Back to Journals » Journal of Hepatocellular Carcinoma » Volume 9
SLC39A1 Overexpression is Associated with Immune Infiltration in Hepatocellular Carcinoma and Promotes Its Malignant Progression
Authors Ma X, Zhuang H, Wang Q , Yang L, Xie Z, Zhang Z, Tan W, Tang C , Chen Y, Shang C
Received 30 November 2021
Accepted for publication 25 January 2022
Published 15 February 2022 Volume 2022:9 Pages 83—98
DOI https://doi.org/10.2147/JHC.S349966
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Imam Waked
Xiaowu Ma,1,2,* Hongkai Zhuang,1,2,* Qingbin Wang,1,2 Lei Yang,1,2 Zhiqin Xie,1,2 Ziyu Zhang,1,2 Wenliang Tan,1,2 Chenwei Tang,1,2 Yajin Chen,1,2 Changzhen Shang1,2
1Department of Hepatobiliary Surgery, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, 510080, People’s Republic of China; 2Guangdong Provincial Key Laboratory of Malignant Tumor Epigenetics and Gene Regulation, Sun Yat-sen Memorial Hospital, Sun Yat-Sen University, Guangzhou, 510080, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yajin Chen; Changzhen Shang, Department of Hepatobiliary Surgery, Sun Yat-sen Memorial Hospital, Sun Yat-sen University, Guangzhou, 510080, People’s Republic of China, Tel +86-2034070701, Email [email protected]; [email protected]
Background: Solute carrier family 39 member 1 (SLC39A1) has been identified as a zinc ion transport protein that possesses oncogenic properties in various types of cancers. However, its potential function in hepatocellular carcinoma (HCC) remains unknown. This study aimed to investigate the expression profile and potential mechanisms of SLC39A1 in HCC.
Methods: SLC39A1 expression was analyzed using multiple databases. The clinical significance and associated biological pathways of SLC39A1 were investigated using bioinformatics analysis. Potential correlations between SLC39A1 expression and tumor immunity in HCC were also evaluated using single-sample gene set enrichment analysis (GSEA). Sixty paired HCC samples were used to verify the expression pattern of SLC39A1. In vitro studies were performed to investigate the oncogenic effects of SLC39A1 in HCC. Western blot analysis was conducted to further investigate the possible involved signaling pathways.
Results: The overexpression of SLC39A1 in HCC was determined by bioinformatics analysis and was confirmed in tissues from our center. SLC39A1 overexpression was also significantly correlated with worse prognosis, advanced TNM stage, and histological grade. GSEA analysis demonstrated that SLC39A1 overexpression was involved in various tumor-related pathways, such as the cell cycle and Wnt signaling pathway. SLC39A1 knockdown repressed the proliferation, invasion, and migration abilities of HCC cells. Furthermore, SLC39A1 knockdown decreased the expression of the tumor progression-related proteins (eg, cyclin D1 and MMP2) and Wnt signaling pathway-related proteins (eg, Wnt3A and β-catenin). In addition, SLC39A1 overexpression may be associated with impaired tumor immunity in HCC, as evidenced by the increased infiltration of Th2 cells and reduced infiltration of cytotoxic cells.
Conclusion: These findings preliminarily suggested the crucial effect of SLC39A1 overexpression on HCC tumor progression and immunosuppression, suggesting its potential as a novel prognostic and therapeutic target in HCC.
Keywords: SLC39A1, hepatocellular carcinoma, prognosis, Wnt signaling pathway, immune infiltration
Introduction
The predominant malignant liver tumor, HCC, is one of the most common causes of cancer-related mortality globally.1 Despite the rapid development of curative therapies such as partial hepatectomy, liver transplantation, radiofrequency ablation, and targeted therapy, the overall survival of HCC patients is still poor due to tumor heterogeneity and the specific tumor microenvironment.2 Based on some immune checkpoints, specifically expressed on T cells, scientists have provided some novel therapies for HCC patients. For instance, patients receiving atezolizumab plus bevacizumab had longer progression-free survival compared with those receiving sorafenib.3 Another study showed that HCC patients receiving the combination therapy of sintilimab and a bevacizumab biosimilar (IBI305) had a longer median progression-free survival than those receiving sorafenib.4 However, approximately 75% of HCC patients show a weak response to immunotherapies, and the reason remains unknown.5 Therefore, a comprehensive investigation of the underlying molecular mechanisms involved in HCC progression and the associated immune microenvironment is increasingly needed for developing more effective therapeutic strategies.
