Back to Journals » OncoTargets and Therapy » Volume 15
SPINDOC is Highly Expressed in Pan-Cancer Samples and Can Promote the Proliferation, Invasion and Migration of Hepatocellular Carcinoma Cells by Activating Wnt/β-Catenin Signaling Pathway
Authors Tong W, Yang L, Liu L, Liu X, Luo N
Received 4 December 2021
Accepted for publication 26 April 2022
Published 18 May 2022 Volume 2022:15 Pages 555—570
DOI https://doi.org/10.2147/OTT.S348843
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Arseniy Yuzhalin
Wangxia Tong,1 Lilan Yang,1 Li Liu,1 Xudong Liu,1 Ning Luo2
1The Medical Department of Hepatology, Ruikang Hospital Affiliated to Guangxi University of Chinese Medicine, Nanning, 530011, People’s Republic of China; 2RuiKang Hospital Affiliated to Guangxi University of Chinese Medicine, Nanning, 530011, People’s Republic of China
Correspondence: Ning Luo, RuiKang Hospital Affiliated to Guangxi University of Chinese Medicine, 10 Huadong Road, Nanning, People’s Republic of China, 530011, Tel +86 0771-2183191, Fax +86 0771-4733943, Email [email protected]
Purpose: As a novel genetic biomarker, little information is available about the possible role of SPINDOC in different malignant tumors and in hepatocellular carcinoma (HCC).
Methods: Based on The Cancer Genome Atlas (TCGA) database, the expression level of SPINDOC in pan-cancer and hepatocellular carcinoma samples was first determined using bioinformatics analysis. The potential relationship between the expression level as well as the clinical characteristics and the molecular mechanisms through which SPINDOC can promote the proliferation, invasion and migration of HCC cells was evaluated. In addition, cell-based studies and in vivo experiments were used to verify the bioinformatics analysis results.
Results: Bioinformatics analysis showed that SPINDOC expression was significantly increased in 18 human malignancies and the gene expression level was positively correlated with poor clinical prognosis in kidney renal papillary cell carcinoma (KIRP), pheochromocytoma and paraganglioma (PCPG) and liver hepatocellular carcinoma (LIHC). The main type of genetic variation of SPINDOC was amplification, and the increase of SPINDOC mRNA expression level was directly related to the amplification of this gene. The expression level of SPINDOC in patients with primary HCC was positively correlated with poor clinical prognosis, as well as the clinical grade and stage of carcinoma. Gene set enrichment analysis (GSEA) analysis showed that high expression of SPINDOC could activate Wnt/β-catenin signaling pathway. Moreover, in vitro and in vivo experiments showed that SPINDOC gene silencing significantly inhibited the proliferation, migration and invasion of Huh7 and SK-HEP-1 cells and decreased the expression of SPIN1, Wnt1, β-catenin and cyclin D1 but increased the expression of AXIN2.
Conclusion: SPINDOC is highly expressed in pan-cancer and HCC samples. This factor can effectively promote the invasion and migration of hepatocellular carcinoma (HCC) cells by activating Wnt/β-catenin signaling pathway, and thus can serve as a potential therapeutic target for HCC management.
Keywords: SPINDOC, SPIN1, Wnt/ß-catenin, pan-cancer, hepatocellular carcinoma
Introduction
Liver cancer is a lethal malignancy, among which hepatocellular carcinoma (HCC) constitutes the main type. HCC displays the characteristics of high malignancy and rapid metastasis and has recurrent rate after treatment.1,2 The clinical symptoms of early HCC have been found to be insidious, and approximately 70% to 80% of patients have metastasized tumors when they are initially diagnosed. Although there are several treatment options such as surgical liver resection, radiotherapy as well as chemotherapy available for the management of this malignancy, the prognosis of HCC patients still remains very poor.1–3 Targeted therapy represented by sorafenib as an important treatment modality for HCC has achieved promising clinical effects to some extent.4 However, there is an urgent need to identify novel and efficient targeted therapy biomarkers to improve clinical treatment outcomes.
Spindlin1 (SPIN1) is an important member of the SPIN/SSTY gene family.5 A number of previous studies have shown that SPIN1 can promote tumor progression by activating Wnt signaling pathway in breast cancer, non-small cell lung cancer and glioma.6–8 Based on SPIN1ʹs tumor-promoting effects in breast cancer,9 gastric cancer10 and liver cancer,11 SPIN1 is considered function as a tumor-promoting factor. As an interaction partner of SPIN1, SPIN1 docking protein (SPINDOC) plays an important biological role by binding to SPIN1 protein.12 SPINDOC can block the potential interaction between SPIN1 and methylated histone tails, thus inhibiting the activation of SPIN1 on the various downstream pathways and molecules. SPINDOC has been considered as a negative regulator of SPIN1. However, it was also found that SPINDOC was highly expressed in a variety of tumors, and the mutant SPINDOC could activate the Wnt signaling cascade.13 Since SPINDOC has been poorly studied in cancer, the role of this factor in cancer remains unclear. Therefore, we have designed this study to investigate the potential role of SPINDOC HCC progression.
Bioinformatics analysis was used to observe the expression of SPINDOC and to decipher the relationship between the expression level of SPINDOC in pan-cancer and HCC samples of different age, gender, tumor clinical grade, prognosis stage, etc. Subsequently, we employed the Gene Set Enrichment Analysis (GSEA) method to observe the pathway of SPINDOC enrichment in the tumors to elucidate the underlying mechanisms through which SPINDOC can promote the progression of HCC. Additionally, these results were further verified by in vitro experiments in two different HCC cell lines and xenograft mouse model.
