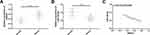Back to Journals » Cancer Management and Research » Volume 12
SNHG1 Promotes Malignant Progression of Glioma by Targeting miR-140-5p and Regulating PI3K/AKT Pathway
Authors Cai RD, Zhang CC, Xie LL, Wang PC, Huang CX, Chen JL, Lv HT
Received 26 June 2020
Accepted for publication 15 October 2020
Published 24 November 2020 Volume 2020:12 Pages 12011—12020
DOI https://doi.org/10.2147/CMAR.S269572
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
This paper has been retracted.
Ren-Duan Cai, 1 Chao-Cai Zhang, 1 Li-Li Xie, 2 Peng-Cheng Wang, 3 Chui-Xue Huang, 3 Jian-Long Chen, 3 Hong-Tao Lv 4,*
1Department of Neurosurgery, Hainan General Hospital/Hainan Affiliated Hospital of Hainan Medical University, Haikou, Hainan Province, People’s Republic of China; 2Department of Neurology, Dalian Central Hospital, Dalian, Liaoning Province, People’s Republic of China; 3Department of Neurosurgery, Hainan People’s Hospital, Haikou, Hainan Province, People’s Republic of China; 4Department of Neurosurgery, The First Affiliated Hospital of Dalian Medical University, Dalian, Liaoning Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hong-Tao Lv
Department of Neurosurgery, The First Affiliated Hospital of Dalian Medical University, No. 222 Zhongshan Road, Xigang District, Dalian 116011, Liaoning Province, People’s Republic of China
Tel +86-18098876125
Email [email protected]
Purpose: To explore the regulatory mechanism of long non-coding RNA small nucleolar RNA host gene 1 (SNHG1) in glioma.
Materials and Methods: The expression of SNHG1 and miR-140-5p in glioma tissues and glioma cell lines (LN-18, KNS-81, and KALS-1) was determined, and the effect of the two on cell proliferation, invasion, and PI3K/AKT pathway was analyzed.
Results: SNHG1 was overexpressed in glioma tissues, while miR-140-5p was underexpressed in them, and there was a significant negative correlation between SNHG1 and miR-140-5p. In addition, both down-regulation of SNHG1 and up-regulation of miR-140-5p significantly inhibited the malignant proliferation and invasion of glioma, intensified the apoptosis, and also significantly suppressed the activation of the PI3K/AKT pathway. The dual-luciferase reporter assay, RNA pull-down assay, and RIP determination all confirmed that there was a targeting relationship between SNHG1 and miR-140-5p, and there was no difference between KNS-81 and KALS-1 cells transfected with SNHG1+mimics and si-SNHG1+inhibitor and those in the si-NC group with unrelated sequences in terms of cell malignant progression.
Conclusion: SNHG1/miR-140-5p axis and its regulation on PI3K/AKT pathway might be a novel therapeutic direction to curb the malignant progression of glioma.
Keywords: SNHG1, glioma, miR-140-5p, PI3K/AKT pathway
Introduction
Glioma, as a malignant brain tumor, is a common cause of human brain cancer-related death.1,2 According to the epidemiological statistics of glioma, the annual incidence of glioma is approximately 3–8 cases per 100,000 individuals, and the disease is more common in men and people aged 50–69.3,4 Glioma is mainly manifested as class II epileptic seizure, and the age of patients is a potential risk factor for the increase of glioma grade.5 Patients with glioma are mostly already in the advanced stage at the time of diagnosis, and they are usually treated with surgical resection, radiotherapy, and chemotherapy that is the most common treatment for it.6 However, glioma is located in the sensitive central nervous system, so it is difficult for conventional chemotherapy to break through the blood–brain barrier and exert its medicinal properties, which appeals for new treatment strategies.7 We would explore the potential therapeutic direction by studying the molecular mechanism of glioma, which is of great value for the treatment of glioma.