Solute carrier family 39 members (SLC39As) have been identified as zinc metal ion transporting proteins, and their dysregulation has been proven to induce cellular zinc dyshomeostasis and exhibit various oncogenic properties in multiple malignancies, such as lung, cervical, and pancreatic cancers.6–11 For instance, SLC39A4 accelerates epithelial mesenchymal transition (EMT), and induces resistance to cisplatin therapy in lung cancer cells. It also induces phosphorylation of focal adhesion kinase (FAK) and paxillin, thus promoting tumor progression in pancreatic cancer cells.9,10 Conversely, SLC39A8 effectively inhibits the development of clear cell renal cell carcinoma by blocking EMT progression, whereas knockdown of SLC39A7 attenuates the metastatic ability of cervical cancer.11,12 As the first member of SLC39As, SLC39A1 was mainly expressed on the plasma membrane and involved in the zinc absorption activity.7,13 Oncogenic studies about SLC39A1 have been conducted in several malignancies, such as glioma and prostate cancer. Wang et al demonstrated that SLC39A1 overexpression promotes tumor progression and correlates with unfavorable clinical outcomes in glioma.14 In addition, downregulation of SLC39A1 impairs the accumulation of zinc in tumor cells, thereby promoting the malignant phenotype of prostate cancer.15,16 However, little is known about the regulatory mechanism of SLC39A1 in HCC progress.
In the current study, the expression levels of SLC39A1 and its clinical significance in HCC were comprehensively investigated using multiple bioinformatics analyses tools. In addition, the oncological role of SLC39A1 in HCC was determined through functional enrichment analysis and in vitro studies.
Materials and Methods
Public Databases Analyses
The Oncomine database (http://www.oncomine.org) is an oncogene database that provides different data sets for meta-analysis. We used the Oncomine database to evaluate the gene expression of SLC39As.17 TIMER database (https://cistrome.shinyapps.io/timer/) was used to assess the expression levels of SLC39A1 in pan-cancer.18,19 HCCDB, an HCC expression atlas database, collects 15 HCC expression datasets (approximately 4000 samples) to be used publicly for analysis. We used HCCDB to analyze the expression pattern of SLC39A1 in different HCC datasets.20 The sequencing data of mRNA and the clinical data of HCC (including 50 normal and 374 tumor tissues) were collected from The Cancer Genome Atlas (TCGA) for further analysis. MethSurv (https://biit.cs.ut.ee/methsurv/), an interactive portal for the evaluation of prognosis based on DNA methylation status, was used to analyze the DNA methylation sites of SLC39A1 and its prognostic value in HCC.21 Finally, the open access portal, cBioportal database (http://cbioportal.org), which provides comprehensive cancer genomics datasets analysis, was used to analyze the altered expression of SLC39A1 and its effect on the prognosis of HCC patients.22
Correlation Analysis of SLC39A1 Expression and Clinicopathological Features in HCC
The relationship between the clinicopathological characteristics of HCC patients and SLC39A1 expression was analyzed using Student’s t-test or the Wilcoxon signed-rank sum test using the ggplot2 package. Survminer and survival packages were used for survival and Cox analyses, and the time ROC package was used to draw the receiver operating characteristic curve (ROC).
Functional Enrichment Analysis
The DESeq2 package was used to identify the differentially expressed genes (DEGs) in the two groups of patients having high and low SLC39A1 expression patterns.23 Genes with absolute fold changes (FC) larger than 1.5 and adjusted P value < 0.05 were screened for further analysis. The ClusterProfiler package was used for Gene Ontology (GO) enrichment analysis of SLC39A1-related DEGs.24 The functions with adjusted P values < 0.05, and FDR < 0.2 indicated significance. GSEA was used to investigate the biological pathways mediated by high SLC39A1 expression.25 The enriched pathways with adjusted P value < 0.05, and FDR < 0.25 indicated significance after 1000 permutations. The R package ClusterProfiler was used for statistical analysis and graphical plotting.
Immune Infiltration Analysis
Single-sample gene set enrichment analysis (ssGSEA) from the gene set variation analysis (GSVA) package was used to quantify the relative infiltration levels of 24 immune cells.26 The Spearman correlation analysis and Wilcoxon rank sum test were used to assess the immune infiltration associated with SLC39A1 expression in HCC.
