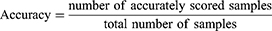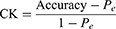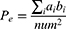Back to Journals » Nature and Science of Sleep » Volume 14
Sleep Stage Classification Based on Multi-Centers: Comparison Between Different Ages, Mental Health Conditions and Acquisition Devices
Authors Xu Z , Zhu Y , Zhao H, Guo F, Wang H, Zheng M
Received 24 December 2021
Accepted for publication 16 May 2022
Published 24 May 2022 Volume 2022:14 Pages 995—1007
DOI https://doi.org/10.2147/NSS.S355702
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Ahmed BaHammam
Ziliang Xu,1,* Yuanqiang Zhu,1,* Hongliang Zhao,1,* Fan Guo,1 Huaning Wang,2 Minwen Zheng1
1Department of Radiology, Xijing Hospital, Fourth Military Medical University, Xi’an, Shaanxi, 710032, People’s Republic of China; 2Department of Psychiatry, Xijing Hospital, Fourth Military Medical University, Xi’an, Shaanxi, 710032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Minwen Zheng, Department of Radiology, Xijing Hospital, Fourth Military Medical University, 127# Changle West Road, Xi’an 710032, People’s Republic of China, Email [email protected] Huaning Wang, Department of Psychiatry, Xijing Hospital, Fourth Military Medical University, 127# Changle West Road, Xi’an 710032, People’s Republic of China, Email [email protected]
Purpose: To investigate the general sleep stage classification performance of deep learning networks, three datasets, across different age groups, mental health conditions, and acquisition devices, comprising adults (SHHS) and children without mental health conditions (CCSHS), and subjects with mental health conditions (XJ), were included in this study.
Methods: A long short-term memory (LSTM) network was used to evaluate the effect of different ages, mental health conditions, and acquisition devices on the sleep stage classification performance and the general performance.
Results: Results showed that the age and different mental health conditions may affect the sleep stage classification performance of the network. The same acquisition device using different parameters may not have an obvious effect on the classification performance. When using a single dataset and two datasets for training, the network performed better only on the training dataset. When training was conducted with three datasets, the network performed well for all datasets with a Cohen’s Kappa of 0.8192 and 0.8472 for the SHHS and CCSHS, respectively, but performed relatively worse (0.6491) for the XJ, which indicated the complexity effect of different mental health conditions on the sleep stage classification task. Moreover, the performance of the network trained using three datasets was similar for each dataset to that of the network trained using a single dataset and tested on the same dataset.
Conclusion: These results suggested that when more datasets across different age groups, mental health conditions, and acquisition devices (ie, more datasets with different feature distributions for each sleep stage) are used for training, the general performance of a deep learning network will be superior for sleep stage classification tasks with varied conditions.
Keywords: sleep stage classification, deep learning network, electroencephalogram, time-frequency spectrum
Introduction
Sleep is an important element of one’s daily routine and occupies about one-third of the average human’s life.1 However, a substantial number of people get inadequate sleep and have various sleep disorders, as the result of stressful life-events, the 24/7 rhythm of the modern world, shift-work, entertainment, and media consumption before sleep.2 Sleep disturbances are a hallmark of most, if not all, neurological diseases, and can contribute to a range of health problems including obesity, depression, and anxiety, and so on.3
The American Academy of Sleep Medicine (AASM) proposed that sleep can be split into five stages: the wake stage, three non-rapid eye movement sleep stages (N1 stage, N2 stage, and N3 stage), and the rapid eye movement (REM) stage. These stages are distinguished according to the amplitude, frequency, or other characteristics of neural electrical signals acquired via electroencephalography (EEG) or polysomnography (PSG) during sleep.4 Many studies have demonstrated that the features of these electrical signals extracted from different sleep stages, such as frequency, amplitude, duration, and specific waves, and so on, are strongly associated with attention,5,6 memory,7 and cognition.8 As a result, these factors have been widely used to examine the mechanisms of mental and neurological disorders such as Parkinson’s disease,9 Alzheimer’s disease,10 and schizophrenia.11,12
To ensure the precision of these specific sleep features, accurate sleep stage classification is very important. However, as a whole night sleep often contains about 6–8 hours EEG or PSG data, manual sleep stage classification is very time consuming, and physicians may make inter- or intra-observer errors due to the fatigue and the difference in experience.13 Currently, new technological developments have enabled researchers to propose methods for automatic sleep stage classification.14–16 Particularly, many automatic sleep stage classification methods based on artificial intelligence (AI) have been proposed. For instance, Biswal et al proposed a novel SLEEPNET framework for sleep stage classification, consisting of a feature extraction mode, sleep stage classification mode, performance evaluation mode, and deployment mode. This system achieved an accuracy of 0.8576 and a Cohen’s Kappa of 0.7946 for the dataset created at Massachusetts General Hospital Sleep Laboratory.17 Sun et al proposed a hierarchical neural network for automatic sleep stage classification comprising a feature extraction network and a recurrent neural network. This system had an accuracy of 0.878 and an F1-score of 0.818 on the Montreal Archive of Sleep Studies database.18 Also, our team conducted a previous study in which we proposed an automatic sleep stage classification method based on a long short-term memory (LSTM) network that used time-frequency spectra extracted from sleep EEG signals as input. When we used our proposed method to estimate sleep stages given data from the Sleep and Heart Health Study dataset, it achieved an accuracy of 0.876 and a Cohen’s Kappa of 0.8256.19 Although many promising AI-based automatic sleep stage classification methods have been proposed, due to the different scoring patterns by different physicians/clinics and lacking clinical certification and transparency, these methods are not yet widely used in clinical practice. But all these problems can be summarized into one main problem, namely that these studies' networks were trained using only a single dataset, which limited their application in terms of general sleep stage classification.