Materials and Methods
Data Sources and Bioinformatics Analysis
The mRNA expression and clinical data of SPINDOC in 33 tumors were obtained from UCSC Xena online database (https://xena.ucsc.edu/). We only extracted the tumor types from the tumors with the number of normal control samples greater than or equal to 1 for analysis, and the specific number of samples has been shown in Supplementary Table S1–3 Genetic variation information from biological portal (https://www.cbioportal.org/).14,15 “Liver” and “Liver Hepatocellular Carcinoma (TCGA, Firehose Legacy, 442)” were selected in “Query”, and then the gene change characteristics of SPINDOC was searched. The results of SPINDOC mutation frequency, mutation type and CAN (copy number change) in TCGA database were observed in the module “Tumor Type Summary”. The cancer samples were then divided into low and high groups according to the median value of the SPINDOC expression level. We used the gene set enrichment analysis (GSEA) method to compare the various gene expression profiles of HCC specimens with SPINDOC low expression as well as SPINDOC high expression. CIBERSORT was then used to examine the relative fractions of the 22 infiltrating immune cell types in each tumor sample and to determine the variations among the various immune cells present in the different groups. In addition, the “survival” Receiver Operating Characteristic (ROC) software package verified the validity of SPINDOC expression levels in evaluating patient outcomes by calculating Area Under Curve (AUC).16,17
Cell Culture
HCC cell lines Huh7, HepG2, SK-HEP-1 and immortalized human liver cell line LO2 were obtained from School of Life Sciences, Sun Yat-sen University (Guangzhou, CHN). The cells were cultured in dulbecco’s modified eagle medium (DMEM) (Gibco, USA) with 10% fatal bovine serum (FBS) (HyClone, USA) and 1% penicillin and streptomycin (Sigma, USA) at 37°C with 5% of CO2.
Cell Transfection
Huh7 cells were transfected with siRNA NC and SPINDOC siRNA-1, SPINDOC siRNA-2, SPINDOC siRNA-3 (OLIGOBIO, CHN) by liposome transient transfection method. According to the Lipofectamine™ RNAiMAX specification (Invitrogen, USA), the cell transfection concentration was in the range of 40%–50%. The siRNA concentration used was 30 nmol/L. The transfection was observed under fluorescence microscope. shRNA targeting SPINDOC and shRNA-NC (OLIGOBIO, CHN) were stably transfected into Huh7 cells. Spindoc-plvx-shRNA2-ZSGreen-T2A-Puro was introduced into 293T cells to package the lentivirus. 293T cells were co-transfected with Lipofectamine 2000 (Invitrogen, USA) according to the manufacturer’s instructions. Eight hours after transfection, they were replaced with the complete culture medium. After 48 h of culture, the cell supernatant rich in lentivirus particles was collected and concentrated to obtain the concentrated lentivirus with high titer. The virus titers were thereafter measured and calibrated in 293T cells. siRNA sequences for SPINDOC are shown below: SPINDOC-siRNA-1 (Sense, 5’-GCCGCACCAUGGCCCUAAATT-3ʹand anti-sense, UUUAGGGCCAUGGUGCGGCTT); SPINDOC-siRNA-2 (Sense,5’-CGUUCUCAUUUACAUCUGUTT-3ʹand anti-sense, ACAGAUGUAAAUGAGAACGTT); SPINDOC-siRNA-3 (Sense,5’-GAGUGACCCAACAGGAGAATT-3’ and anti-sense, UUCUCCUGUUGGGUCACUCTTT).
Quantitative Real-Time PCR
Total RNA was extracted by Trizol (Invitrogen, USA) method and RNA concentration was then determined. According to the transcription response kit instructions (Invitrogen, USA), 1μL of RNA was taken from each sample for reverse transcription reaction to synthesize cDNA. Using cDNA as the template, PCR reaction was performed using 20 μL system for gene detection of each sample. The expression of SPINDOC was calculated by the 2−ΔΔCt method. The target-specific primer sequences for the RT-PCR used were as follows: SPINDOC (forward primer, GAGGTGACGCTGAAATGC and reverse primer, ACTCCTGCTCCCAAGACA); GAPDH (forward primer, GGAGCGAGATCCCTCCAAAAT and reverse primer, GGCTGTTGTCATACTTCTCATGG).
Western Blot Analysis
The proteins were extracted by using Radio-Immunoprecipitation Assay (RIPA) (Beyotime Institute of Biotechnology, CHN) lysis buffer adding with PMSF (KeyGEN, CHN) and protease inhibitor cocktail. Forty micrograms of protein lysates were separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel (KeyGEN, CHN) and transferred to a polyvinylidene fluoride (PVDF) membrane (KeyGEN, CHN). After blocking with skimmed milk for 1 hour, the membrane was incubated with the specific primary antibodies overnight. The membranes were then washed and incubated with secondary antibodies (ZSGB-BIO, CHN) at room temperature for 2 hours. Antibodies against Wnt1 (1500, ab85060), cyclin D1 (1100, ab16663), SPINDOC (1:50, ab220962) were purchased from Abcam (UK). β-catenin (1:500, 610153), GAPDH (1:5000, 60004-1-Ig) were purchased from BD sciences (USA). AXIN2 (1:50, K006954P) and SPIN1 (1:50, K006346P) were purchased from Solarbio (CHN).