Clinical Specimens
Fresh-frozen HCC tissues and paired normal adjacent tissues of 60 HCC patients were collected after hepatectomy at the Sun Yat-sen Memorial Hospital between 2016 and 2019. None of the patients received any therapy such as chemotherapy or radiotherapy prior to surgery. After surgical resection, all tissues were immediately stored at −80 °C or liquid nitrogen until RNA extraction. This study was approved by the Medical Ethics Committee of Sun Yat-sen Memorial Hospital, and written informed consent was obtained from all participants before participation in the study. The clinical information of the participants is summarized in Supplementary Table S1.
Cell Culture and Plasmid Transfection
Human Huh7 cells, obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco BRL, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS; Procell Life Science & Technology Co., Ltd., Wuhan, China) in an incubator with 5% CO2 at 37 °C. Plasmids containing the negative control sequence (shNC) or the SLC39A1 knockdown sequence (sh#1-3) were obtained from Kidan Biosciences (Guangzhou, China). Briefly, cells were seeded in 6-well plates, cultured to reach an appropriate confluence, and then transfected with different plasmids using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. After 48 h of incubation, transfected cells were harvested to be used for quantitative real-time polymerase chain reaction (qRT-PCR), cell function experiments, and Western blot analysis. SLC39A1 knockdown sequences were as follows: sh#1, 5’-GGGTATGGACAGGTCATAAACCTCGAGGTTTATGACCTGTCCATACCC-3’; sh#2, 5’-CCTGTTGGTCCTAGCCCATTTCTCGAGAAATGGGCTAGGACCAACAGG-3’; sh#3, 5’-GCAGATCACACTGGCTTACAACTCGAGTTGTAAGCCAGTGTGATCTGC-3’.
RNA Extraction and qRT-PCR
Total RNA was isolated from tissues and cell lines using the EZ-press RNA Purification Kit (EZBioscience, Roseville, USA) and then was reverse-transcribed to cDNA with Evo M-MLV RT Premix for qPCR (Accurate Biotechnology Co., Ltd., Hunan, China). qRT-PCR was performed to evaluate the SLC39A1 expression levels in cell lines and tissues using the 2−ΔΔCT method. The following primers were used: SLC39A1, 5’-GAACAAGAGATGGTCAAGTC-3’ (forward), and 5’-ATGTGAGCCTGTCCTTATG-3’ (reverse); actin, 5’-CTACCTCATGAAGATCCTCACCGA-3’ (forward), and 5’-TTCTCCTTAATGTCACGCACGATT-3’ (reverse).
Western Blot
Radioimmunoprecipitation assay (RIPA) lysis buffer (CWBIO, Beijing, China) with phosphatase and protease inhibitors (CWBIO, Beijing, China) was used to lyse cultured cells, followed by centrifugation at 14,000 rpm for 30 min at 4°C. A bicinchoninic acid assay (CWBIO, Beijing, China) was used to measure protein concentrations. After proteins were separated on SDS–PAGE gels (EpiZyme, Shanghai, China), the measured proteins were transferred onto PVDF membranes (Roche Applied Science, Germany). Appropriate antibodies were then used to carry out immunoblot analysis of all related proteins. Finally, bands were visualized using an Omni-ECLTM Femto Light Chemiluminescence Kit (EpiZyme, Shanghai, China) with a G: BOX Chemi XT4 imager (Syngene, UK). The antibodies for the relative proteins mentioned in this study are listed in Supplementary Table S2.
Cell Counting Kit-8 (CCK-8) Assay
The Cell Counting Kit-8 (APExBIO, Houston, Texas, USA) was used to examine the cellular viability. Briefly, Huh7 cells transfected with different plasmids (shNC, sh#1, sh#2, and sh#3) were seeded in 96-well plates at a density of 2×103 cells/well. At days 1–6, 10 µL CCK-8 reagent were mixed with 100 µL DMEM, and the mixed solution was added to each well and re-incubated for 2 h, followed by measurement of the absorbance at a wavelength of 450 nm using a microplate reader (Thermo MK3, Thermo Fisher Scientific, USA).