Recently, joint training is now being conducted using multi-center datasets. For instance, Patanaik et al proposed an end-to-end convolutional neural network-based sleep stage classification method that was trained and tested using four datasets. The network had good performance for two of the training datasets and sub-optimal performance for the other two datasets.20 Other researchers have also proposed multi-center dataset-based automatic sleep stage classification systems.21,22 In 2021, Perslev et al used 23 datasets from 16 clinical studies to investigate the sleep stage classification performance of U-sleep net.23 Although 23 datasets were used, their study did not investigate the relationship between the number of datasets used for training and the general classification performance of the network. Thus, to systematically investigate this relationship, Olesen et al used five big public datasets to conduct joint training.24 They found that the general sleep stage classification performance was better when more datasets were used for training. To the best of our knowledge, these two studies are currently the largest investigation of automatic sleep stage classification in terms of data size and diversity. However, almost all of the subjects in these datasets were normal subjects with sleep disorders. Thus, the general performance is still unverified based on different groups of subjects. Stephansen et al used sleep stage scoring to diagnose narcolepsy,21 and found evidence of differences among sleep stages between healthy subjects and those with mental health conditions. Moreover, factors such as age and acquisition device may also affect sleep features.
To address this, in the present study, we used three datasets, across different age groups, mental health conditions, and acquisition devices, to train a deep learning network. A LSTM network, which has a relatively simple network structure and a smaller number of training parameters compared to other types of deep learning networks,25 and which can consider the time information among signals, was used to perform the sleep stage classification task.
Methods
Datasets
Three datasets were used in this study. The first two datasets were from the Sleep Heart Health Study (SHHS)26,27 and the Cleveland Children’s Sleep and Health Study (CCSHS),26,28 and were accessed via the National Sleep Research Resource (NSRR) database, which contains PSG data collected from subjects with sleep disorders and children with normal sleep, respectively. The SHHS is a large dataset with two subsets, named SHHS visit 1 (SHHS1) and SHHS visit 2 (SHHS2). The SHHS1 comprises PSG data from one night of sleep in 5793 subjects (aged 40–89) recruited between 1995 and 1998. The SHHS2 comprises PSG data from one night of sleep from 2651 of the subjects from the SHHS1, collected between 2001 and 2003. The CCSHS is a relatively small dataset, and contains PSG data from one night of sleep in 515 children (aged 16–19). The third dataset (XJ dataset) contains PSG data collected from patients with mental health conditions (depression, schizophrenia, bipolar affective disorder, and so on) in the department of psychiatry at Xijing Hospital. Specifically, the XJ comprises PSG data from one night of sleep in 8325 patients (aged 6–91) between 2010 and 2018.
We split each dataset into a training and testing group according to the acquisition device and parameters. For the SHHS dataset, as SHHS2 was a second visit of SHHS1, and the acquisition parameters had some differences between two datasets, data from the SHHS1 and SHHS2 subsets were used for training and testing, respectively. For the XJ datasets, considering the updating of software system and the acquisition device, we used the data from the XJ dataset collected between 2010 and 2016 for training and the rest of the data for testing. For the CCSHS dataset, as the acquisition device and parameters were all the same, according to the training to testing ratio of SHHS and XJ datasets (about 2:1), the data from the first 360 subjects were used for training and the other data were used for testing.
