.ejca.2016.10.011
23. Oh HY, Leem J, Yoon SJ, Yoon S, Hong SJ. Lipid raft cholesterol and genistein inhibit the cell viability of prostate cancer cells via the partial contribution of EGFR-Akt/p70S6k pathway and down-regulation of androgen receptor. Biochem Biophys Res Commun. 2010;393(2):319–324. doi:10.1016/j.bbrc.2010.01.133
24. Nagaraja R, Olaharski A, Narayanaswamy R, et al. Preclinical toxicology profile of squalene epoxidase inhibitors. Toxicol Appl Pharmacol. 2020;401:115103. doi:10.1016/j.taap.2020.115103
25. Li L, Zhang Q, Wang X, et al. Squalene epoxidase-induced cholesteryl ester accumulation promotes nasopharyngeal carcinoma development by activating PI3K/AKT signaling. Cancer Sci. 2020;111(7):2275–2283. doi:10.1111/cas.14426
26. Helms MW, Kemming D, Pospisil H, et al. Squalene epoxidase, located on chromosome 8q24.1, is upregulated in 8q+ breast cancer and indicates poor clinical outcome in stage I and II disease. Br J Cancer. 2008;99(5):774–780. doi:10.1038/sj.bjc.6604556
27. Stopsack KH, Gerke TA, Sinnott JA, et al. Cholesterol metabolism and prostate cancer lethality. Cancer Res. 2016;76(16):4785–4790. doi:10.1158/0008-5472.CAN-16-0903
28. Abudurexiti M, Zhu W, Wang Y, et al. Targeting CPT1B as a potential therapeutic strategy in castration-resistant and enzalutamide-resistant prostate cancer. Prostate. 2020;80(12):950–961. doi:10.1002/pros.24027
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.