5-Ethynyl-20-Deoxyuridine (EdU) Assay
EdU Imaging Kits (Cy3) (APExBIO, Houston, Texas, USA) were used to evaluate the cellular proliferation. Briefly, Huh7 cells were seeded into 24-well plates at a density of 7×104 cells/well and incubated for 24 h. After re-incubation with the EdU reagent for 2h, the cells were fixed and stained for 30 min. Nucleic acid was stained with Hoechst 33342. Representative images were obtained using a fluorescence microscope.
Transwell Assay
Transfected cells were seeded into each upper transwell chamber (Corning, NY, USA) with or without Matrigel (Corning, NY, USA) at a density of 5×104 cells/well and incubated in 200 µL serum-free DMEM. The lower chambers contained 500 µL DMEM supplemented with 100 µL FBS. Then, cells from the upper compartments were removed after 48 h, and the migrated cells were fixed with paraformaldehyde for 15 min, followed by staining with 0.3% crystal violet for 10 min at room temperature. Cell counting was performed using a microscope.
Statistical Analysis
All experimental data were analyzed using SPSS software (version 25; Chicago, IL, USA) and GraphPad Prism software (version 8.0; San Diego, CA, USA). All bioinformatics statistical analyses were conducted using R software version 4.0.5 (https://www.r-project.org/). Statistical significance was defined as P < 0.05, unless otherwise specified.
Results
SLC39A1 Was Upregulated in HCC
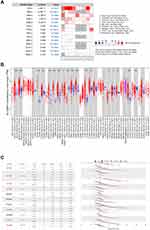
SLC39As expression in HCC was evaluated using the Oncomine database. Results indicated that SLC39A1 expression was notably upregulated in HCC tissues compared with that in adjacent normal tissues (Figure 1A). SLC39A1 pan-cancer expression was investigated using the TIMER database. As shown in Figure 1B, SLC39A1 expression levels were markedly elevated in most cancers, including stomach adenocarcinoma, hepatocellular carcinoma, cholangiocarcinoma, esophageal carcinoma, rectum adenocarcinoma, colon adenocarcinoma, glioblastoma multiforme, bladder urothelial carcinoma, and breast invasive carcinoma. However, SLC39A1 was markedly downregulated in pheochromocytoma, paraganglioma, thyroid carcinoma, prostate adenocarcinoma, and kidney chromophobe. To further verify the overexpression of SLC39A1 in HCC, we assessed the mRNA levels of SLC39A1 in different HCC cohorts from the HCCDB database, which showed that SLC39A1 expression levels in HCC were also markedly upregulated compared to that in adjacent normal tissues (Figure 1C).
SLC39A1 Overexpression Was Associated with Unfavorable Clinical Outcomes in HCC
As shown in Table 1, high SLC39A1 expression was markedly correlated with poor clinicopathological characteristics such as TNM stage, histological grade, and alpha fetoprotein (AFP) levels. HCC patients with a higher TNM stage (P = 0.014), advanced histologic grades (P < 0.001), and higher AFP levels (P < 0.001) had higher expression levels of SLC39A1 (Figure 2A–C). In addition, Kaplan–Meier survival analysis demonstrated that HCC patients with lower SLC39A1 expression had a superior overall survival (OS), progression-free survival and disease specific survival time when compared with those having higher SLC39A1 expression (Figure 2D–F). ROC analysis indicated that the area under curves (AUC) of SLC39A1 expression for 1-, 3-, and 5-year OS were 0.753, 0.636, and 0.646, respectively, suggesting a reliable prognostic predictive discrimination for SLC39A1 (Figure 2G). Univariate and multivariate Cox analyses indicated that SLC39A1 expression was an independent unfavorable prognostic biomarker for HCC patients (Figure 3A and B).