We re-split the training and testing groups in the SHHS and XJ datasets into several subsets according to age. For the SHHS dataset, three subsets were created: the 40–60 years group, 60–80 years group, and >80 years group. For the XJ dataset, four subsets were created: the 6–20 years group, 20–40 years group, 40–60 years group, and 60–80 years group. As the number of subjects in the >80 group in the XJ dataset was only 5 (training + testing), these subjects were not used for further analysis of the effects of age on sleep stage classification. As all of the subjects in the CCSHS dataset were aged 16–19 years, this dataset was not re-split.
Additionally, we also re-split the training and testing groups in the XJ dataset into several subsets according to mental health conditions. Considering the number of training samples, the subsets of the training group with at least 100 subjects were used for further analysis. Finally, four subsets were created: the anxiety group, bipolar affective disease group, depression group, and schizophrenia group.
The SHHS and CCSHS datasets can be requested from the NSRR (https://www.sleepdata.org/). The use of the XJ dataset in this study was approved by the institutional review board of Xijing Hospital, which is affiliated with the Fourth Military Medical University.
PSG Data Pre-Processing
PSG data pre-processing was performed using methods similar to those used in our previous study.19 Specifically, a channel check was first performed for five PSG signal channels, including two EEG channels (C3 and C4), two EOG channels (left and right), and one EMG channel, to assess potential electrode dropping problems. The threshold was set at the half of the maximum acquisition amplitude for each channel, which could be acquired from the header of the EDF files. If the mean absolute signal value of a channel was bigger than the threshold, this channel was considered to have an electrode dropping problem, and the data from this channel were not used for further analysis. Considering that the electrode dropping problem is common in realistic sleeping situations, we excluded a subject if two EEG channels, two EOG channels, or the EMG channel were dropped.
After the channel check, we performed zero-phase band-pass filtering. The filtering frequency was set at 0.3–45 Hz, 0.3–12 Hz, and 0.3–20 Hz for the EEG, EOG, and EMG channels, respectively. A 50th order Hamming window-based finite impulse response was used. Finally, all filtered channels were resampled to 100 Hz.
Feature Extraction
We used short-time Fourier transform (STFT) to extract the time-frequency (TF) spectra from the pre-processed PSG data,20 which finally resulted in a 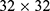 TF spectrum for each 30-s signal block. The amplitude of the frequency at each time point in the TF spectra was normalized into 0 to 1.
TF spectrum for each 30-s signal block. The amplitude of the frequency at each time point in the TF spectra was normalized into 0 to 1.
TF spectra from four channels, named EEG, EOG L, EOG R, and EMG, were used as the input of the network. For the EEG channel, the mean of the signal from the C3 and C4 was used for TF spectra calculation. If one of these two channels had an electrode dropping problem, the signal from the normal one was used. For the EOG L and EOG R channel, if one of these two channels had an electrode dropping problem, the TF spectra of these two channels were all calculated using the signal from the normal one.
According to the sleeping stage scoring standards of the AASM, there should not be any overlap between each consecutive 30-s signal block. However, this would result in a loss of information. Thus, we made a 30-s block every 10 s (ie, there was a 20-s overlap between each consecutive 30-s signal block), which might address the problem of lost information to some extent. The value of 10s was chosen for two reasons. The first is that an insufficient value will introduce large-scale redundancy to the training sample, with may lead to network overfitting. The second is that we wanted to expand the size of the training sample to be as large as possible.
Classification Model
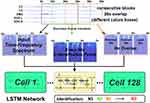
Considering the relatively simple network structure and a smaller number of trainable parameters compared to other types of deep learning networks, such AlexNet,29 VGG,30 GoogLeNet,31 or ResNet,32 the LSTM network that could also capture time information among the TF spectra was used to build the sleep stage classification model. Instead of decreasing the number of output nodes in the LSTM from 512 to 128 (eg, one LSTM hidden layer with 128 output nodes), other model settings were similar to that in our previous study.19 Specifically, we used three TF spectra with no overlap as an input. This kind of input could consider the time information of the previous 30 s, current 30 s, and subsequent 30 s in the TF spectrum and was used to predict the sleep stage of the current time point. We also combined the TF spectra from the four channels into one big TF spectrum along with the frequency axis. Thus, the size of the TF spectra that was inputted to the LSTM network was 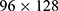 .
.
To evaluate the effects of age on sleep stage classification, we used each of the age subsets (3 subsets for SHHS, 1 for CCSHS, and 4 for XJ) to train the network in turn, and tested the network for all age subsets. As the three datasets were generated using different devices for data acquisition, we were also able to analyze the effects of device on sleep stage classification performance. To examine the effects of mental health conditions, we used each mental health condition subset in the XJ dataset in turn for network training, and tested the networks for all mental health condition subsets, and in the SHHS and CCSHS datasets.