Long non-coding RNA (lncRNA) is a therapeutic direction for glioma. As an endogenous RNA molecule that regulates gene expression at post-transcriptional level, lncRNA is abnormally expressed in cases with one of various diseases including glioma. Dynamic monitoring on it is helpful for us to understand the progress of diseases.8–10 For example, urothelial carcinoma-associated 1 (UCA1) can act as a carcinogen for non-small cell lung cancer and promotes its development by targeting miR-193a-3p, and dysregulation of growth-arrest-specific transcript 5 (GAS5) is involved in the resistance mechanism of trastuzumab in human epidermal growth factor receptor 2 (HER2) in patients with breast cancer. In addition, lncRNA colon cancer-associated transcript1 (CCAT1) is reported to be able to down-regulate miR-181b to promote the malignant progression of glioma.11–13 LncRNA is also widely used in drug resistance and clinical diagnosis of glioma. For instance, down-regulation of lncRNA PVT1 is beneficial to increasing the anti-drug resistance of paclitaxel in patients with glioma, and lncRNA miR210HG has the potential to be a serum indicator for diagnosis of glioma.14,15 Small nucleolar RNA host gene 1 (SNHG1), as a member of the lncRNA family, is an oncogene of glioma. Its up-regulation often indicates malignant progression of patients with glioma.16 Studies have revealed that miR-140-5p, as a cancer suppressor, can inhibit the malignant behavior of glioma by partially targeting JAG1.17 The SNHG1/miR-140-5p axis mediates the regulatory mechanism of malignant expansion of esophageal cancer cells, but the potential pathological mechanism of the two in glioma has not yet been clarified.18 The PI3K/AKT signaling pathway, as one of the classical regulatory pathways of human cancer, is also involved in angiogenesis and cell growth of glioma.19–21
We suspected that the regulatory network of SNHG1/miR-140-5p axis may be involved in the malignant progression of glioma by medicating PI3K/AKT signaling pathway, and we verified it by detecting the expression of SNHG1 and miR-140-5p.
Materials and Methods
Tissue Sample Collection
Normal brain tissues and glioma tissues were sampled from patients who had signed informed consent forms. The sampled glioma tissue specimens were assigned to a glioma group (n=40), including 22 cases in grade I/II and 18 cases in grade III/IV, and sampled normal brain tissue specimens were as assigned to a normal group (n=30). The normal brain tissues were non-tumor brain tissues, which were sampled from patients with craniocerebral injury who had undergone partial resection of brain tissues. Those specimens were sampled from December 2016 to December 2018, and the general data including sex and age of the two groups were comparable (P<0.05). The experiment was approved by the Ethics Committee of Hainan People’s Hospital, and operations and tissue storing were carried out in accordance with corresponding standards.
Cell Culturing
Human glioma cell lines (LN-18, KNS-81, and KALS-1) and normal glial cells (HEB) (C0786, C1648, GD-C0098632A65190, and DA-C5504, Guandao Biological Engineering Co., Ltd., Shanghai, China) were cultured in dulbecco’s modified eagle (DMEM) (BH-S3208, Bohu Biotechnology Co., Ltd., Shanghai, China) supplemented with 10% phosphate buffer saline (PBS, 120882, Chreagen Biotechnology Co., Ltd., Beijing, China), 100 U/mL penicillin (07500, Yihui Biological Technology Co. Ltd., Shanghai, China), and 100 mg/mL streptomycin (YSH106-01, Yanjin Biological Co., Ltd., Shanghai, China) under 5% CO2 at 37°C.
Cell Transfection
MiR-140-5p overexpression sequence (mimics), miR-140-5p inhibition sequence (inhibitor), miR negative control (miR-NC), targetedly inhibited SNHG1 RNA (si-SNHG1), targetedly overexpressed SNHG1 RNA (SNHG1), and negative control RNA (si-NC) were transfected into human glioma cells by a Lipofectamine™ 2000 Kit (11668, Biomics Biotechnologies Co., Ltd., Nantong, China) in strict accordance with the kit instructions.
qRT-PCR Assay
Total RNA was extracted from collected glioma tissues and cells with a TRIzol Kit (KL058, Kanglang Biotechnology Co., Ltd., Shanghai, China), and cDNA was synthesized using a Bio-Rad Ssofast EvaGreen Supermix Kit (1725202, Yihui Biological Technology Co. Ltd., Shanghai, China) and Stepone Plus fluorescence ration PCR instrument (Biocytocare Biotechnology Co., Ltd., Beijing, China). All primers were designed and synthesized by Shanghai Qiantu Biotechnology Co., Ltd. U6 and β-Actin were taken as internal references for miRNA and mRNA, respectively, and data of this experiment were analyzed using 2-ΔΔct.