Flow Cytometry
After the transfected Huh7 and SK-HEP-1 cells were harvested, fluorescein isothiocyanate (FITC) and Annexin V (BestBio, CHN) were used to stain the cells according to the manufacturer’s protocol (FITC Annexin V Apoptosis Detection Kit, BD Biosciences, USA) and a flow cytometric analysis was performed.
Colony Formation Assay
Twenty-four hours after transfection, the cells were digested with 0.25% trypsin (BestBio, CHN), neutralized and centrifuged. The cells were then inoculated at a density of 300 cells/2mL/well in a 6-well plate (Corning, USA) and incubated at 37°C in a 5% CO2 incubator (ShangHai KunCheng Scientific Instrument Co., LTD, CHN). On the 5th day, 500μL FBS was added to each well in the plate and the colony formation was closely observed. When the colony formation was visible to the naked eye, the supernatant was discarded, the cells were fixed with 4% paraformaldehyde and then stained with 0.1% crystal violet dye (Beijing Dingguo Changsheng Biotechnology, CHN), and the colony number of >50 cells was counted.
MTT Colorimetric Assay
MTT colorimetric assay was used to measure the cell viability. The cells (1×104) were seeded in 96-well plates (Corning, USA). Huh7 and SK-HEP-1 cells were inoculated with 96-well plates in an incubator (ShangHai KunCheng Scientific Instrument Co., LTD, CHN) containing 5% CO2 and saturated humidity at 37°C for 24 h. The complete medium was added and the cells were cultured for 48 h, after which the supernatant was removed. The wavelength of 490 nm was measured by the microplate analyzer (Olympus, JPN) to determine the absorbance A, and the inhibition rate of the cell proliferation was calculated.
Invasion and Migration Experiments
Matrigel matrix glue (BD Biosciences, USA) was diluted in the serum-free medium at 1:6, and 50ul was evenly spread into the upper chamber of Transwell chamber (Corning, USA). The chamber was placed in a 24-well plate and incubated at 37°C for 4h to cause it to gelatinize. The cell density was adjusted to 2×105 cells/mL, and 100μL per well was inoculated into the upper chamber of transwell chamber. The cells in the upper chamber were removed after 24h, fixed with methanol and 0.1% crystal violet (Beijing Dingguo Changsheng Biotechnology, CHN) for 20min, stained for 10min, and washed with PBS twice. The count was carried out under an inverted optical microscope (Olympus, JPN).
Scratch Wound-Healing Assay
The Huh7 and SK-HEP-1 cells were seeded in 6-well culture plates (Corning, USA) at 95–100% confluence. The cells were harvested 48 hours after transfection. The scratch was created using a 200 µL pipette tip. PBS was used to wash the plates for three times to remove the cellular debris. Images for the wound closure were photographed at the different times (12, 24, 36 and 48 h) by microscope (Olympus, JPN).
Animal Experiments
Female BALB/c nude mice (4–6 weeks of age) were purchased from GemPharmatech Co., Ltd. [Experimental Animal Production License No: SCXK (Su) 2019–0007]. After successful implantation of tumor, nude mice were placed in experimental Animal Center of Guangxi University of Chinese Medicine. The mice were divided into sh-NC group and SPINDOC-shRNA group. SPINDOC-shRNA and negative control (sh-NC) were stably transfected into Huh7 cells. The cells infected with fluorescent virus were sub-cultured to the required number and collected. In total, 1×107 transfected cells were suspended in 100 μL serum-free RPMI 1640 culture medium and were injected into the axilla of nude mice. After inoculation, the nude mice were raised in a sterile environment. The long diameter and short diameter of the tumor were measured with vernier calipers at an interval of 2–3 days. The tumor volume was calculated according to the formula 0.5×length×width2 and the growth curve was drawn. When subcutaneous tumor volume >50mm3, the tumor formation reached the standard size. Until day 33, all the mice were sacrificed and tumor xenografts were collected and weighed. All the animal experiments were approved by the Animal Experiment Ethics Committee of Guangxi University of Chinese Medicine (DW20220310-032). National Institutes of Health guide for the care and use of laboratory animals was strictly followed by us.
Immunohistochemistry
Fresh nude mouse tumor specimens were converted into wax blocks by routine fixation, and standardized operation was carried out according to the kit instructions (Beijing Dingguo Changsheng Biotechnology, CHN). The primary antibody was added and incubated at 4°C overnight. The specific information of primary antibodies used is as follows: Wnt1, 1:500, ab85060, Abcam, (UK); cyclin D1, 1:100, ab16663, Abcam, (UK); SPINDOC, 1:50, ab220962, Abcam, (UK); β-catenin,1:500, 610153, BD sciences, (USA); AXIN2, 1:50, K006954P, Solarbio, (CHN); SPIN1, 1:50, K006346P, Solarbio, (CHN). The blocks were washed with PBS 3 times, 2min each, and biotin-labeled sheep anti-rabbit IgG solution (1:100, Z021006Y, ZSGB-BIO, CHN), was added followed by incubation at room temperature for 30min. Thereafter, DAB chromogenic solution was added after PBS washing, and the blocks were placed under the microscope for observation. The reaction time was controlled, and the blocks were then washed with the distilled water to stop the reaction. Hematoxylin (Beijing Dingguo Changsheng Biotechnology, CHN) was added for redyeing, followed by washing, routine dehydration, spin drying, and observation of the results under the microscope after sealing with the neutral adhesive.
