Back to Journals » Infection and Drug Resistance » Volume 13
Spoligotype and Drug Susceptibility Profiles of Mycobacterium tuberculosis Complex Isolates in Golestan Province, North Iran
Authors Mansoori N, Vaziri F, Amini S, Khanipour S, Pourazar Dizaji S, Douraghi M
Received 28 March 2020
Accepted for publication 18 June 2020
Published 1 July 2020 Volume 2020:13 Pages 2073—2081
DOI https://doi.org/10.2147/IDR.S255889
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Noormohamad Mansoori,1 Farzam Vaziri,2,3 Sirus Amini,4 Sharareh Khanipour,2,3 Shahin Pourazar Dizaji,2,3 Masoumeh Douraghi1,5
1Division of Microbiology, Department of Pathobiology, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran; 2Department of Mycobacteriology and Pulmonary Research, Pasteur Institute of Iran, Tehran, Iran; 3Microbiology Research Center (MRC), Pasteur Institute of Iran, Tehran, Iran; 4Regional Tuberculosis Reference Laboratory, Tehran University of Medical Sciences, Tehran, Iran; 5Food Microbiology Research Center, Tehran University of Medical Sciences, Tehran, Iran
Correspondence: Masoumeh Douraghi
Division of Microbiology, Department of Pathobiology, School of Public Health, Tehran University of Medical Sciences, PO Box: 14155-6446, Tehran, Iran
Tel +98 21 42933152
Fax +98 21 88954913
Email [email protected]
Introduction: Despite the moderate incidence of tuberculosis (TB) in many parts of Iran, Golestan province had a permanently higher TB incidence rate than the national average. Moreover, Golestan province receives immigrants, mainly from TB-endemic areas of Iran and neighbor countries. Here, we aimed to characterize the circulating Mycobacterium tuberculosis complex (MTBC) isolates in terms of the spoligotype and drug resistance patterns, across Golestan province.
Materials and Methods: A set of 166 MTBC isolates was collected during July 2014 to July 2015 and subjected to drug susceptibility testing for first- and second-line anti-TB drugs and spoligotyping.
Results: Of 166 MTBC isolates, 139 (83.7%) isolates were assigned to 28 spoligotype international types (SITs). The most frequent SITs were SIT127/Ural-2 (n=25, 15.1%), followed by SIT1/Beijing (n=21, 12.7%) and SIT3427/Ural-2 (n=18, 10.8%). The set of 18 isolates (10.8%) showed resistance to at least one drug, which mainly belonged to SIT1/Beijing (n=7, 38.9%), orphan patterns (n=4, 22.2%) and SIT357/CAS1-Delhi (n=3, 16.7%). In addition, four isolates (2.4%) were resistant to pyrazinamide. The analysis of mutation corresponded to resistance to rifampin and isoniazid showed that two isolates had Ser531Leu substitution in rpoB, four isolates had Ser315Thr substitution in katG and one isolate had [C(− 15)T] in inhA locus.
Conclusion: High diversity in spoligotypes of the MTBC isolates and lack of dominant genotype might be due to residence of immigrants in this region and consequent reactivation of latent infection. In addition, due to the presence of extensively drug-resistant (XDR) isolates in Golestan province, it is important to conduct future studies to determine transmission pattern of drug-resistant isolates in this region.
Keywords: Mycobacterium tuberculosis, tuberculosis, spoligotyping, genotyping, transmission
Introduction
Mycobacterium tuberculosis (MTB) and Mycobacterium bovis are the most important members of M. tuberculosis complex (MTBC).1 They are known as causative agents of tuberculosis (TB), which infects almost one third of the global population.2 The TB incidence is moderate in Iran as a fairly large country (14 per 100,000 people), but is high in neighbor countries such as Turkmenistan, Afghanistan and Pakistan.2,3 Running the national TB control programs in the recent decades, incidence of TB has decreased dramatically in Iran, but it has remained high in some regions such as Golestan province (32.1 per 100,000).4 Emergence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) isolates can result in complicated as well as costly treatment regimens and is attributed to the failure of the implementation of proper TB control strategies.5 In this situation, reliable and timely drug susceptibility testing using conventional and molecular methods have an essential role for diagnosis of MDR isolates.6
Pyrazinamide (PZA) is one of the most critical drugs for treatment of drug-susceptible and MDR-TB. Despite including PZA as a first-line drug for TB treatment, assessment of its in vitro activity against MTB isolates is technically difficult because PZA is only active at acidic pH. Hence, the pyrazinamidase (PZase) test, also known as the Wayne test, was introduced as simple method for PZA drug susceptibility testing (DST).7
Studying the molecular epidemiology of MTBC isolates could assist TB control programs through understanding the predominant MTBC genotypes and their transmission dynamics within the population.8 Spacer oligonucleotide typing (spoligotyping) is an approach that is widely used for exploring the genotypic structure of MTBC isolates because it is simple, rapid, and requires a very low amount of DNA.9,10
Golestan province is situated in the north of Iran in the vicinity of the Caspian Sea and Turkmenistan country. We previously reported that Golestan province has received a large number of immigrants by inter-province movement and from neighbor countries such as Afghanistan and Turkmenistan.11 Within this context, the present study aimed to explore the circulating spoligotypes, drug resistance rate to PZA and to identify the mutations conferring resistance to rifampin (RMP) and isoniazid (INH) in MTBC isolates in Golestan province.