Western Blotting Assay
Total RNA was extracted from cells in each group that were cultured in RIPA lysate (PS0033, Zhenyu Biotechnology Co., Ltd., Shanghai, China), and its concentration was detected using a BCA Kit (LCB004, Junrui Biotechnology Co., Ltd., Shanghai, China), and adjusted to 4 μg/μL. Subsequently, the RNA was separated through 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (LM0053A, LMAI Bioengineering Co., Ltd., Shanghai, China), and then transferred to a polyvinylidene fluoride (PVDF) membrane (ISEQ00011, Chreagen Biotechnology Co., Ltd. Beijing, China). The membrane was sealed with 5% skim milk (N/A-433, Lianshuo Biological Technology Co., Ltd., Shanghai, China) for 4 h, and then added with AMPK, p-AMPK, PI3K, and p-AKT (1:1000) and β-Actin primary antibody (1:1000), and sealed at 4°C overnight. The membrane was washed to remove the primary antibody, and then it was added with horseradish peroxidase-labeled goat anti-rabbit secondary antibody (1:2000), and cultured at 37°C for 1 h. Antibodies were all purchased from Beijing Future Biotechnology Co., Ltd. The chemiluminescence was detected using an electrochemiluminescence (ECL) kit (KL-16664, Kalang Biotechnology Co., Ltd., Shanghai, China) and Bio-Rad ChemiDoc MP Imaging System (17001402, Yihui Biological Technology Co. Ltd., Shanghai, China), and the grey value was analyzed using Quantity One.
Cell Proliferation Assay
The proliferation of glioma cells was determined using a MTT Kit (EY-19002, Yiyan Biotechnology Co., Ltd., Shanghai, China). The cells were seeded into 96-well plates at 5×103 cells/well, and then the plates were added with 20 μL 5 μmg/mL MTT solution, and cultured at 37°C for 24, 48, 72 and 96 h, separately. The plates were added with 200 μL dimethyl sulfoxide at each culturing time point, and then the optical density (OD) of cells in each group at 450 nm absorbance was determined using a V-1200 spectrophotometer (Hengfei Biotechnology Co., Ltd., Shanghai, China).
Cell Apoptosis Assay
The transfected cells were digested by 0.25% trypsin (T4049-500ML, Bei Nuo Biotechnology Co., Ltd., Shanghai, China). After digestion, the cells were washed with PBS twice, added with 100 μL binding buffer to prepare 1×106 cells/mL suspension. The suspension was added with AnnexinV-FITC and PI in order, incubated at room temperature in the dark for 5 min, and finally detected using a flow cytometer (AMG0002051, Image Trading Co., Ltd., Beijing, China). The experiment was repeated three times, and the results were averaged.
Cell Invasion Assay
Glioma cells transfected for 24 h were collected and seeded into 24-well plates at 3×104 cells/well. The cells were digested with trypsin, and transferred to the upper compartment. The upper compartment was added with 200μL RPMI 1640 culturing solution, and the lower compartment was added with 500mL RPMI 1640 supplemented with 10% FBS. The plate was incubated at 37°C for 48 h. The substrate and cells that did not penetrate the membrane surface in the upper compartment were wiped off. The plates were washed with PBS three times, immobilized with paraformaldehyde for 10 min, and then washed with double-distilled water three times. Finally, the plates were stained with 0.5% crystal violet after being dried naturally. The invasion of the cells was evaluated using a microscope.
Dual-Luciferase Reporter (DLR) Assay
The fragment for predicting binding locus of miR-140-5p from SNHG1 was cloned into the pmirG10 dual-luciferase miRNA target expression vector (Promega) to generate the report vector SNHG1-wild type (SNHG1-Wt). With the aim of mutating the putative binding loci of miR-140-5p in SNHG1, the sequence of the putative binding loci was replaced as indicated and named SNHG1 mutant (SNHG1-Mut).
RNA Pull-Down Assay and RNA-Binding Protein Immunoprecipitation (RIP) Assay
A RNeasy Mini Kit (DXT-74126, Kemin Biotechnology Co., Ltd., Shanghai, China) was adopted to purify biotinylated SNHG1. Then, 3μg purified SNHG1 was incubated with 1 mg KNS-81 or KALS-1 cell lysate at 25°C for 1 h, and then recovered by streptavidin-agarose beads. Finally, RNA in the pull-down materials was quantitatively determined by qRT-PCR.