Hematoxylin-Eosin (HE) Staining
The tissue sections were washed with the distilled water and stained with hematoxylin (Beyotime, CHN). The staining time was controlled (about 5min) according to the condition of the tissue and dye. After rinsing with the distilled water, 1% hydrochloric acid ethanol (Beyotime, CHN) was added. After rinsing with the distilled water, eosin staining was performed (about 2s~8s). After rinsing with the distilled water again, anhydrous ethanol (Beyotime, CHN) was added to dehydrate, and neutral gum (Beyotime, CHN) was added. After sealing, the tablets were observed under a microscope and photographed (Olympus, JPN).
Statistical Analysis
Bioinformatics analysis was performed based on R programming (version 4.0.2, https://www.r-project.org/), statistical analysis was performed by using SPSS statistical software (version 22.0, Chicago, USA) and GraphPad Prism V7.0 software (GraphPad Software, Inc., California, USA). All the data were expressed as mean ± standard deviation (SD), and t-test was used to compare the mean values between the two groups. The values of more than two groups were compared using analysis of variance (ANOVA). Chi-square test was used to analyze the relationship between SPINDOC expression and clinical features. Kaplan Meier method was used for survival analysis, and long rank test was used for comparison. When P<0.05, the difference was considered as statistically significant.
Results
SPINDOC Expression in Pan-Cancer Samples Was Correlated with Adverse Clinical Outcomes
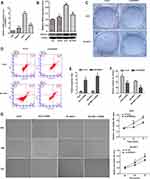
A total of 24 tumor types were included in the statistical analysis by analyzing specific tumor types with a normal sample number greater than or equal to 1 (Supplementary Table S1). SPINDOC expression was elevated in 18 of these tumors compared with the adjacent normal tissues (P<0.05) (Figure 1A and B) and there was no difference in expression as compared to the normal controls in the other 6 tumors (Supplementary Table S2). We divided the expression level of SPINDOC into high and low groups and observed the effect of different expression levels on the prognosis of patients in 24 tumor types. SPINDOC was highly expressed in KIRP (Kidney renal papillary cell carcinoma), PCPG (Pheochromocytoma and Paraganglioma), (Figure 1D and E) and Liver hepatocellular carcinoma (LIHC) (Figure 3A) resulted in the poor prognosis. The findings of univariate analysis indicated that SPINDOC can be used as a potential risk factor affecting the prognosis of patients in LIHC, KIRP and PCPG tumor types (Figure 1C and Supplementary Table S3). These results indicated that SPINDOC can act as a tumor-promoting factor in a variety of human tumors. To investigate the potential association between SPINDOC and primary liver cancer, we decided to further analyze its oncogenic role in HCC models.
Genetic Variation and Methylation Analysis of SPINDOC
We observed the genetic changes of SPINDOC in LIHC samples from the TCGA cohort, and the results showed that SPINDOC was genetically altered in 8.9% (34/382) of HCC samples. High mRNA expression (7.74%) displayed the highest frequency among various variations, followed by mutation (0.86%), and amplification and deep deletion both accounted for 0.29% (Figure 2A). According to the type, site and number of cases of SPINDOC gene mutation, the most common SPINDOC gene mutation was missense mutation (0.61%) occurred in R197C, and the type was copy number deletion. The second was missense mutation (0.28%) in R53T, with two alleles. The mutation of P247Hfs*147 was intra-frame insertion or deletion (0.09%), and the type was two alleles. Missense mutation occurred in D234G (0.07%), and the type was copy number increase (Figure 2B). The increase of SPINDOC mRNA expression in the tumors was mainly caused by gain and amplification (Figure 2C). In terms of the relationship between mutation and survival prognosis of patients, mutation was found to lead to poor prognosis of patients (P<0.01) (Figure 2D). From the methylation data of SPINDOC in liver cancer, the mRNA expression level of SPINDOC was found to be significantly negatively correlated with methylation level (Cor =−0.68, P=3.98E-52) (Figure 2E). The above indicated that the increased mRNA expression level of SPINDOC was related to the amplification of this gene and was negatively correlated with the methylation level.
SPINDOC Was Highly Expressed in HCC Samples and Was Correlated with Adverse Clinical Outcomes
We observed that high SPINDOC expression was associated with poor overall survival in patients with HCC in above analysis (P<0.001) (Figure 3A). To further clarify the potential relationship between SPINDOC high expression and clinical characteristics of patients, we analyzed SPINDOC expression level with patients’ gender (male, female), clinical Stage (Stage I, II, III, IV), histological Grade (Grade G1, G2, G3, G4) and T Stage (T1, T2, T3, T4). We observed that the SPINDOC was highly expressed in patients younger than 60 years old (P=0.015) (Figure 3B), and the expression level of SPINDOC increased with an increase in the clinical stage, histological grade and T stage (P<0.01) (Figure 3C–E), thereby indicating that the high expression of SPINDOC was closely related to the poor clinical prognosis. The AUC of ROC curve at SPINDOC expression levels was observed to be 0.64 (Supplementary Figure S1A), thereby indicating a relatively better diagnostic performance. Multiple ROC curves showed that the prediction value of SPINDOC expression for 1-year, 3-year and 5-year survival was 0.71, 0.64 and 0.597, respectively (Supplementary Figure S1B).