Materials and Methods
Isolates Collection
The study proposal was approved by the ethics committee of Tehran University of Medical Sciences. A cross-sectional epidemiological study was carried out for the pulmonary TB suspected cases living in Golestan province, from July 2014 to July 2015. The patients who were culture positive for MTB were included in this study. All isolates were collected during routine procedures at the Tuberculosis Reference laboratory, Golestan, Iran, and no samples were specifically collected for this research. Patients with extrapulmonary TB and culture negative pulmonary TB cases were excluded. The 11,807 clinical specimens were collected from diagnosed cases during this study and 166 clinical isolates were recovered from new (n=164) and previously treated (n=2) TB cases in Golestan province (one isolate per patient) and included in this study. The identification of the isolates was performed according to the standard microbiologic procedures.12
Drug Susceptibility Testing
DST to first-line anti-TB drugs; rifampin (RMP; 40 μg/mL), isoniazid (INH; 0.2 μg/mL), ethambutol (ETL; 2 μg/mL) and streptomycin (STM; 4 μg/mL) was performed for all isolates on Lowenstein-Jensen (LJ) medium using proportional method. MDR isolates were further subjected for second-line anti-TB drugs; ciprofloxacin (CIP; 2 μg/mL), ofloxacin (OFX; 4 μg/mL), ethionamide (ETD; 40 μg/mL), kanamycin (KAN; 30 μg/mL), capreomycin (CAP; 40 μg/mL), cycloserin (CYN; 30 μg/mL), para-aminosalicylic acid (PAS; 1 μg/mL) (all from Sigma, USA) and amikacin (AMK; 30 μg/mL) (Exir, Iran) on the LJ medium via the proportional method and susceptibility testing for levofloxacin (LVX; 1 μg/mL) (Sigma, USA) performed on Middlebrook 7H10 agar media (BBL Microbiology Systems, Cockeysville, MD, USA).12,13 Reference strain MTB H37Rv was used for quality control.
PZase (Wayne) Test
PZase activity was determined using Wayne technique,7,14 in Middlebrook 7H10 medium containing albumin dextrose catalase (ADC) growth supplement (BBL Microbiology Systems, Cockeysville, MD, USA), 100 μg/mL PZA and 2 mg/mL sodium pyruvate (Sigma, United States). Briefly, isolates cultured in the LJ medium, then a heavy suspension of each isolate inoculated onto the two tube of Middlebrook 7H10 agar, and incubated at 37°C. One milliliter of freshly prepared 1% ferrous ammonium sulfate solution (Merck, Germany) was introduced into one of the test tubes after 4 days of incubation. A positive PZase activity appeared as a pink-brown band on the agar surface.15 In the case of the absence of the pink-brown ring, the second tube was tested after additional two days of incubation. The MTB strain H37Rv and M. bovis used as PZase positive and negative control, respectively.
PCR and Sequencing
The three loci (rpoB, katG and inhA) were amplified by PCR using locus-specific primers, followed by agarose gel electrophoresis of amplicons.5,16 The amplicons were subjected to sequencing (Macrogen, Korea) and the sequences were compared with the deposited sequences for MTB strain H37Rv using the BLAST. The sequences have been deposited in GenBank and provided accession numbers were documented.