A Magna RIP reagent kit (17–700, Fushen Biotechnology Co, Ltd., Shanghai, China) was adopted for RIP assay. The properties of antibody beads, including connection and configuration, were evaluated. The thawed antibody beads were suspended and mixed with samples, and incubated at 4°C overnight. The suspension was placed on a magnetic frame and washed with buffer solution. After immunoprecipitation, the co-immunoprecipitated products were harvested. RNA was extracted and purified, and the abundance of target RNA was determined.
Statistical Analyses
In this study, the collected data were analyzed statistically and visualized into required figures using GraphPad 6. Inter-group comparison was carried out by the independent t test, and multi-group comparison was carried out by the one-way ANOVA, expressed by F. Post hoc pairwise comparison was carried out using the LSD-t test, and comparison in expression at different time points was carried out using the repeated measures analysis of variance, and expressed by F, and Bonferroni post hoc test was adopted. In addition, Pearson’s correlation analysis was carried out. P < 0.05 indicates a significant difference.
Results
Clinical Value of SNHG1 and miR-140-5p in Glioma
Glioma tissues showed significantly higher SNHG1 expression and significantly lower miR-140-5p expression than normal brain tissues (both P<0.05). Further analysis of the correlation between SNHG1 and miR-140-5p showed that SNHG1 was significantly negatively correlated with miR-140-5p (r=−0.773, P<0.001). Figure 1.
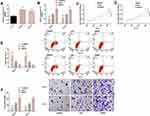
Effect of SNHG1 on the Malignant Progression of Glioma Cells
Significantly higher SNHG1 expression was found in KNS-81 and KALS-1 cells, so KNS-81 and KALS-1 cells were selected for transfection analysis. It came out that KNS-81 and KALS-1 cells transfected with si-SNHG1 showed decreased SNHG1 expression and those transfected with SNHG1 showed increased SNHG1 expression. Cell behavior studies revealed that compared with KNS-81 and KALS-1 cells transfected with si-NC, those transfected with SNHG1 showed significantly increased proliferation and invasion and significantly decreased apoptosis, while compared with KNS-81 and KALS-1 cells transfected with si-NC, those transfected with si-SNHG1 showed significantly decreased proliferation and invasion and significantly increased apoptosis (all P<0.05). Figure 2.
Effect of miR-140-5p on the Malignant Progression of Glioma Cells
Significantly lower miR-140-5p expression was found in KNS-81 and KALS-1 cells, so KNS-81 and KALS-1 were selected for transfection analysis. It came out that KNS-81 and KALS-1 cells transfected with mimics showed increased miR-140-5p expression and those transfected with inhibitor showed decreased miR-140-5p expression. Cell behavior studies revealed that compared with KNS-81 and KALS-1 cells transfected with miR-NC, those transfected with inhibitor showed significantly increased proliferation and invasion and significantly decreased apoptosis, while compared with KNS-81 and KALS-1 cells transfected with miR-NC, those transfected with mimics showed significantly decreased proliferation and invasion and significantly increased apoptosis (P<0.05). Figure 3.
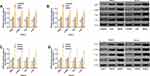
Effects of SNHG1 and miR-140-5p on the PI3K/AKT Pathway
Cells transfected with SNHG1 or inhibitor showed significantly up-regulated AMPK, p-AMPK, PI3K, and p-AKT, while those transfected with si-SNHG1 or mimics showed a significant down-regulation of them. Figure 4.
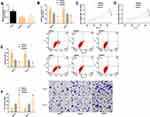
Identification of Target Genes of SNHG1
We found through starBase website that there were targeting binding locus between SNHG1 and miR-140-5p, and the DLR assay revealed that up-regulation of miR-140-5p significantly lowered the luciferase activity of SNHG13ʹUTR-Wt (P<0.05), but exerted no effect on that of SNHG13ʹUTR-Mut (P>0.05). In the RNA pull-down assay, miR-140-5p was pulled down only by SNHG13ʹUTR-Wt (P< 0.05), but not affected by SNHG13ʹUTR-Mut (P> 0.05). RIP analysis showed that under the action of miR-140-5p mimics, SNHG1 in KNS-81 and KALS-1 cells was significantly up-regulated (P< 0.05). In addition, real-time PCR detection revealed that KNS-81 and KALS-1 cells transfected with si-SNHG1 showed significantly increased miR-140-5p expression (P< 0.05). Figure 5.