Thereafter, in the univariate analysis of clinical characteristics and the possible correlation between SPINDOC gene expression and the survival status, as well as the poor prognosis of the patient was related to the relatively higher expression of SPINDOC (HR: 1.10; P=4.89E-05), and the clinical stage of the tumor (HR: 1.865; P=8.07E-07), T stage (HR: 1.804; P=4.73E-07) and M stage (HR: 3.850; P=0.023). In multivariate analysis, the high expression of SPINDOC (HR: 1.076; P=0.005) was found to be significant as an independent predictor for patients with primary hepatocellular carcinoma (Table 1). We also analyzed whether there were differences in immune cell invasion between SPINDOC high and low expression groups in the liver cancer tissues, and CIBERSORT analysis showed that there were no substantial differences in immune cell invasion between SPINDOC high–low expression groups in liver cancer tissues (Supplementary Figure S1C).
 |
Table 1 Univariate and Multivariate Analysis of the Clinical Characteristics of HCC Patients and the Possible Correlation Between SPINDOC Gene Expression and Patient’s Survival Status |
GSEA Revealed the Potential Effects of SPINDOC on the Various Oncogenic Signaling Pathways That Can Regulate Tumor Proliferation and Metastasis
To explore the molecular mechanisms underlying the ability of SPINDOC in promoting the occurrence and development of HCC, we used the gene set enrichment analysis (GSEA) method to compare the various gene expression profiles of HCC specimens with SPINDOC low expression as well as SPINDOC high expression. According to the median of SPINDOC expression level, they were divided into SPINDOC low expression group and SPINDOC high expression group. GSEA analysis showed that high SPINDOC expression was significantly related to the various cancer-related signaling pathways, such as those of WNT, cell cycle, adhesion junctions, etc. thereby suggesting that these pathways might play a potential role in the tumorigenesis and metastasis of SPINDOC (Figure 3F–I, Supplementary Table S4).
SPINDOC Levels Were Significantly Up-Regulated in HCC Cells, and Its Knockdown Can Inhibit the Proliferation and Promote Apoptosis
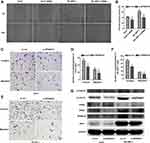
The cell-based experiments were next used to verify the different biological functions of SPINDOC in hepatocellular carcinoma cells. We employed qRT-PCR to detect the expression level of SPINDOC in the normal liver cell lines (LO2) and the different HCC cell lines (Huh7, HepG2, SK-HEP-1). The results showed that mRNA and protein expression levels of SPINDOC in Huh7, HepG2 and SK-HEP-1 cells were significantly increased compared with those in LO2 cells, and among all the cell lines tested, Huh7 and SK-HEP-1 showed the maximal expression (P<0.01) (Figure 4A and B). We designed the corresponding primers to knock out the SPINDOC gene and selected the target sequence and Huh7 and SK-HEP-1 cell lines with lowest expression for the next step.
We used MTT assay to determine the cell viability of Huh7 and SK-HEP-1 and it was found that SPINDOC gene knockout could significantly reduce the survival rate of Huh7 and SK-HEP-1 cells at 48 and 72 h, respectively (P<0.01) (Figure 4G). Additionally, we also used colony formation experiments to further analyze the proliferation of Huh7 and SK-HEP-1 cells. The findings showed that the colony forming ability of Huh7 and SK-HEP-1 cells was significantly reduced after SPINDOC gene was effectively knocked out (Figure 4C and F). Moreover, SPINDOC gene knockout promoted a substantial increase in the apoptosis rate of HCC cells (Figure 4D and E). The above results suggested that SPINDOC can functionally promote the proliferation of HCC cells.
SPINDOC Can Effectively Promote the Migration and Invasion and Can Activate the WNT/β-Catenin Signaling Pathway in HCC Cells
To study the possible effects of SPINDOC on the migration and invasion of HCC cells, wound healing assay and transwell invasion assays were employed. The analysis of wound healing assay clearly showed that knocking down the expression of SPINDOC gene can significantly inhibit the wound healing ability (Figure 5A and B). In addition, knockout of SPINDOC gene markedly reduced the invasion of HCC cells (Figure 5C and D) and the number of migrating cells (Figure 5E and F). In summary, these results indicated that SPINDOC can significantly promote the migratory and invasive abilities of both Huh7 and SK-HEP-1 cells.
In addition, previous GSEA analysis has revealed that high SPINDOC expression was significantly related to the various cancer-related signaling pathways, such as Wnt signaling cascade. Hence, after knocking down SPINDOC gene, we used Western blot analysis to detect the expression of the various key proteins involved in the Wnt/β-catenin signal pathway. These proteins included Wnt1 that can activate Wnt pathway, Wnt signaling pathway activator marker β-catenin, downstream target genes of β-catenin such as cyclin D1, and AXIN2 that negatively regulates this pathway. At the same time, in view of the close relationship between SPIN1 and SPINDOC, we also analyzed the expression of SPINDOC also. The results showed that the expression of SPIN1, Wnt1, β-catenin and cyclin D1 proteins in Huh7 and SK-HEP-1 cells after SPINDOC gene knockdown decreased markedly, but the expression of AXIN2 was increased (see Figure 5G and Supplementary Figure S1 D–I). These results clearly demonstrated that SPINDOC can effectively promote the proliferation, migration and invasion of HCC cells by activating Wnt/β-catenin signaling pathway.