Moreover, we used three training strategies to systematically evaluate the general sleep stage classification performance of the LSTM network: 1) training the model using each dataset individually; 2) training the model using two datasets (3 groups, 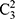 ); and 3) training the model using all three datasets. The workflow of this study is displayed in Figure 1.
); and 3) training the model using all three datasets. The workflow of this study is displayed in Figure 1.
Experimental Setup
We approached sleep stage classification as a five-class classification problem, with the following classes: 0 for the waking state, 1 for the N1 stage, 2 for the N2 stage, 3 for the N3 stage, and 4 for REM sleep. As we calculated a 30-s TF spectrum block every 10 s, each 30-s PSG data had three identification points (0 s, 10 s, and 20 s). The sleep stage with the maximum occurrence time was considered to be the stage of that 30-s data. If a 30-s PSG data had three different stages predicted by the LSTM network, we considered the stage that had the maximum predictive probability to be the stage of this 30-s PSG data.
For model training, we used an Adam optimizer with 0.9 and 0.999 exponential decay rates for the first and second moments, respectively, for updating the model weights. Considering the huge number of training samples, we randomly chose about a quarter of the training samples to train the model in every epoch. The performance of the model trained using this strategy was similar to that of the model trained using all of the training samples in every epoch, and this strategy could save a lot of training time. Considering the unbalanced number of samples in each sleep stage, we used a weighted softmax cross entropy logit function as the cost function for each network.33 The maximum number of epochs was set to 200. The initial learning rate was set at 0.001, and this was divided by 10 every 20 epochs. Training was stopped under the following conditions: 1) if the number of epochs reached the maximum number; 2) if the accuracy of the testing group decreased across five successive epochs; 3) if an absolute accuracy difference of less than 1e-5 occurred between two successive testing epochs on five successive occasions. In total, the LSTM network had 132,229 trainable parameters. To further avoid the effects of random parameter initialization on performance, we trained each LSTM network 5 times, and chose the best performance as the final performance for each model.
Data pre- and post-processing (10-s strategy) and the creation of input signal blocks were implemented using MATLAB software (https://www.mathworks.com/). Training and testing of the LSTM network was implemented using the Tensorflow package with GPU support on a Python 3.5 platform (https://www.python.org/).
Evaluation Index
We used the model accuracy and Cohen’s Kappa (CK) coefficient to evaluate the performance of our LSTM-based sleep stage classification model. We used the accuracy index to assess the overall sleep stage classification precision of the model, and the CK coefficient to assess the overall performance of the model, ie, higher classification precision in each stage was associated with a higher CK coefficient. Detailed definitions of these two indices are as follows:
where 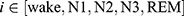 ;
; 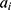 represents the real number of samples in a sleep stage i;
represents the real number of samples in a sleep stage i; 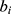 represents the number of samples predicted by the model in a sleep stage i; num represents the total number of samples.
represents the number of samples predicted by the model in a sleep stage i; num represents the total number of samples.
Results
Data Structure for Each Dataset
Tables 1 and 2 show the data structures of each dataset according to age group. In the two datasets in which the subjects had no mental health conditions (SHHS for adults and CCSHS for children, Table 1), the total sleep time and the N3 ratio monotonically decreased with age, and the N1 ratio and N2 ratio monotonically increased with age. Although the sleep efficiency and REM ratio monotonically decreased with age in the SHHS dataset, this was not the case when the SHHS and CCSHS were combined, perhaps because of the different acquisition devices used. In the XJ dataset, the age-related trend was the same as in the SHHS and CCSHS, but the ratio of time in each sleep stage was very different (especially for N3), which may reflect the effects of mental health conditions on sleep.
 |
Table 1 The Detailed Information of SHHS and CCSHS Datasets According to Age |
 |
Table 2 The Detailed Information of XJ Datasets According to Age |
Table 3 shows the data structures of each dataset according to the different mental health conditions. The age in the anxiety and depression groups was higher than those in the bipolar affective disease and schizophrenia groups. The N3 ratio and REM ratio in the anxiety and depression groups were lower than those in the bipolar affective disease and schizophrenia groups. These results indicate that anxiety and depression may be more common in middle-aged people, and that subjects with these mental conditions may not sleep deeply compared with subjects with bipolar affective disease and schizophrenia.
 |
Table 3 The Detailed Information of XJ Datasets According to Mental Condition |
The Effect of Age on Performance of the Model
Tables 4 and 5 show the performance of the LSTM network for different age subsets. Overall, the LSTM had good performance for subsets from the same dataset, and poorer performance for subsets from different datasets. This may have been related to the use of different acquisition devices. For the SHHS dataset, the networks trained using each age subset performed similarly for the 40–60 and 60–80 years groups, but the performance was slightly decreased for the >80 group. This trend was also observed for the XJ dataset (eg, similar performance for the 20–40 and 40–60 years groups, slight decrease in performance for the 6–20 and 60–80 years groups).