Co-Transfection Experiment
KNS-81 and KALS-1 cells transfected with SNHG1+mimics or si-SNHG1+inhibitor were not different from those transfected with si-NC in cell invasion, proliferation, and apoptosis, and the PI3K/AKT pathway-related protein levels (all P>0.05), but showed significantly stronger proliferation and invasion, significantly weaker apoptosis, and significantly higher AMPK, p-AMPK, PI3K, and p-AKT protein levels than those transfected with si-SNHG1, and also showed significantly weaker proliferation and invasion, significantly stronger apoptosis, and significantly lower levels of proteins than those transfected with SNHG1 (all P<0.05). Figure 6.
Discussion
SNHG1 is a regulatory factor with dynamic expression changes in various cancers, and is widely used for diseases including the above-mentioned glioma, prostate cancer, hepatic carcinoma, and colon cancer.22,23 An increasing number of researchers show enthusiasm for studying the role of SNHG1 in glioma, and many research results appear one after another. For example, one study by Li et al24 has pointed out that SNHG1 can regulate the miR-154-5p/miR-376b-3p-FOXP2-KDM5B network to affect the behavior of glioma cells. One other study by Liu et al25 has reported that SNHG1 is overexpressed in glioma tissues and cell lines, and it is involved in the pathological progress of glioma through negative regulation on miR-194 and PHLDA1. Moreover, there are many reports about the regulatory effects of miR-140-5p on glioma. MiR-140-5p can not only targetedly inhibit ADAM9 and further regulate glioma growth and metastasis but also accept the regulation by IncRNA HOXA11 to affect the cell proliferation, cycle and apoptosis.26,27
In this study, both SNHG1 and miR-140-5p showed abnormal expression in glioma tissues. The expression of SNHG1 was relatively high, while that of miR-140-5p was opposite, implying that they might participate in the progression of glioma. The analysis on the correlation between SNHG1 and miR-140-5p showed that SNHG1 was significantly negatively correlated with miR-140-5p, suggesting that the two may play antagonistic roles in the pathological process of glioma. The subsequent in vitro experiments revealed that the expression of SNHG1 and miR-140-5p in glioma cell lines was consistent with that of them in tissue samples, and was also consistent with the previous research results. We selected two glioma cell lines (KNS-81 and KALS-1) with the most significant differential expression to carry out transfection research, and up-regulated and down-regulated SNHG1 and miR-140-5p in them, respectively. It came out that transfection of sh-SNHG1 or inhibitor strongly intensified the proliferation and invasion of KNS-81 and KALS-1 cells, and strongly weakened the apoptosis of them, while transfection of si-SNHG1 or mimics exerted opposite effects on the cells. It indicates that down-regulating SNHG1 or up-regulating miR-140-5p might become potential therapeutic targets for glioma, and help to inhibit malignant progression of it. Previous studies have shown that the PI3K/AKT pathway plays a part in the regulation mechanism of invasion, proliferation, and apoptosis of human glioma cells. AMPK, p-AMPK, PI3K and p-AKT are important PI3K/AKT pathway-related factors, of which AMPK and p-AMPK are upstream regulators of the PI3K/AKT pathway, and inhibition on related proteins of this pathway can exert anti-tumor activity.28 Wei et al29 have reported that activating the PI3K/AKT pathway promotes the malignant progression of glioma and stimulates the tumorigenic ability of glioma stem cells, which is one of the mechanisms of action. Therefore, we also investigated the regulation of SNHG1 and miR-140-5p on the PI3K/AKT pathway. It came out that both down-regulation of SNHG1 and up-regulation of miR-140-5p could effectively inhibit the activation of the PI3K/AKT pathway, which was manifested as significantly down-regulation of PI3K/AKT pathway-related proteins such as PI3K, p-AKT, AMPK, and p-AMPK. The results suggest that SNHG1 and miR-140-5p have important regulatory effects on the activation of the PI3K/AKT pathway.