Spoligotyping
Genomic DNA for 166 MTBC isolates was extracted from fresh sub-cultures.17 Spoligotyping was carried out using the commercially available kit (Mapmygenome, India) according to the standard protocol.18 MTB strain H37Rv and M. bovis strain BCG were used as positive control and distilled water as a negative control. Distribution of the isolates into different genotypes was performed using the SITVITWEB database (http://www.pasteur-guadeloupe.fr:8081/SITVIT2/batch.jsp).19
Definitions
In the SITVITWEB database, spoligotype international type (SIT) represents spoligotype shared by two or more isolates, whereas patterns reported for a single isolate designates as “orphan”.19 Isolates sharing identical spoligotyping patterns were categorized as “clusters” and the pattern was called “unique”, if the spoligotype patterns correspond to only one isolate.11 Isolate that was resistant to at least one anti-TB drugs was defined as “any drug resistant”. MDR-TB is defined as isolates that resistant to both INH and RMP, while XDR-TB is defined as MDR-TB that is resistant to any fluoroquinolone and at least one of injectable second-line drugs (AMK, KAN or CAP).5
Statistical Analysis
Data analysis was performed using SPSS ver. 22 (SPSS Inc., Chicago, IL, USA). Descriptive analysis of the data was conducted using frequencies (counts). For comparison of the categorical variables, the Fisher’s exact test was used at p <0.05.
Results
Demographic Information
A total of 166 clinical MTBC isolates (164 MTB and 2 M. bovis) were obtained from pulmonary TB patients and included in this study. The majority of isolates (n=150, 90.4%) were recovered from sputum while 15 (9%) were from bronchoalveolar lavage and only one isolate (0.6%) was obtained from gastric juice. The average age of patients were 50.4 ± 19.5 years and 59.6% (n= 99) were male. The 158 (95.2%) patients were Iranian whereas 8 (4.8%) were foreign immigrants.
Drug Susceptibility Patterns
The set of 148 (89.2%) isolates were susceptible to all first-line anti-TB drugs, and 18 (10.8%) were found to be any drug resistant, including; two (1.2%) MDR, three (1.8%) INH monoresistant, 12 (7.2%) STM monoresistant, and one isolate (0.6%) that resistant to INH and STM. In addition to the phenotypic DST for second-line anti-TB drugs was completed on two MDR isolates and both were resistant to ETL, but one of them showed resistance to CIP, OFX, LVX, AMK and KAN.
Resistance to PZA
As M. bovis is naturally resistant to the PZA, then PZase test was conducted for 164 MTB isolates. Among them, four (2.4%) were resistant to PZA, including two MDR and two non-MDR isolates.
Mutations in rpoB, katG and inhA Loci
The analysis for rifampin resistance-determining region (RRDR) of rpoB loci for two MDR isolates showed that both had Ser531Leu substitution (TCG→TTG) (GenBank accession nos. MG490375 and MG697230). We analyzed inhA and katG loci for six INH resistant isolates, four isolates (66.7%) had Ser315Thr substitution (AGC→ACC) in katG gene (GenBank accession nos. MG461957, MG461959, MG461960 and MG490376) and one (16.7%) cytosine-to-thymine transition [C(−15)T] was detected at the nucleotide positioned 15 bases upstream of the start codon within the inhA promoter region (GenBank accession no. MH373357). One INH-resistant isolate had not mutation at katG and inhA loci, and termed as wild type. Identified mutations and relevant accession nos. in GenBank were summarized in Table 1.
 |
Table 1 Identified Mutations for rpoB, katG and inhA Loci in Mycobacterium tuberculosis Complex Isolates, and the Relevant GenBank Accession Numbers |
Spoligotyping Family Distribution
A total of 139 (83.7%) isolates were represented by 28 SITs. The most frequent genotypes were; SIT127/Ural-2 (n=25, 15.1%), SIT1/Beijing (n=21, 12.7%), SIT3427/Ural-2 (n=18, 10.8%), SIT21/CAS1-Kili (n=11, 6.6%), SIT25/CAS1-Delhi (n=10, 6%), SIT357/CAS1-Delhi (n=10, 6%), and 27 isolates (16.3%) displayed orphan patterns (Figure 1). MDR isolates shared SIT-1/Beijing spoligotype and the detailed information about assigned spoligotypes with respect to drug resistance pattern were described in Table 2.
 |
Table 2 Association Between Identified Mycobacterium tuberculosis Complex Genotypes and Drug Resistance Patterns of 166 M. tuberculosis Isolates in Golestan Province |
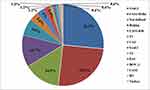 |
Figure 1 The Mycobacterium tuberculosis complex genotypes identified in the studied population. |
Among the 166 MTBC isolates, a total of 49 different patterns were detected; 136 isolates were distributed into 19 clusters and the remaining (n=30) had unique patterns. The largest cluster corresponded to the SIT127 (n=25), followed by SIT1 (n=21) and SIT127 (n=18).