 |
Table 4 The Accuracy of LSTM Network Between Different Age Subsets |
 |
Table 5 The CK Coefficient of LSTM Network Between Different Age Subsets |
The Effect of Mental Health Condition on Performance of the Model
Tables 6 and 7 show the performance of the LSTM network for the different mental health condition subsets. Overall, the LSTM performed well for each mental health condition subset in the XJ dataset, but performed worse for the SHHS and CCSHS datasets. The performance of the network trained using the anxiety and bipolar affective disease subsets was similar to that of the network trained using the schizophrenia and depression subsets, respectively.
 |
Table 6 The Accuracy of LSTM Network Between Different Mental Condition Subsets |
 |
Table 7 The CK Coefficient of LSTM Network Between Different Mental Condition Subsets |
General Performance of Model
When trained using a single dataset, for each dataset, the classification performance was better only when tested using the data from that dataset and poorer for the data from the other two datasets (Table 8). For each dataset used for training, the testing accuracy and CK coefficient were 0.8685 and 0.8115, 0.8762 and 0.8294, and 0.7902 and 0.6377 for SHHS, CCSHS, and XJ, respectively; however, for the other two datasets, the testing accuracy and CK coefficient were decreased severely.
 |
Table 8 The Performance of LSTM Network Trained Using Single Dataset |
When trained using two datasets, the classification performance of the network was still only better for data from the dataset used for training (Table 9), which was different from the results in Olesen’s study, that the general classification performance of the network would improve with the increasing number of datasets used for training.24
 |
Table 9 The Performance of LSTM Network Trained Using Two Datasets |
Table 10 shows the sleep stage classification of the LSTM network trained using all datasets. The performance of the network was better for all datasets. The testing accuracy and CK coefficient were 0.8680 and 0.8102, 0.8825 and 0.8380, and 0.7923 and 0.6402 for SHHS, CCSHS, and XJ, respectively, which was very close to that of the network trained using a single dataset and tested on the same dataset.
 |
Table 10 The Performance of LSTM Network Trained Using All Datasets |
Performance of Model Using 10-s Strategy
The network general performance was evaluated using both original 30-s and 10-s strategy. From Tables 8–10, the 10-s strategy could only improve the sleep stage classification performance with about 0.5%. Thus, we suggested that the original 30-s strategy was enough for sleep stage classification. However, the 10-s strategy was a good way to expand the size of the training sample, because the specific sleep features or waves might appear at any time points in a 30-s window.
Discussion
In this study, we used an LSTM network for the sleep stage classification. We used three datasets to evaluate the model classification performance between different age, acquisition device, and mental health conditions, and the general performance (SHHS and CCSHS for subjects without mental health conditions, XJ for subjects with mental health conditions). We also proposed a 10-s sleep stage identification method to correct the classification results obtained by the LSTM network. Our results showed that the network performed better on the training dataset but worse on the remaining dataset(s) when we used a single dataset or two datasets for training. In all-datasets training situations, the network performed well for all datasets. However, the age, the acquisition device and presence of mental health conditions could have led to differences in sleep features, which may have affected the sleep stage classification performance of the LSTM network. The corresponding results are discussed in detail below.
From Tables 4 and 5, for the SHHS dataset, networks trained using each age subset performed similarly for the 40–60 and 60–80 years groups, but performance slightly decreased for the >80 group. A similar trend was found for the XJ dataset. These results suggest that sleep features may change with age. However, the relatively small difference in performance between the subsets in the same dataset indicates that the effect of age on sleep stage classification may be small. Additionally, we obtained the following interesting results. First, when testing with different datasets, the networks trained using the age subsets from the SHHS dataset performed relatively well with the XJ dataset compared with the CCSHS dataset. Second, the networks trained using the subsets from the XJ dataset performed best with the >80 years group from the SHHS dataset. These results indicate that the sleep features of younger subjects with mental health conditions may be similar to those of elderly subjects with no mental health conditions.
For the XJ dataset, the networks trained using each mental health condition subset performed similarly for the anxiety and schizophrenia groups and for the bipolar affective disorder and depression groups. Thus, sleep features may be similar between these two pairs of mental health conditions. We also found that networks trained using each mental health condition subset performed relatively well with the anxiety subset compared with the other subsets. This may indicate that anxiety has a relatively small effect on sleep features compared with the other mental health conditions. Still, networks trained using the mental health condition subsets performed relatively well with the SHHS dataset compared with the CCSHS dataset, which may further suggest that the sleep features of subjects with mental health conditions are similar to those of elderly subjects with no mental health conditions.