On the other hand, we verified the targeted relationship between SNHG1 and miR-140-5p through a DLR assay. The results revealed that knock-down of SNHG1 strongly lowered the luciferase activity of miR-140-5p3ʹUTR-Wt, but exerted no effect on that of miR-140-5p3ʹUTR-Mut, and transfection of si-SNHG1 significantly increased the expression of miR-140-5p. In addition, in the RNA pull-down assay, only SNHG13ʹUTR-Wt obviously pulled down miR-140-5p. RIP analysis showed that under the action of miR-140-5p mimics, SNHG1 in KNS-81 and KALS-1 cells was significantly up-regulated. The above verification experiments of gene relationship all indicate that SNHG1 can specifically bind to miR-140-5p, and has negative regulation on the expression of miR-140-5p. We also performed co-transfection experiments, and found that KNS-81 and KALS-1 cells transfected with SNHG1+mimic and si-SNHG1+inhibitor were not different from those in the si-NC group with unrelated sequences in cell malignant progression, but showed significantly stronger proliferation and invasion, significantly weaker apoptosis, and significantly higher levels of PI3K/AKT-related proteins than those transfected with si-SNHG1, and also showed significantly weaker proliferation and invasion, significantly stronger apoptosis, and significantly lower levels of proteins than those transfected with sh-SNHG1. Those results once again confirmed the existence of a targeted regulatory relationship between SNHG1 and miR-140-5p. The above results implied that SNHG1 could promote the malignant progression of glioma by targeting miR-140-5p and activating the PI3K/AKT pathway, and down-regulating SNHG1 or up-regulating miR-140-5p may be potential targets for inhibiting malignant progression of glioma.
Conclusion
The regulatory network of the SNHG1/miR-140-5p axis is a novel potential therapeutic direction to curb the malignant progression of glioma. However, there is still a room for improvement in this study. First of all, we can supplement the research on potential drug resistance of SNHG1 and miR-140-5p in glioma. Secondly, we can supplement an analysis on the value of both in the diagnosis of pathological parameters of patients. In addition, we can explore the downstream target mRNA of miR-140-5p.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Miller AM, Shah RH, Pentsova EI, et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature. 2019;565(7741):654–658.
2. Ludwig K, Kornblum HI. Molecular markers in glioma. J Neurooncol. 2017;134(3):505–512. doi:10.1007/s11060-017-2379-y
3. Zhang N, Shuai K, Cheng J, Yang W, Kan Z. LncRNA linc01116 prometes glioma cell migration and invasion by modulation of radixin targeted by miR-31. Int J Clin Exp Pathol. 2019;12(3):1078–1086.
4. Amirian ES, Armstrong GN, Zhou R, et al. The glioma international case-control study: a report from the genetic epidemiology of glioma international consortium. Am J Epidemiol. 2016;183(2):85–91.
5. Rasmussen BK, Hansen S, Laursen RJ, et al. Epidemiology of glioma: clinical characteristics, symptoms, and predictors of glioma patients grade I-IV in the the Danish Neuro-Oncology Registry. J Neurooncol. 2017;135(3):571–579. doi:10.1007/s11060-017-2607-5
6. Ma CC, Xiong Z, Zhu GN, et al. Long non-coding RNA ATB promotes glioma malignancy by negatively regulating miR-200a. J Exp Clin Cancer Res. 2016;35(1):90. doi:10.1186/s13046-016-0367-2
7. Kundu M, Das S, Dhara D, Mandal M. Prospect of natural products in glioma: a novel avenue in glioma management. Phytother Res. 2019;33(10):2571–2584. doi:10.1002/ptr.6426
8. Reon BJ, Anaya J, Zhang Y, et al. Expression of lncRNAs in low-grade gliomas and glioblastoma multiforme: an in silico analysis. PLoS Med. 2016;13(12):e1002192. doi:10.1371/journal.pmed.1002192
9. Hao NB, He YF, Li XQ, Wang K, Wang RL. The role of miRNA and lncRNA in gastric cancer. Oncotarget. 2017;8(46):81572–81582. doi:10.18632/oncotarget.19197