 |
Table 1 Correlation Between SLC39A1 Expression and Clinicopathological Features of HCC Patients from TCGA |
Prognostic Value of SLC39A1 DNA Methylation and Genetic Alteration in HCC
As shown in Figure 4A, nine CpG methylated sites were found in SLC39A1, among which four were identified as unfavorable prognostic factors (cg16414271, HR = 1.735, P = 0.0047; cg10624480, HR = 1.556, P = 0.033; cg14747580, HR = 1.542, P = 0.026; and cg11411904, HR = 1.509, P = 0.049), and one was identified as a favorable prognostic factor (cg09079613, HR = 0.505, P = 0.00068; Figure 4B–F). In addition, Spearman correlation analysis demonstrated that the four CpGs of SLC39A1 were negatively correlated with SLC39A1 expression (cg11073883: Cor = −0.214, P < 0.001; cg08623548: Cor = −0.271, P < 0.001; cg09079613: Cor = −0.290, P < 0.001; cg0546481: Cor = −0.143, P = 0.006; Figure S1a–d). We further investigated the prognostic value of SLC39A1 genetic alterations in HCC. Results indicated that approximately 13% of HCC patients had SLC39A1 genetic alterations, most of which were amplified. The most common nucleotide change was C > T transversion (Figure 4G). SLC39A1 genetic alterations in HCC patients resulted in a poor OS (P < 0.05; Figure 4H).
Functional Enrichment Analysis of SLC39A1 Expression in HCC
To further elucidate the potential mechanism of SLC39A1 in HCC, GO enrichment analysis and GSEA were conducted. GO enrichment analysis results indicated that SLC39A1 might have a regulatory effect on various cellular components and molecular functions, including the catenin complex, transmembrane transporter complex, serine-type peptidase activity, DNA-binding transcriptional activator activity, RNA polymerase II-specific, metal ion transmembrane transporter activity, and receptor ligand activity. As predicted, biological processes (BP) associated with zinc regulation were significantly enriched in the high SLC39A1 group, including cellular zinc ion homeostasis, cellular response to zinc ions, zinc ion homeostasis, and response to zinc ions (Figure 5A). In addition, GSEA demonstrated that SLC39A1 overexpression may be involved in many tumor progression-related pathways, such as the Wnt signaling pathway, MAPK signaling pathway, cell cycle, DNA replication, NOD-like receptor signaling pathway, and TGF-β signaling pathway (Figure 5B–G). These results suggest that SLC39A1 overexpression promotes HCC tumorigenesis and metastasis.
Correlation Between Immune Infiltration and SLC39A1 Expression
We performed ssGSEA to investigate the potential association between SLC39A1 expression levels and the infiltration levels of 24 types of immune cells in HCC. Results revealed that Th2 cells were positively associated with SLC39A1 expression (Cor > 0.3, P < 0.05), while cytotoxic cells were negatively associated with SLC39A1 expression (Cor < −0.3, P < 0.05; Figure 6A–C). Tumors having higher SLC39A1 expression exhibited higher infiltration levels of Th2 cells but lower infiltration levels of cytotoxic cells (Figure 6D and E). These findings indicate that SLC39A1 overexpression may be involved in the immune suppression of HCC.
Validation of SLC39A1 Overexpression in HCC in the Study’s Cohort
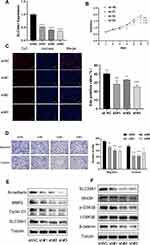
Given that SLC39A1 is upregulated in multiple HCC databases, qRT-PCR experiments and genomic sequencing were performed to validate the overexpression of SLC39A1 in tumor tissues. As shown in Figure 7A, SLC39A1 mRNA levels were significantly overexpressed in 60 HCC tissues compared to their paired adjacent non-tumorous tissues (P < 0.0001). The 60 patients were then stratified into three groups; over-expression, down-expression, and no-change under the thresholds of P < 0.05, and FC > 1.5 or < 0.67. Among the 60 HCC samples from our center, 38 (63%) exhibited SLC39A1 upregulation in tumor tissues, while six (10%) exhibited SLC39A1 downregulation in tumor tissues (Figure 7B and C). Furthermore, genomic sequencing of 20 pairs of HCC samples showed that SLC39A1was significantly overexpressed in tumor tissues than in paired adjacent non-tumorous tissues (P < 0.0001; Figure 7D).