Discussion
Using spoligotyping method, the large variety of MTBC genotypes was identified in Golestan province. The overall population structure of MTBC isolates in Golestan province is dominated by the CAS family (including CAS1-Delhi, CAS1-Kili, CAS and CAS2), that corresponded to one-third of the studied isolates. As mentioned in the previous study,11 Golestan province receives large numbers of immigrants, especially from Sistan and Baluchestan province (south of Iran) and Afghanistan country. Moreover, demographic investigations showed that most of the CAS family isolates were obtained from patients who migrated from Sistan and Baluchestan province. In 2013, Haeili et al,20 reported that 57% of isolates in Sistan and Baluchistan province belonged to the CAS family. It is important to mention that the CAS/Delhi genotype is ubiquitously prevalent in Iran’s neighboring countries such as, Saudi Arabia, India, Pakistan, and Afghanistan.21,25 Therefore, it is likely that the CAS/Delhi family was disseminated to Iran through frequent migration and travel between the boundaries.
Ural-2 was found as the second most common genotype in the current study, that was assigned as NEW-1 genotype, based on the MIRU-VNTRplus database.26 Ural-2 has earlier been reported with high prevalence from Tehran (capital of Iran) and Khorasan (eastern neighbor of Golestan province),27,28 indicating that this genotype was abundant in Iran and reported with a much lesser extent in Kyrgyzstan and Kazakhstan countries.29,30 The current study confirms the previous theory that mentioned Ural-2 is endemic for Iran.31
The Beijing family was found to be particularly prevalent in Golestan province, and has been associated with any drug resistance pattern as well as two MDR isolates. It has been shown that Beijing isolates were associated with MDR and XDR-TB in several studies.32,33 Beijing is known as common genotype circulating in the countries located in northern Iran (Kazakhstan, Georgia, and Russia) and countries located in Central and Eastern Central Asia.29,32,34,35 Therefore, the presence of this genotype in Golestan province can be attributed to the migration and traffic of the people from the mentioned countries to this geographical region.
In this study, Ural-1 and T1 family were found with a lower percentage. In Iran, Ural-1 and T1 family, were previously found with percentages of 1–16% and 2.1–13% in various regions of Iran,20,27,28,36,38 which could indicate a relatively uniform scattering of them in different parts of the country.
In this study we find orphan isolates belonged to cluster that we named as Orphan-4, which shared by three clinical isolates. Their fingerprints were closely related to SIT127, but differ in spoligospacers four and five. Thus, to the best of our knowledge, this could be evidence for active evolution of SIT127/Ural-2 in Golestan province. In addition to Orphan-4, in this study we found four new orphan clusters (Orphan-1, Orphan-3, and Orphan-5). Surprisingly, we found that Orphan-3 was associated with drug resistance, as two out of three isolates were INH resistant but other clusters were pansusceptible.
In the present study, spoligotyping patterns of 27 isolates did not match the patterns available in SITVITWEB database, suggesting the high evolutionary pressure on the circulating isolates (as mentioned for Orphan-4) and/or presence of new spoligotypes specially for Orphan-1, Orphan-3, and Orphan-5 clusters in this region.
Golestan province is one of the main ranch areas in Iran, and finding two M. bovis isolates is a reminder for zoonotic nature of bovine TB. Despite the efforts that made for M. bovis eradication, it has not yet been completely controlled in Iran. As one of M. bovis isolates was resistant to INH, a comprehensive study must be conducted in order to assess the antibiotic resistance pattern and epidemiology of bovine TB in this province.
In this study, gene regions with resistance-associated mutations of katG, inhA and rpoB were amplified using locus-specific primers. The results of sequencing for INH resistant isolates showed that 83% of them had mutations in the codon 315 of katG or promoter region of inhA genes. According to the results of the present study, mutation in the codon 315 was reported in the previous studies conducted in Iran and other countries with the percentages of 27– 97.5%.5,16,39,42 Then, it is considered that mutations in the katG are the most important reason for the INH resistance in MTBC isolates, and the mutation in the inhA gene has a minor role. Moreover, one isolate was wild type at both loci, which may have other mechanisms for INH resistance, such as mutation at the oxyR-ahpC promoter regions, kasA or ndh loci.