10. Wang W, Zhao Z, Yang F, et al. An immune-related lncRNA signature for patients with anaplastic gliomas. J Neurooncol. 2018;136(2):263–271. doi:10.1007/s11060-017-2667-6
11. Nie W, Ge HJ, Yang XQ, et al. LncRNA-UCA1 exerts oncogenic functions in non-small cell lung cancer by targeting miR-193a-3p. Cancer Lett. 2016;371(1):99–106. doi:10.1016/j.canlet.2015.11.024
12. Li W, Zhai L, Wang H, et al. Downregulation of LncRNA GAS5 causes trastuzumab resistance in breast cancer. Oncotarget. 2016;7(19):27778–27786. doi:10.18632/oncotarget.8413
13. Cui B, Li B, Liu Q, Cui Y. lncRNA CCAT1 promotes glioma tumorigenesis by sponging miR-181b. J Cell Biochem. 2017;118(12):4548–4557. doi:10.1002/jcb.26116
14. Song T, Yan L, Cai K, Zhao T, Xu M. Downregulation of long noncoding RNA PVT1 attenuates paclitaxel resistance in glioma cells. Cancer Biomark. 2018;23(3):447–453. doi:10.3233/CBM-181573
15. Min W, Dai D, Wang J, et al. Long noncoding RNA miR210HG as a potential biomarker for the diagnosis of glioma. PLoS One. 2016;11(9):e0160451. doi:10.1371/journal.pone.0160451
16. Wang Q, Li Q, Zhou P, et al. Upregulation of the long non-coding RNA SNHG1 predicts poor prognosis, promotes cell proliferation and invasion, and reduces apoptosis in glioma. Biomed Pharmacother. 2017;91:906–911. doi:10.1016/j.biopha.2017.05.014
17. Yang HL, Gao YM, Zhao JA. miR1405p inhibits human glioma cell growth and invasion by targeting JAG1. Mol Med Rep. 2017;16(3):3634–3640. doi:10.3892/mmr.2017.6951
18. Zhang K, Chen J, Song H, Chen LB. SNHG16/miR-140-5p axis promotes esophagus cancer cell proliferation, migration and EMT formation through regulating ZEB1. Oncotarget. 2018;9(1):1028–1040. doi:10.18632/oncotarget.23178
19. Mayer IA, Arteaga CL. The PI3K/AKT pathway as a target for cancer treatment. Annu Rev Med. 2016;67:11–28. doi:10.1146/annurev-med-062913-051343
20. Zhu Y, Zhang X, Qi L, et al. HULC long noncoding RNA silencing suppresses angiogenesis by regulating ESM-1 via the PI3K/Akt/mTOR signaling pathway in human gliomas. Oncotarget. 2016;7(12):14429–14440. doi:10.18632/oncotarget.7418
21. Zhang L, Liang X, Li Y. Long non-coding RNA MEG3 inhibits cell growth of gliomas by targeting miR-93 and inactivating PI3K/AKT pathway. Oncol Rep. 2017;38(4):2408–2416. doi:10.3892/or.2017.5871
22. Li J, Zhang Z, Xiong L, et al. SNHG1 lncRNA negatively regulates miR-199a-3p to enhance CDK7 expression and promote cell proliferation in prostate cancer. Biochem Biophys Res Commun. 2017;487(1):146–152. doi:10.1016/j.bbrc.2017.03.169
23. Zhang M, Wang W, Li T, et al. Long noncoding RNA SNHG1 predicts a poor prognosis and promotes hepatocellular carcinoma tumorigenesis. Biomed Pharmacother. 2016;80:73–79. doi:10.1016/j.biopha.2016.02.036
24. Li H, Xue Y, Ma J, et al. SNHG1 promotes malignant biological behaviors of glioma cells via microRNA-154-5p/miR-376b-3p- FOXP2- KDM5B participating positive feedback loop. J Exp Clin Cancer Res. 2019;38(1):59. doi:10.1186/s13046-019-1063-9
25. Liu L, Shi Y, Shi J, et al. The long non-coding RNA SNHG1 promotes glioma progression by competitively binding to miR-194 to regulate PHLDA1 expression. Cell Death Dis. 2019;10(6):463. doi:10.1038/s41419-019-1698-7
26. Liu X, Wang S, Yuan A, Yuan X, Liu B. MicroRNA-140 represses glioma growth and metastasis by directly targeting ADAM9. Oncol Rep. 2016;36(4):2329–2338. doi:10.3892/or.2016.5007
27. Cui Y, Yi L, Zhang MM, et al. WITHDRAWN: knockdown of LncRNA HOXA11 antisense promotes glioma cell apoptosis via sponging MiR-140-5p. Oncol Res. 2017. doi:10.3727/096504017X14972669062547
28. Tian Y, Nan Y, Han L, et al. MicroRNA miR-451 downregulates the PI3K/AKT pathway through CAB39 in human glioma. Int J Oncol. 2012;40(4):1105–1112.
29. Wei Y, Jiang Y, Zou F, et al. Activation of PI3K/Akt pathway by CD133-p85 interaction promotes tumorigenic capacity of glioma stem cells. Proc Natl Acad Sci U S A. 2013;110(17):6829–6834. doi:10.1073/pnas.1217002110
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.