Knockdown of SPINDOC Inhibited HCC Growth and Wnt/β-Catenin Signaling Pathway in Xenograft Mouse Model
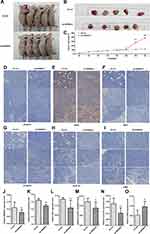
Finally, we verified these results in a xenograft mouse HCC model. We selected stably transfected Huh7 cell lines for subcutaneous inoculation of the nude mice. The results showed that subcutaneous tumors grew in both the control group and the SPINDOC knockout group. The subcutaneous tumors in the SPINDOC knockout group were significantly smaller than those in the control group on day 26 (P=0.016), and there was a significant statistical difference observed on day 29 (P=0.009) (Figure 6A–C). In addition, compared with the control group, the HE staining pathological sections of the tumor showed loose arrangement and obvious necrosis after knocking down of SPINDOC (Supplementary Figure S1J). Immunohistochemical staining showed that the expression of SPINDOC (Figure 6D and J), SPIN1 (Figure 6E and K), Wnt1 (Figure 6F and L), β-Catenin (Figure 6G and M), Cyclin D1 (Figure 6H and N) in SPINDOC knockout group was significantly lower than that in control group, whereas the expression of AXIN2 (Figure 6I and O) was markedly higher than that in control group.
Discussion
In this study, we reported an important oncogenic factor SPINDOC that has not been extensively studied in tumors. We observed the role and mechanism of SPINDOC in pan-cancer and HCC samples. We found that SPINDOC is highly expressed in a variety of malignant tumors and primary liver cancer, as an oncogenic factor can effectively promote the proliferation, invasion and migration of HCC by activating Wnt/β-catenin signaling pathway.
It has been reported by Bae et al that SPINDOC is located at chr11 and consists of 1945bp. SPINDOC can block the potential interaction between SPIN1 and methylated histone tails, it was thereafter named based on its function in the complex.12 SPINDOC can interfere with the binding of SPIN1 to the chromatin and negatively regulate spin1-mediated transcriptional activity, thereby largely neutralizing SPIN1ʹs activation of its target genes.12 Spindlin1 (SPIN1) is the main member of the SPIN/SSTY gene family. This gene family is highly conserved and has been found to be expressed in the mature gametes and early embryos.18 SPIN1 is a tumor-domain protein, which contains three tandem repeats of tumor domains and can specifically recognize trimethylation of the fourth lysine of histones (H3K4me3) to bind to the histones. The human SPIN1 gene is located in interphase nuclei and mitotic chromosomes and encodes proteins involved in the cell cycle regulation which can influence epigenetic regulation.19,20 SPIN1 plays a role in early embryonic development and tumorigenesis by modulating the activation of PI3K/AKT pathway. SPIN1 has been found to display tumor-promoting effects in a variety of tumors by activating Wnt/β-catenin signaling pathway.21
It is speculated that SPINDOC is a transcription corepressor of SPIN1 due to its tumor-promoting effect,12,13 but the results also show a paradoxical effect. As an inhibitor of SPIN1, SPINDOC controls the chromatin load of SPIN1, and overexpression of SPINDOC can lead to an increase in SPIN1 level. At the same time, SPINDOC knockout can lead to SPIN1 instability.13 These findings suggest a potential synergistic rather than antagonistic relationship between SPINDOC and SPIN1. In addition, previous data has shown that SPINDOC was highly expressed in a variety of tumors. However, it is not clear whether SPINDOC can function as a tumor suppressor or a tumor progenitor in tumors? With this question, we observed the expression of SPINDOC in pan-cancer and liver cancer samples and its potential relationship between the tumors and SPINDOC.
In this study, the bioinformatics analysis was used to observe the potential expression of SPINDOC in pan-cancer and HCC based on the TCGA database. The results showed that the expression of SPINDOC in variety of tumors and in HCC was significantly higher than that in normal controls (P<0.05). Additionally, the findings of bioinformatics analysis indicated that the expression of this factor in variety of tumors and in HCC was positively correlated with the poor prognosis of patients. In view of the high incidence and hazard degree of liver cancer in the population, we chose liver cancer for further analysis. Based on this observation, we speculated that this factor might act as a tumor-promoting factor in variety of tumors and HCC. Thereafter, analysis of the possible relationship between the SPINDOC gene expression and the patient’s age, gender, clinical grade and stage of tumors revealed that the expression SPINDOC in was positively correlated with young patients, and clinical grade, stage and lymphatic stage of the tumor, thereby indicating that a high expression of this factor in HCC can significantly promote the clinical progression of tumors. In other words, SPINDOC can significantly promote the proliferation, invasion and metastasis of HCC cells. In order to explore the underlying mechanisms through which SPINDOC can promote the progression of HCC, we used the GSEA method to observe the different pathways of SPINDOC enrichment in the tumors. It was found that SPINDOC was enriched in a wide variety of oncogenic pathways, such as those of Wnt, cell cycle and cellular adhesion pathways, which can regulate the proliferation, invasion and metastasis of the tumors.
A number of previous studies have reported that overexpression of SPINDOC can effectively reduce Wnt signaling, while SPINDOC mutants can activate Wnt signaling. These results suggested that the various phenotypic differences may result from gene disruption and alteration of genetic content.22 We also analyzed the genetic variation of SPINDOC. SPINDOC was found to be genetically altered in 8.9% (34/382) of the samples. High mRNA expression (7.74%) and mutation (0.86%) were the main types of SPINDOC genetic changes observed in HCC. SPINDOC gene had the most missense mutation (0.61%) in R197C, and the type was copy number deletion. Based on the proportion of different mutations in the genetic alteration of SPINDOC, we speculated that SPINDOC itself might have the function of activating the Wnt/β-catenin signaling pathway in HCC, independent of the small number of SPINDOC mutations at the overall level. In addition, amplification of genetic alteration of SPINDOC resulted in increased mRNA levels of SPINDOC, while methylation resulted in decreased mRNA levels of SPINDOC.