It was well established that mutations within the RRDR of rpoB locus occur in more than 95% of RMP-resistant isolates. In this study, both MDR isolates had Ser531Leu mutations in rpoB gene. Mutation in codon 531 was frequently reported by studies from Iran, Pakistan, and Egypt, as the most predominant mutation responsible for RMP-resistance.5,43,44 In addition, studies conducted in other countries have shown that mutation in codon 531 corresponded with 29–79% of RMP-resistant phenotype.16,45,46 Furthermore, we found Thr481Ser substitution that was never reported previously, and further studies are required to definitively characterize its role in RMP resistance.
In this study, we found an MDR isolate that was simultaneously resistant to fluoroquinolones and second-line injectable drugs, determined as XDR-TB. To the best of our knowledge, this was the first report of XDR-TB in Golestan province. DST for second-line drugs was studied in different parts of Iran and resistance varied from 11.1–100%.5,47,49 Although the use of first-line drugs was effective for treatment of TB, the emergence of drug-resistant isolates has reduced the efficacy of the standard regime and becomes a major constraint for TB control programs. Emergence of MDR and XDR isolates is a significant barrier for TB control, and it is important to ensure that patients receive effective treatment. Moreover, future epidemiological studies must be designed for determining the transmission root of drug-resistant isolates in this region.
Assessment of PZase activity of MTB isolates using the Wayne method showed that the resistance to PZA was low in the drug susceptible isolates, but high in MDRs. Results of a meta-analysis study showed that resistance to PZA in non-MDR isolates was up to 9%, but it ranged from 31– 89% in MDR isolates,50 which is consistent with the current study. PZA is a critical drug, which is active in acidic pH (5 to 5.5) and able to eradicate semidormant forms of MTB. Using PZA, duration of TB treatment shortened from 9–12 months to six months.51,52 Currently, DST in the Mycobacteria Growth Indicator Tube (MGIT) and BACTEC 460 systems are considered as the reference method for PZA susceptibility testing.13 Unfortunately, such systems are not available in resource-limited countries; and they could not routinely perform DST for PZA. The Wayne test is a simple and relatively inexpensive method. Considering the advantages of this method, it could be proposed as alternative method of detection of PZA resistance. It is important to mention that the Wayne test is mainly correlated with the activity of pyrazinamidase enzyme, but other mechanisms might be implicated in the resistance to PZA. Molecular detection of mutations in pncA locus is another method for determining the resistance to PZA that could be used in such condition. Due to the limited number of MDR isolates in this study, future studies using a large number of MDR isolates is necessary for determining the resistance to PZA in MDR isolates circulating in this region.
This study has some limitations. First, we had a limited number of MDR isolates for PZA susceptibility testing and DST for second-line drugs in this study. Second, results for the Wayne test must be confirmed using MGIT960, BACTEC 460 systems or molecular method.
Conclusion
This study showed the presence of highly diverse spoligotyping patterns in Golestan province. The observed genetic diversity of MTBC isolates in this region reflects the role of immigration plus active evolution of isolates circulating in Golestan province. Migration and presence of XDR-TB in this region must be noted as epidemiological and clinical concerns that could be a threat to TB control programs in Iran. Detection of isolates with reduced PZase activity implies the possibility for the presence of initial resistance of MTB isolates recovered from new cases. We recommend further studies for identifying genetic relation between MTBC isolates using whole-genome sequencing method, also such information could be used for detection of mutations responsible for resistance to first- and second-line anti-TB drugs.
Ethical Statement
All studied isolates were collected during routine procedures at Tuberculosis Reference laboratory, Golestan, Iran, and no samples were specifically collected for this research.
Acknowledgments
This research was supported by grants from Tehran University of Medical Sciences and Health Services (grant number: 30242), Iran. We thank all the staff of Mycobacteriology and Pulmonary Research Department, Pasteur Institute of Iran and Tehran Tuberculosis Regional Reference Laboratory for their assistance in this project.
Disclosure
The authors declare that there is no conflict of interest.
References
1. Mansoori N, Douraghi M, Rajabloo AA, Taziki M, Yaseri M, Vaziri F. Mycobacterium tuberculosis complex drug resistance in a high tuberculosis incidence area from the WHO eastern mediterranean region. J Pharm Pharm Sci. 2017;20(1):428–434. doi:10.18433/J3J64H
2. WHO. Global Tuberculosis Report 2018. Geneva, Switzerland: WHO; 2018.
3. http://www.who.int/tb/country/data/profiles.