SLC39A1 Promotes HCC Cellular Proliferation, Migration, and Invasion via the Wnt/β-Catenin Pathway
To further determine the underlying biological mechanism of SLC39A1 in HCC, Huh7 cells were transfected with sh-SLC39A1 plasmid. A significant reduction in SLC39A1 expression was observed in the shRNA group (P < 0.0001) (Figure 8A). Using CCK-8 and EdU assays, knockdown of SLC39A1 significantly suppressed the Huh7 cellular proliferation (Figure 8B and C). Transwell assays demonstrated that the cellular migration and invasion abilities were significantly inhibited in the sh-SLC39A1 group compared to those in the shNC group (Figure 8D). In addition, upon knocking down SLC39A1, a significant reduction was observed in the protein levels of cyclin D1, N-cadherin, and MMP2, which contribute to tumor proliferation, migration, and invasion (Figure 8E). Furthermore, Wnt/β-catenin signaling pathway-related proteins, including Wnt3A, phosphorylation-GSK3β (p-GSK3β), and β-catenin, were also markedly decreased in the sh-SLC39A1 groups (Figure 8F). These findings preliminarily illustrate that cellular proliferation, migration, and invasion of HCC are markedly suppressed by SLC39A1 knockdown via the Wnt/β-catenin pathway.
Discussion
Multiple zinc transporters in the SLC39A family have been demonstrated to mediate the dysregulation of zinc accumulation in tumor cells and are associated with the malignant phenotype of multiple cancers, such as gastric, lung, and breast cancers.27–29 The current study revealed that SLC39A1, one of the SLC39A family members, exerts a pro-tumorigenic effect in HCC. Results also showed that SLC39A1 overexpression was notably observed in HCC tissues and correlated with advanced TNM stage, histologic differentiation, AFP levels, and poor prognosis of HCC patients. This suggests an unfavorable predictive prognostic role of SLC39A1 in HCC.
GSEA analysis demonstrated that various oncogenic BP were mediated by SLC39A1 overexpression, such as cell cycle, DNA replication and Wnt signaling pathway, which contribute to tumor proliferation and metastasis.30–35 This was also evidenced by the fact that SLC39A1 downregulation notably repressed HCC cellular proliferation, migration, and invasion. Furthermore, SLC39A1 knockdown also decreased the expression of cyclin D1, N-cadherin, and MMP2, which are critical for the cellular proliferation and metastasis of the tumor.36–41 Of note, downregulation of some members of Wnt signaling pathway was also observed upon knocking down SLC39A1, indicating the potential role of SLC39A1 in the Wnt signaling pathway. Taken together, these findings suggest a critical pro-tumor effect of SLC39A1 overexpression on HCC tumor proliferation and metastasis. However, in-depth experimental studies are needed to elucidate the underlying mechanisms of SLC39A1 in hepatocellular carcinogenesis.
Reduced cellular zinc content affects the balance between Th1 and Th2 cytokine shifts and the activation of many immune cells.42,43 This study also demonstrated the specific effect of SCL39A1 on the tumor immune infiltration of HCC. Higher SLC39A1 expression was significantly associated with higher infiltration of Th2 cells but lower infiltration of cytotoxic cells. Th2 cells have been widely considered as tumor-promoting immune cells, leading to unfavorable clinical outcomes in HCC.44,45 Higher levels of Th2 cytokines (eg, interleukin [IL]-4 and IL-10) were observed in metastatic HCC, whereas Th1 cytokines (eg, interferon [IFN]-γ and IL-1) were notably decreased.46,47 IL-4 and IL-10 have been reported to restrain IFN-γ production and impair the cytotoxic cell-mediated anti-tumor immunity,48–51 suggesting that an increased shift from Th1 cells to Th2 cells promotes tumor progression and immunosuppression. In addition, aberrant Wnt signaling pathway in the tumor microenvironment also impairs the cytotoxic T cell infiltration and anti-tumor function, inducing therapeutic resistance to immune checkpoint inhibitors.33,52–54 Therefore, blocking the Wnt signaling pathway through targeting SLC39A1 may be a promising therapeutic strategy to improve the current HCC immunotherapeutic approaches. Further studies should be performed to elucidate the underlying mechanism of SLC39A1 overexpression in immune cell infiltration in HCC.
The results described in this study may provide new insights into the malignant phenotype and the modulation of immune infiltration in HCC. However, this study had several limitations. First, the findings of this study were dependent on the quality of the data retrieved from the publicly-available databases. Second, further in vivo studies are required to explore and validate the molecular mechanisms of SLC39A1 in HCC.
Conclusions
In conclusion, this study identified SLC39A1 as an unfavorable prognostic biomarker for HCC. SLC39A1 overexpression may promote tumor progression and pro-tumor immunity via the Wnt/β-catenin signaling pathway, which may be a novel therapeutic approach in HCC.