As for the effect of acquisition device on sleep stage classification, we found that in datasets that used the same acquisition device, different acquisition parameters did not appear to have a strong effect on the classification task. As mentioned above, the LSTM performed well on subsets from the same dataset (age subsets in the SHHS and XJ and mental health condition subsets in the XJ, Tables 4–7). However, when we trained a network using one dataset and tested it using the other two datasets, the performance was dramatically decreased. Although the acquisition device was different between the three datasets, as none of them contained EEG or PSG data acquired using different devices, the decreased performance maybe not necessarily caused by the use of different acquisition devices. Indeed, different scoring patterns used by different physicians/clinics could also impact the performance. Thus, future studies with datasets containing data acquired using different devices are needed to further investigate the effects of acquisition device on sleep stage classification.
When we used a single dataset to train the LSTM network, the network performed well only on the trained dataset. When the number of datasets used for training was increased to two, the classification performance of the LSTM network for the remaining dataset was similar to that observed in the single dataset training situation (Table 9 vs Table 8). The overfitting problem34 might explain these results. However, the network performed well with the datasets used for training, especially after joint training with the SHHS and XJ dataset (which comprised data from subjects without and with mental health conditions, respectively), and joint training with the CCSHS and XJ datasets (which were dramatically different in terms of sample size and also contained data from two different groups of subjects). These results suggested that the LSTM network indeed could identify the sleep stage features across different datasets. However, it still performed worse on the remaining dataset. Thus, we assumed that the difference of TF spectrum features for each sleep stage among three datasets might be the main reason. When two datasets were used for training, the deep learning network could only capture the information from the two training datasets, and therefore exhibited poorer performed on the remaining dataset.
When we used all of the datasets to train the network, the LSTM network performed well on all three datasets. Moreover, the performance of the network trained using three datasets was similar for each dataset to that of the network trained using a single dataset and tested on the same dataset (Table 10 vs Table 8). These results further proved our assumption above. Thus, our results were partly consistent with Olesen’s finding24 that, when the number of datasets used for network training is increased, the general classification performance of the network improves. However, this only holds when all of the datasets have similar feature distributions for each sleep stage (almost all of the subjects in Olesen’s study were subjects with sleep disorders). Thus, as a supplement to Olesen’s study, our results suggest that using more datasets across different age groups, mental health conditions, and acquisition devices and parameters for training will enhance the general performance of the deep learning network, where age and mental health conditions are relatively important. Conversely, networks trained using only datasets from subjects with similar conditions will perform poorly on datasets from subjects with new conditions. Besides, the LSTM network performed well on all datasets after joint training, which also indicates that a deep learning network has the ability to identify the sleep feature difference among subjects with different ages and mental health conditions, and therefore has great feature generalization ability.
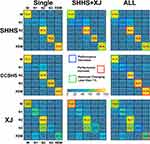
After comparing the classification sensitivity for each sleep stage among the three datasets, we found some interesting differences between models prior to correction by our proposed sleep stage identification strategy (as this strategy was a post-processing operation, the classification performance corrected by this strategy cannot represent the performance of the LSTM network itself). First, as shown in Figure 2, the sensitivity to the wake, N1, N3, and REM stages in the single-dataset training situation substantially differed between the SHHS and XJ datasets. If this situation is considered in isolation, it is possible to imagine that these results might have been caused by the difference in the sample distribution in the training samples between these two datasets (Tables 1 and 2). During joint training, the number of training samples in each sleep stage had been extremely supplemented; however, the sensitivity to these four sleep stages was still similar to that in the single-dataset training situation, although there existed some increases or decreases in performance. Thus, these results suggest that the PSG signals associated with the wake, N1, N3, and REM sleep stages may differ substantially between subjects with (XJ) and without (SHHS) mental health conditions. Second, all of the sleep stages had similar sensitivity in the SHHS and CCSHS datasets during both single-dataset training and joint-dataset training. This means that the PSG sleep signals may have been similar between children (CCSHS) and adults (SHHS) without mental health conditions. Third, when comparing the misclassified ratio for each sleep stage among the three datasets, the N1 stage may be more clearly different from the wake and N2 stages in the subjects with mental health conditions, and the N3 and REM stages may be more similar to the N2 and the N1 stages, respectively, in these subjects. Moreover, differences in the performance of networks trained using different subsets could also suggest that sleep features vary according to experimental conditions (eg, similar performance reflects similar sleep features between conditions, and vice versa). All of these results indicate that as long as the design of a scientific problem is reasonable, a deep learning model can be used to investigate the mechanism of the problem without analyzing the “black box” of the model itself. This notion may stimulate new ideas regarding mechanism research using deep learning-based methods. However, this kind of design is also a big challenge. Sleep stage classification is a special case, and additional research will be needed to apply this type of design to other clinical problems.