4. http://tb-lep.behdasht.gov.ir/TBsituationinIran.aspx., 2019.
5. Sakhaee F, Ghazanfari M, Ebrahimzadeh N, et al. A comparative study of phenotypic and genotypic first- and second-line drug resistance testing of Mycobacterium tuberculosis. Biologicals. 2017;49:33–38. doi:10.1016/j.biologicals.2017.07.003
6. WHO. WHO Treatment Guidelines for Drug-Resistant Tuberculosis 2016 Update; 2016.
7. Wayne LG. Simple pyrazinamidase and urease tests for routine identification of mycobacteria. Am Rev Respir Dis. 1974;109(1):147–151. doi:10.1164/arrd.1974.109.1.147
8. Chawla K, Kumar A, Shenoy VP, Chauhan DS, Sharma P. Genetic diversity of Mycobacterium tuberculosis in south coastal Karnataka, India, using spoligotyping. Indian J Med Res. 2018;147(3):278–286. doi:10.4103/ijmr.IJMR_2026_16
9. Monteserin J, Paul R, Gravina E, et al. Genotypic diversity of Mycobacterium tuberculosis in Buenos Aires, Argentina. Infect Genet Evol. 2018;62:1–7. doi:10.1016/j.meegid.2018.04.006
10. Jagielski T, Minias A, van Ingen J, et al. Methodological and clinical aspects of the molecular epidemiology of Mycobacterium tuberculosis and Other Mycobacteria. Clin Microbiol Rev. 2016;29(2):239–290. doi:10.1128/CMR.00055-15
11. Mansoori N, Yaseri M, Vaziri F, Douraghi M. Genetic diversity of Mycobacterium tuberculosis complex isolates circulating in an area with high tuberculosis incidence: using 24-locus MIRU-VNTR method. Tuberculosis. 2018;112:89–97. doi:10.1016/j.tube.2018.08.003
12. Rieder HL, Chonde TM, Myking H, et al. The Public Health Service National Tuberculosis Reference Laboratory and the National Laboratory Network; Minimum Requirements, Role and Operation in a Low-Income Country. Paris: International Union against Tuberculosis and Lung Disease (IUATLD); 1998:110 p.
13. WHO. Updated interim critical concentrations for first-line and second-line DST Geneva, Switzerland 2012. Available from: http://www.stoptb.org/wg/gli/assets/documents/Updated%20critical% 20concentration%20table_1st% 20and%202nd% 20line%20drugs.pdf..
14. Stottmeier KD, Beam RE, Kubica GP. Determination of drug susceptibility of mycobacteria to pyrazinamide in 7H10 agar. Am Rev Respir Dis. 1967;96(5):1072–1075. doi:10.1164/arrd.1967.96.5.1072
15. Aono A, Hirano K, Hamasaki S, Abe C. Evaluation of BACTEC MGIT 960 PZA medium for susceptibility testing of Mycobacterium tuberculosis to pyrazinamide (PZA): compared with the results of pyrazinamidase assay and Kyokuto PZA test. Diagn Microbiol Infect Dis. 2002;44(4):347–352. doi:10.1016/S0732-8893(02)00471-6
16. Campbell PJ, Morlock GP, Sikes RD, et al. Molecular detection of mutations associated with first- and second-line drug resistance compared with conventional drug susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2011;55(5):2032–2041. doi:10.1128/AAC.01550-10
17. van Soolingen D, De Haas PE, Hermans PW, van Embden JD. DNA fingerprinting of Mycobacterium tuberculosis. Methods Enzymol. 1994;235:196–205.