Data Sharing Statement
The original contributions presented in the study are included in the article and Supplementary Materials. Further inquiries can be directed to the corresponding authors, Yajin Chen, [email protected] and Changzhen Shang, [email protected].
Ethical Standards
The study was reviewed and approved by the Ethics Committee of Sun Yat-sen Memorial Hospital. The patients/participants provided their written informed consent to participate in this study. The study was completed in accordance with the Declaration of Helsinki.
Acknowledgments
The authors would like to give their sincere appreciation to the reviewers for their helpful comments on this article and research groups for the TCGA, which provided data for this collection.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work. Xiaowu Ma and Hongkai Zhuang contributed equally to this manuscript. Co-corresponding authors: Yajin Chen and Changzhen Shang.
Funding
This work was supported by grants from the National Natural Science Foundation of China (nos. 81972263, 82072714 and 82103221), the program of Guangdong Provincial Clinical Research Center for Digestive Diseases (2020B1111170004) and China Postdoctoral Science Foundation (2020M683094).
Disclosure
All authors declare that they have no conflicts of interest for this work.
References
1. Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi:10.3322/caac.21660
2. Villanueva A, Longo DL. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–1462. doi:10.1056/NEJMra1713263
3. Finn RS, Qin S, Ikeda M, et al. Atezolizumab plus bevacizumab in unresectable hepatocellular carcinoma. N Engl J Med. 2020;382(20):1894–1905. doi:10.1056/NEJMoa1915745
4. Ren Z, Xu J, Bai Y, et al. Sintilimab plus a bevacizumab biosimilar (IBI305) versus sorafenib in unresectable hepatocellular carcinoma (ORIENT-32): a randomised, open-label, phase 2–3 study. Lancet Oncol. 2021;22(7):977–990. doi:10.1016/s1470-2045(21)00252-7
5. Giraud J, Chalopin D, Blanc JF, Saleh M. Hepatocellular carcinoma immune landscape and the potential of immunotherapies. Front Immunol. 2021;12:655697. doi:10.3389/fimmu.2021.655697
6. Schweigel-Rontgen M. The families of zinc (SLC30 and SLC39) and copper (SLC31) transporters. Curr Top Membr. 2014;73:321–355. doi:10.1016/B978-0-12-800223-0.00009-8
7. Jeong J, Eide DJ. The SLC39 family of zinc transporters. Mol Aspects Med. 2013;34(2–3):612–619. doi:10.1016/j.mam.2012.05.011
8. Wang J, Zhao H, Xu Z, Cheng X. Zinc dysregulation in cancers and its potential as a therapeutic target. Cancer Biol Med. 2020;17(3):612–625. doi:10.20892/j.issn.2095-3941.2020.0106
9. Wu DM, Liu T, Deng SH, Han R, Xu Y. SLC39A4 expression is associated with enhanced cell migration, cisplatin resistance, and poor survival in non-small cell lung cancer. Sci Rep. 2017;7(1):7211. doi:10.1038/s41598-017-07830-4
10. Liu M, Yang J, Zhang Y, et al. ZIP4 promotes pancreatic cancer progression by repressing ZO-1 and claudin-1 through a ZEB1-dependent transcriptional mechanism. Clin Cancer Res. 2018;24(13):3186–3196. doi:10.1158/1078-0432.CCR-18-0263
11. Liu L, Hou Y, Hu J, et al. SLC39A8/zinc suppresses the progression of clear cell renal cell carcinoma. Front Oncol. 2021;11:651921. doi:10.3389/fonc.2021.651921
12. Wei Y, Dong J, Li F, Wei Z, Tian Y. Knockdown of SLC39A7 suppresses cell proliferation, migration and invasion in cervical cancer. EXCLI J. 2017;16:1165–1176. doi:10.17179/excli2017-690
13. Kambe T, Taylor KM, Fu D. Zinc transporters and their functional integration in mammalian cells. J Biol Chem. 2021;296:100320. doi:10.1016/j.jbc.2021.100320
14. Wang P, Zhang J, He S, Xiao B, Peng X. SLC39A1 contribute to malignant progression and have clinical prognostic impact in gliomas. Cancer Cell Int. 2020;20(1):573. doi:10.1186/s12935-020-01675-0