This study had several limitations. First, we only used three datasets in this study. Future studies will use more datasets, especially those containing subjects with mental health conditions, to validate the results of this study and further improve the sleep stage classification performance of the LSTM network. Second, due to the sample size, we did not further split each mental health condition subset of the XJ dataset into several age subsets. Further studies will collect more data to investigate in detail the effect of age and mental health conditions on sleep stage classification.
In summary, this study investigated the sleep stage classification performance of a deep learning network trained using three datasets. Our results indicate that using more datasets across different age groups, mental health conditions, and acquisition devices and parameters for training may enhance the general performance of the network for sleep stage classification tasks under varied conditions. Deep learning-based methods can also be used to investigate the mechanisms of a scientific problem without analyzing the “black box” of the model itself. However, an appropriate experimental design is needed.
Acknowledgment
We thank Sydney Koke, MFA, from Liwen Bianji (Edanz) (https://www.liwenbianji.cn), for editing the English text of a draft of this manuscript.
We thank the National Sleep Research Resource (https://www.sleepdata.org) team for their work in sharing SHHS (https://www.sleepdata.org/datasets/shhs) and CCSHS datasets (https://www.sleepdata.org/datasets/ccshs) used in this study.
Funding
This study was financially supported by the National Natural Science Foundation of China under grant 81801772 and 82071917.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Cirelli C, Tononi G. Is sleep essential? PLoS Biol. 2008;6(8):e216. doi:10.1371/journal.pbio.0060216
2. Geiker NRW, Astrup A, Hjorth MF, Sjödin A, Pijls L, Markus CR. Does stress influence sleep patterns, food intake, weight gain, abdominal obesity and weight loss interventions and vice versa? Obes Rev. 2018;19:81–97. doi:10.1111/obr.12603
3. Iranzo A. Sleep and neurological autoimmune diseases. Neuropsychopharmacology. 2020;45(1):129–140. doi:10.1038/s41386-019-0463-z
4. Berry RB, Albertario CL, Harding SM, et al. for the American Academy of Sleep Medicine. The AASM manual for the scoring of sleep and associated events: rules, terminology and technical specifications. Darien, IL: American Academy of Sleep Medicine. 2018.
5. Furrer M, Jaramillo V, Volk C, et al. Sleep EEG slow-wave activity in medicated and unmedicated children and adolescents with attention-deficit/hyperactivity disorder. Transl Psychiatry. 2019;9(1):324. doi:10.1038/s41398-019-0659-3
6. Diep C, Garcia-Molina G, Jasko J, et al. Acoustic enhancement of slow wave sleep on consecutive nights improves alertness and attention in chronically short sleepers. Sleep Med. 2021;81:69–79. doi:10.1016/j.sleep.2021.01.044
7. Ferrarelli F, Kaskie R, Laxminarayan S, et al. An increase in sleep slow waves predicts better working memory performance in healthy individual. Neuroimage. 2019;191:1–9. doi:10.1016/j.neuroimage.2019.02.020
8. Fernandez LMJ, Lüthi A. Sleep spindles: mechanisms and functions. Physiol Rev. 2020;100(2):805–868. doi:10.1152/physrev.00042.2018
9. Pushpanathan ME, Loftus AM, Thomas MG, et al. The relationship between sleep and cognition in Parkinson’s disease: a meta-analysis. Sleep Med Rev. 2016;26:21–32. doi:10.1016/j.smrv.2015.04.003
10. Zhang F, Zhong R, Li S, et al. Alteration in sleep architecture and electroencephalogram as an early sign of Alzheimer’s disease preceding the disease pathology and cognitive decline. Alzheimers Dement. 2019;15(4):590–597. doi:10.1016/j.jalz.2018.12.004
11. Bartsch U, Simpkin AJ, Demanuele C, et al. Distributed slow-wave dynamics during sleep predict memory consolidation and its impairment in schizophrenia. NPJ Schizophr. 2019;5(1):18. doi:10.1038/s41537-019-0086-8