18. Kamerbeek J, Schouls L, Kolk A, et al. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35(4):907–914. doi:10.1128/JCM.35.4.907-914.1997
19. Demay C, Liens B, Burguiere T, et al. SITVITWEB–a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol. 2012;12(4):755–766. doi:10.1016/j.meegid.2012.02.004
20. Haeili M, Darban-Sarokhalil D, Fooladi AA, et al. Spoligotyping and drug resistance patterns of Mycobacterium tuberculosis isolates from five provinces of Iran. Microbiologyopen. 2013;2(6):988–996. doi:10.1002/mbo3.139
21. Sharma P, Chauhan DS, Upadhyay P, et al. Molecular typing of Mycobacterium tuberculosis isolates from a rural area of Kanpur by spoligotyping and mycobacterial interspersed repetitive units (MIRUs) typing. Infect Genet Evol. 2008;8(5):621–626. doi:10.1016/j.meegid.2008.05.002
22. Chatterjee A, Mistry N. MIRU-VNTR profiles of three major Mycobacterium tuberculosis spoligotypes found in western India. Tuberculosis. 2013;93(2):250–256. doi:10.1016/j.tube.2012.10.004
23. Arora J, Singh UB, Suresh N, et al. Characterization of predominant Mycobacterium tuberculosis strains from different subpopulations of India. Infect Genet Evol. 2009;9(5):832–839. doi:10.1016/j.meegid.2009.05.008
24. Ali A, Hasan Z, Jafri S, Inayat R, Hasan R. Mycobacterium tuberculosis Central Asian Strain (CAS) lineage strains in Pakistan reveal lower diversity of MIRU loci than other strains. Int J Mycobacteriol. 2014;3(2):108–116. doi:10.1016/j.ijmyco.2014.03.002
25. Al-Hajoj S, Varghese B, Al-Habobe F, Shoukri MM, Mulder A, van Soolingen D. Current trends of Mycobacterium tuberculosis molecular epidemiology in Saudi Arabia and associated demographical factors. Infection, Genetics and Evolution. 2013;16:362–368. doi:10.1016/j.meegid.2013.03.019
26. Allix-Beguec C, Harmsen D, Weniger T, Supply P, Niemann S. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2008;46(8):2692–2699. doi:10.1128/JCM.00540-08
27. Ravansalar H, Tadayon K, Mosavari N, Derakhshan M, Ghazvini K. Genetic Diversity of Mycobacterium tuberculosis Complex Isolated from Patients in the Northeast of Iran by MIRU-VNTR and Spoligotyping. Jundishapur J Microbiol. 2017;10(4):e39568.
28. Feyisa SG, Haeili M, Zahednamazi F, et al. Molecular characterization of Mycobacterium tuberculosis isolates from Tehran, Iran by restriction fragment length polymorphism analysis and spoligotyping. Rev Soc Bras Med Trop. 2016;49(2):204–210. doi:10.1590/0037-8682-0405-2015
29. Skiba Y, Mokrousov I, Ismagulova G, et al. Molecular snapshot of Mycobacterium tuberculosis population in Kazakhstan: a country-wide study. Tuberculosis. 2015;95(5):538–546. doi:10.1016/j.tube.2015.04.012
30. Mokrousov I, Isakova J, Valcheva V, Aldashev A, Rastogi N. Molecular snapshot of Mycobacterium tuberculosis population structure and drug-resistance in Kyrgyzstan. Tuberculosis. 2013;93(5):501–507. doi:10.1016/j.tube.2013.05.008
31. Mokrousov I, Shitikov E, Skiba Y, Kolchenko S, Chernyaeva E, Vyazovaya A. Emerging peak on the phylogeographic landscape of Mycobacterium tuberculosis in West Asia: definitely smoke, likely fire. Mol Phylogenet Evol. 2017;116:202–212. doi:10.1016/j.ympev.2017.09.002
32. Niemann S, Diel R, Khechinashvili G, Gegia M, Mdivani N, Tang YW. Mycobacterium tuberculosis Beijing lineage favors the spread of multidrug-resistant tuberculosis in the Republic of Georgia. J Clin Microbiol. 2010;48(10):3544–3550. doi:10.1128/JCM.00715-10
33. Gandhi NR, Nunn P, Dheda K, et al. Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet. 2010;375(9728):1830–1843. doi:10.1016/S0140-6736(10)60410-2
34. Merker M, Blin C, Mona S, et al. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet. 2015;47(3):242–249. doi:10.1038/ng.3195
35. Balabanova Y, Nikolayevskyy V, Ignatyeva O, et al. Beijing clades of Mycobacterium tuberculosis are associated with differential survival in HIV-negative Russian patients. Infect Genet Evol. 2015;36:517–523. doi:10.1016/j.meegid.2015.08.028
36. Zamani S, Haeili M, Nasiri MJ, Imani Fooladi AA, Javadpour S, Feizabadi MM. Genotyping of Mycobacterium tuberculosis Isolates from Hormozgan Province of Iran Based on 15-Locus MIRU-VNTR and Spoligotyping. Int J Bacteriol. 2016;2016:7146470. doi:10.1155/2016/7146470
37. Torkaman MR, Nasiri MJ, Farnia P, Shahhosseiny MH, Mozafari M, Velayati AA. Estimation of recent transmission of mycobacterium tuberculosis strains among iranian and afghan immigrants: a cluster-based study. J Clin Diagn Res. 2014;8(9):5–8.