Thereafter, various in vitro experiments were used to verify the above hypothesis. Firstly, we found that mRNA and protein expression levels of SPINDOC were significantly higher in HCC cells as compared to the normal liver cells. Secondly, based on the results of cell proliferation, colony formation and apoptosis assay, we observed that the low expression of SPINDOC can markedly inhibit the proliferation and promote the apoptosis of HCC cells, thus indicating that SPINDOC can essentially function as an oncogene. In the subsequent scratch, invasion and migration assays, it was found that low expression of SPINDOC can markedly inhibit the invasion and migration, thereby indicating that this oncogenic factor also has a significant impact on the invasion and metastasis of HCC cells. These results indicated that SPINDOC was highly expressed in HCC, and it can can promote the proliferation, invasion and migration of HCC cells, and can act as a potential tumor promoting factor. Subsequently, we also observed the regulatory effect of SPINDOC on Wnt signaling pathway.
The Wnt signaling cascade is highly conserved in different species and plays a vital role in regulating the early development, organ formation, tissue regeneration and other important physiological processes. An abnormal activation of this signaling pathway can induce the occurrence of a variety of cancers, including HCC.23,24 Wnt/β-catenin cascade can be activated by autocrine or paracrine Wnt ligand and effectively cause β-catenin accumulation. This causes β-catenin in the cytoplasm to enter into the nucleus and bind to the LEF/TCF transcription factor family to initiate the transcription of the various downstream target genes (e.g c-myc, cyclin D1).25
SPIN1 has been shown to promote tumor progression by activating the Wnt/β-catenin signaling system, and the activation of the Wnt/β-catenin signaling pathway can promote tumor progression by stimulating the transcription of multiple oncogenic factors.20,21 Moreover, previous studies have shown that overexpression of SPINDOC in the SPINDOC-spin1 complex can inhibit the activation of the Wnt signaling pathway, but this study also suggested that mutated SPINDOC can activate the Wnt signaling system.13 Thus, SPINDOC-SPIN1 complex may act as the transcriptional co-repressor of the Wnt signaling system, or other unknown proteins and may bind to the C-terminal region of SPINDOC to inhibit the transcriptional activation. When SPINDOC is mutated and cannot bind to SPIN1, it can act as a transcriptional activator of Wnt signaling.13
To observe the potential effect of SPINDOC expression changes on Wnt/β-catenin signaling pathway in HCC, we knocked down SPINDOC expression and detected the key factors downstream of Wnt/β-catenin signaling pathway. The results showed that with the decrease of SPINDOC, the expression of Wnt1 which can activate Wnt signaling pathway, Wnt signaling pathway activation marker β-catenin, and the downstream target genes such as cyclin D1 decreased, whereas that of AXIN2, which can negatively regulate this pathway was increased. These findings suggested that the decrease of SPINDOC might block Wnt signaling pathway. In view of the close relationship between SPIN1 and SPINDOC, we also analyzed the expression of SPIN1. The results showed that the expression of SPIN1 decreased with the decrease of SPINDOC. This result was consistent with the previous findings that the overexpression of SPINDOC can lead to the elevation of SPIN1 level.13 Based on these results, we believe that SPINDOC can itself act as an oncogenic factor that can activate Wnt/β-catenin signaling pathway in HCC cell lines.
Limitations
This study was associated with certain limitations. First, since no mutant group was set in the experiment, we were unable to determine whether the mutant SPINDOC could also activate the Wnt/β-catenin signaling pathway. Second, PCR and WB analysis of SPINDOC, SPIN1 and the factors related to the activation of Wnt/β-catenin signaling pathway in the tissues obtained from xenograft mouse model was not performed, so the activation effect of SPINDOC on Wnt/β-catenin signaling pathway in HCC could not be confirmed from the multiple perspectives.
Conclusion
SPINDOC was highly expressed in a variety of tumors and primary HCC models. This gene can effectively promote the proliferation, invasion and migration of HCC cells by activating the Wnt/β-catenin signaling cascade. Therefore, this oncogenic factor can form the basis of a potential therapeutic target for the management of HCC.
Abbreviations
KIRP, Kidney renal papillary cell carcinoma; PCPG, Pheochromocytoma and Paraganglioma; LIHC, Liver hepatocellular carcinoma; HCC, Hepatocellular Carcinoma; GSEA, Gene Set Enrichment Analysis; SPINDOC, SPIN1 docking protein; TCGA, The Cancer Genome Atlas; FITC, Fluorescein isothiocyanate; BLCA, Bladder Urothelial Carcinoma; BRCA, Breast invasive carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangiocarcinoma; COAD, Colon adenocarcinoma; ESCA, Esophageal carcinoma; GBM, Glioblastoma multiforme; HNSC, Head and Neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LIHC, Liver hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, Prostate adenocarcinoma; READ, Rectum adenocarcinoma; SARC, Sarcoma; SKCM, Skin Cutaneous Melanoma; STAD, Stomach adenocarcinoma; THCA, Thyroid carcinoma; THYM, Thymoma; UCEC, Uterine Corpus Endometrial Carcinoma.
Consent for Publication
All the patients that were involved in the study have given their consent to publish their individual data.