12. Markovic A, Buckley A, Driver DI, et al. Sleep spindle activity in childhood onset schizophrenia: diminished and associated with clinical symptoms. Schizophr Res. 2020;223:327–336. doi:10.1016/j.schres.2020.08.022
13. Boostani R, Karimzadeh F, Nami M. A comparative review on sleep stage classification methods in patients and healthy individuals. Comput Methods Programs Biomed. 2017;140:77–91. doi:10.1016/j.cmpb.2016.12.004
14. Fonseca P, Long X, Radha M, Haakma R, Aarts RM, Rolink J. Sleep stage classification with ECG and respiratory effort. Physiol Meas. 2015;36(10):2027–2040. doi:10.1088/0967-3334/36/10/2027
15. Shi J, Liu X, Li Y, Zhang Q, Li Y, Ying S. Multi-channel EEG-based sleep stage classification with joint collaborative representation and multiple kernel learning. J Neurosci Methods. 2015;254:94–101. doi:10.1016/j.jneumeth.2015.07.006
16. Sousa T, Cruz A, Khalighi S, Pires G, Nunes U. A two-step automatic sleep stage classification method with dubious range detection. Comput Biol Med. 2015;59:42–53. doi:10.1016/j.compbiomed.2015.01.017
17. Biswal S, Kulas J, Sun H, et al. SLEEPNET: automated sleep staging system via deep learning. arXvi. 2017. doi:10.48550/arxiv.1707.08262
18. Sun C, Chen C, Li W, Fan J, Chen W. A hierarchical neural network for sleep stage classification based on comprehensive feature learning and multi-flow sequence learning. IEEE J Biomed Health Inform. 2020;24(5):1351–1366. doi:10.1109/JBHI.2019.2937558
19. Xu ZL, Yang XJ, Sun JB, Liu P, Qin W. Sleep stage classification using time-frequency spectra from consecutive multi-time points. Front Neurosci. 2020;14:14. doi:10.3389/fnins.2020.00014
20. Patanaik A, Ong JL, Gooley JJ, Ancoli-Israel S, Chee MWL. An end-to-end framework for real-time automatic sleep stage classification. Sleep. 2018;41(5):1–11. doi:10.1093/sleep/zsy041
21. Stephansen JB, Olesen AN, Olsen M, et al. Neural network analysis of sleep stages enables efficient diagnosis of narcolepsy. Nat Commun. 2018;9(1):5229. doi:10.1038/s41467-018-07229-3
22. Biswal S, Sun H, Goparaju B, Westover MB, Sun J, Bianchi MT. Expert-level sleep scoring with deep neural networks. J Am Med Informatics Assoc. 2018;25(12):1643–1650. doi:10.1093/jamia/ocy131
23. Perslev M, Darkner S, Kempfner L, Nikolic M, Jennum PJ, Igel C. U-sleep: resilient high-frequency sleep staging. NPJ Digit Med. 2021;4(1):72. doi:10.1038/s41746-021-00440-5
24. Olesen AN, Jørgen Jennum P, Mignot E, Sorensen HBD. Automatic sleep stage classification with deep residual networks in a mixed-cohort setting. Sleep. 2021;44(1):zsaa161. doi:10.1093/sleep/zsaa161
25. Hochreiter S, Schmidhuber J. Long short-term memory. Neural Comput. 1997;9(8):1735–1780. doi:10.1162/neco.1997.9.8.1735
26. Zhang GQ, Cui L, Mueller R, et al. The National Sleep Research Resource: towards a sleep data commons. J Am Med Inform Assoc. 2018;25(10):1351–1358. doi:10.1093/jamia/ocy064
27. Quan SF, Howard BV, Iber C, et al. The sleep heart health study: design, rationale, and methods. Sleep. 1997;20(12):1077–1085.
28. Rosen CL, Larkin EK, Kirchner HL, et al. Prevalence and risk factors for sleep-disordered breathing in 8- to 11-year-old children: association with race and prematurity. J Pediatr. 2003;142(4):383–389. doi:10.1067/mpd.2003.28
29. Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Commun ACM. 2017;60(6):84–90. doi:10.1145/3065386
30. Simonyan K, Zisserman A. Very deep convolutional networks for large-scale image recognition. arXiv. 2015. doi:10.48550/arXiv.1409.1556
31. Szegedy C, Liu W, Jia YQ, et al. Going deeper with convolutions. arXiv. 2014. doi:10.48550/arXiv.1409.4842
32. He K, Zhang XY, Ren SQ, Sun J. Deep residual learning for image recognition. arXiv. 2015. doi:10.48550/arXiv.1512.03385
33. Xie S, Tu Z. Holistically-nested edge detection. Int J Comput Vis. 2017;125(1–3):3–18. doi:10.1007/s11263-017-1004-z
34. Tetko IV, Livingstone DJ, Luik AI. Neural network studies. 1. comparison of overfitting and overtraining. J Chem Inf Comput Sci. 1995;35(5):826–833. doi:10.1021/ci00027a006
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.