38. Sharifipour E, Nasiri M, Farnia P, Mozafari M, Irani S. Evaluation of molecular diversity of Mycobacterium tuberculosis strains by polymorphisms in RD Regions. J Mycobac Dis. 2014;4(153):2161.
39. Lipin MY, Stepanshina VN, Shemyakin IG, Shinnick TM. Association of specific mutations in katG, rpoB, rpsL and rrs genes with spoligotypes of multidrug-resistant Mycobacterium tuberculosis isolates in Russia. Clin Microbiol Infect. 2007;13(6):620–626. doi:10.1111/j.1469-0691.2007.01711.x
40. Hazbon MH, Brimacombe M, Bobadilla Del Valle M, et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2006;50(8):2640–2649. doi:10.1128/AAC.00112-06
41. Doustdar F, Khosravi AD, Farnia P, Masjedi MR, Velayati AA. Molecular analysis of isoniazid resistance in different genotypes of Mycobacterium tuberculosis isolates from Iran. Microbial Drug Resist. 2008;14(4):273–279. doi:10.1089/mdr.2008.0842
42. Bostanabad SZ, Nojoumi SA, Jabbarzadeh E, et al. High level isoniazid resistance correlates with multiple mutation in the katG encoding catalase proxidase of pulmonary tuberculosis isolates from the frontier localities of Iran. Tuberk Toraks. 2011;59(1):27–35. doi:10.5578/tt.761
43. Yasmin M, Gomgnimbou MK, Siddiqui RT, Refregier G, Sola C. Multi-drug resistant Mycobacterium tuberculosis complex genetic diversity and clues on recent transmission in Punjab, Pakistan. Infect Genet Evol. 2014;27:6–14. doi:10.1016/j.meegid.2014.06.017
44. Abbadi SH, Sameaa GA, Morlock G, Cooksey RC. Molecular identification of mutations associated with anti-tuberculosis drug resistance among strains of Mycobacterium tuberculosis. Int J Infect Dis. 2009;13(6):673–678. doi:10.1016/j.ijid.2008.10.006
45. van Deun A, Aung KJ, Bola V, et al. Rifampin drug resistance tests for tuberculosis: challenging the gold standard. J Clin Microbiol. 2013;51(8):2633–2640. doi:10.1128/JCM.00553-13
46. Suresh N, Singh UB, Arora J, et al. rpoB gene sequencing and spoligotyping of multidrug-resistant Mycobacterium tuberculosis isolates from India. Infect Genet Evol. 2006;6(6):474–483. doi:10.1016/j.meegid.2006.03.001
47. Velayati AA, Masjedi MR, Farnia P, et al. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in iran. Chest. 2009;136(2):420–425. doi:10.1378/chest.08-2427
48. Tasbiti AH, Yari S, Ghanei M, Shokrgozar MA, Fateh A, Bahrmand A. Low levels of extensively drug-resistant tuberculosis among multidrug resistant tuberculosis isolates and their relationship to risk factors: surveillance in Tehran, Iran; 2006 to 2014. Osong Public Health Res Perspect. 2017;8(2):116–123. doi:10.24171/j.phrp.2017.8.2.03
49. Masjedi MR, Farnia P, Sorooch S, et al. Extensively drug-resistant tuberculosis: 2 years of surveillance in Iran. Clin Infect Dis. 2006;43(7):841–847. doi:10.1086/507542
50. Chang KC, Yew WW, Zhang Y. Pyrazinamide susceptibility testing in Mycobacterium tuberculosis: a systematic review with meta-analyses. Antimicrob Agents Chemother. 2011;55(10):4499–4505. doi:10.1128/AAC.00630-11
51. Mitchison DA. The action of antituberculosis drugs in short-course chemotherapy. Tubercle. 1985;66(3):219–225. doi:10.1016/0041-3879(85)90040-6
52. Doustdar F, Khosravi AD, Farnia P. Mycobacterium tuberculosis genotypic diversity in pyrazinamide-resistant isolates of Iran. Microbial Drug Resist. 2009;15(4):251–256. doi:10.1089/mdr.2009.0066
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