Funding
This project was funded by the National Natural Science Foundation of China (No. 82060843), Guangxi Province Science Foundation for Youths (No.2018GXNSFBA281189) and funded by the Youth Science Foundation of Guangxi University of Chinese Medicine (No.2019XK163), and Guangxi Province Science Foundation (No. 2021GXNSFAA196031).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Villanueva A. Hepatocellular Carcinoma. N Engl J Med. 2019;380:1450–1462. doi:10.1056/NEJMra1713263
2. Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314. doi:10.1016/S0140-6736(18)30010-2
3. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi:10.3322/caac.21338
4. Noonan A, Pawlik TM. Hepatocellular carcinoma: an update on investigational drugs in Phase I and II clinical trials. Expert Opin Investig Drugs. 2019;28(11):941–949. doi:10.1080/13543784.2019.1677606
5. Gao Y, Yue W, Zhang P, et al. Spindlin1, a novel nuclear protein with a role in the transformation of NIH3T3 cells. Biochem Biophys Res Commun. 2005;335(2):343–350. doi:10.1016/j.bbrc.2005.07.087
6. Chen X, Wang YW, Xing AY, et al. Suppression of SPIN1-mediated PI3K-Akt pathway by miR-489 increases chemosensitivity in breast cancer. J Pathol. 2016;239:459–472. doi:10.1002/path.4743
7. Song Q, Ji Q, Xiao J, et al. miR-409 inhibits human non-small-cell lung cancer progression by directly targeting SPIN1. Mol Ther Nucleic Acids. 2018;13:154–163. doi:10.1016/j.omtn.2018.08.020
8. Li Y, Ma X, Wang Y, Li G. miR-489 inhibits proliferation, cell cycle progression and induces apoptosis of glioma cells via targeting SPIN1-mediated PI3K/AKT pathway. Biomed Pharmacother. 2017;93:435–443. doi:10.1016/j.biopha.2017.06.058
9. Chen X, Wang YW, Gao P. SPIN1, negatively regulated by miR-148/152, enhances Adriamycin resistance via upregulating drug metabolizing enzymes and transporter in breast cancer. J Exp Clin Cancer Res. 2018;37:100. doi:10.1186/s13046-018-0748-9
10. Lv BB, Ma RR, Chen X, et al. E2F1-activated SPIN1 promotes tumor growth via a MDM2-p21-E2F1 feedback loop in gastric cancer. Mol Oncol. 2020;14:2629–2645. doi:10.1002/1878-0261.12778
11. Zhao M, Bu Y, Feng J, et al. SPIN1 triggers abnormal lipid metabolism and enhances tumor growth in liver cancer. Cancer Lett. 2020;470:54–63. doi:10.1016/j.canlet.2019.11.032
12. Bae N, Gao M, Li X, et al. A transcriptional coregulator, SPIN.DOC, attenuates the coactivator activity of Spindlin1. J Biol Chem. 2017;292:20808–20817. doi:10.1074/jbc.M117.814913
13. Devi MS, Meiguilungpou R, Sharma AL, et al. Spindlin docking protein (SPIN.DOC) interaction with SPIN1 (a histone code reader) regulates Wnt signaling. Biochem Biophys Res Commun. 2019;511(3):498–503. doi:10.1016/j.bbrc.2019.02.096
14. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:l1. doi:10.1126/scisignal.2004088
15. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi:10.1158/2159-8290.CD-12-0095
16. Xie J, Chen L, Cao Y, et al. Single-cell sequencing analysis and weighted co-expression network analysis based on public databases identified that TNC is a novel biomarker for keloid. Front Immunol. 2021;12:783907. doi:10.3389/fimmu.2021.783907
17. Xie J, Zhang X, Zhang K, et al. Construction and validation of the diagnostic model of keloid based on weighted gene co-expression network analysis (WGCNA) and differential expression analysis. J Plast Surg Hand Surg;2022. 1–9. doi:10.1080/2000656X.2021.2024557
18. Staub E, Mennerich D, Rosenthal A. The Spin/Ssty repeat: a new motif identified in proteins involved in vertebrate development from gamete to embryo. Genome Biol. 2002;3:H3.
19. Wang W, Chen Z, Mao Z, et al. Nucleolar protein Spindlin1 recognizes H3K4 methylation and stimulates the expression of rRNA genes. EMBO Rep. 2011;12:1160–1166. doi:10.1038/embor.2011.184
20. Su X, Zhu G, Ding X, et al. Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1. Genes Dev. 2014;28:622–636. doi:10.1101/gad.233239.113
21. Wang JX, Zeng Q, Chen L, et al. SPINDLIN1 promotes cancer cell proliferation through activation of WNT/TCF-4 signaling. Mol Cancer Res. 2012;10:326–335. doi:10.1158/1541-7786.MCR-11-0440
22. Urrutia E, Chen H, Zhou Z, Zhang NR, Jiang Y. Integrative pipeline for profiling DNA copy number and inferring tumor phylogeny. Bioinformatics. 2018;34:2126–2128. doi:10.1093/bioinformatics/bty057
23. He S, Tang S. WNT/beta-catenin signaling in the development of liver cancers. Biomed Pharmacother. 2020;132:110851. doi:10.1016/j.biopha.2020.110851
24. Zhang Y, Wang X. Targeting the Wnt/beta-catenin signaling pathway in cancer. J Hematol Oncol. 2020;13:165. doi:10.1186/s13045-020-00990-3
25. Nusse R, Clevers H. Wnt/beta-catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985–999. doi:10.1016/j.cell.2017.05.016
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.