15. Golovine K, Makhov P, Uzzo RG, Shaw T, Kunkle D, Kolenko VM. Overexpression of the zinc uptake transporter hZIP1 inhibits nuclear factor-kappaB and reduces the malignant potential of prostate cancer cells in vitro and in vivo. Clin Cancer Res. 2008;14(17):5376–5384. doi:10.1158/1078-0432.CCR-08-0455
16. Franklin RB, Feng P, Milon B, et al. hZIP1 zinc uptake transporter down regulation and zinc depletion in prostate cancer. Mol Cancer. 2005;4:32. doi:10.1186/1476-4598-4-32
17. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi:10.1593/neo.07112
18. Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi:10.1186/s13059-016-1028-7
19. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi:10.1158/0008-5472.CAN-17-0307
20. Lian Q, Wang S, Zhang G, et al. HCCDB: a database of hepatocellular carcinoma expression atlas. Genomics Proteomics Bioinformatics. 2018;16(4):269–275. doi:10.1016/j.gpb.2018.07.003
21. Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10(3):277–288. doi:10.2217/epi-2017-0118
22. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi:10.1126/scisignal.2004088
23. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi:10.1186/s13059-014-0550-8
24. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi:10.1089/omi.2011.0118
25. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
26. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. doi:10.1186/1471-2105-14-7
27. Liu L, Yang J, Wang C. Analysis of the prognostic significance of solute carrier (SLC) family 39 genes in breast cancer. Biosci Rep. 2020;40(8). doi:10.1042/BSR20200764
28. Ding B, Lou W, Xu L, Li R, Fan W. Analysis the prognostic values of solute carrier (SLC) family 39 genes in gastric cancer. Am J Transl Res. 2019;11(1):486–498.
29. Zhou H, Zhu Y, Qi H, et al. Evaluation of the prognostic values of solute carrier (SLC) family 39 genes for patients with lung adenocarcinoma. Aging (Albany NY). 2021;13(4):5312–5331. doi:10.18632/aging.202452
30. Williams GH, Stoeber K. The cell cycle and cancer. J Pathol. 2012;226(2):352–364. doi:10.1002/path.3022
31. Boyer AS, Walter D, Sorensen CS. DNA replication and cancer: from dysfunctional replication origin activities to therapeutic opportunities. Semin Cancer Biol. 2016;37–38:16–25. doi:10.1016/j.semcancer.2016.01.001
32. Icard P, Fournel L, Wu Z, Alifano M, Lincet H. Interconnection between metabolism and cell cycle in cancer. Trends Biochem Sci. 2019;44(6):490–501. doi:10.1016/j.tibs.2018.12.007
33. Goldsberry WN, Londono A, Randall TD, Norian LA, Arend RC. A review of the role of Wnt in cancer immunomodulation. Cancers (Basel). 2019;11(6):771. doi:10.3390/cancers11060771
34. Zhan T, Rindtorff N, Boutros M. Wnt signaling in cancer. Oncogene. 2017;36(11):1461–1473. doi:10.1038/onc.2016.304
35. Perugorria MJ, Olaizola P, Labiano I, et al. Wnt-beta-catenin signalling in liver development, health and disease. Nat Rev Gastroenterol Hepatol. 2019;16(2):121–136. doi:10.1038/s41575-018-0075-9
36. Cao ZQ, Wang Z, Leng P. Aberrant N-cadherin expression in cancer. Biomed Pharmacother. 2019;118:109320. doi:10.1016/j.biopha.2019.109320
37. Liao S, Chen H, Liu M, et al. Aquaporin 9 inhibits growth and metastasis of hepatocellular carcinoma cells via Wnt/beta-catenin pathway. Aging (Albany NY). 2020;12(2):1527–1544. doi:10.18632/aging.102698
38. Liu D, Kang H, Gao M, et al. Exosome-transmitted circ_MMP2 promotes hepatocellular carcinoma metastasis by upregulating MMP2. Mol Oncol. 2020;14(6):1365–1380. doi:10.1002/1878-0261.12637
39. Tong H, Liu X, Li T, et al. NR1D2 accelerates hepatocellular carcinoma progression by driving the epithelial-to-mesenchymal transition. Onco Targets Ther. 2020;13:3931–3942. doi:10.2147/OTT.S237804
40. Kaszak I, Witkowska-Pilaszewicz O, Niewiadomska Z, Dworecka-Kaszak B, Ngosa Toka F, Jurka P. Role of cadherins in cancer-a review. Int J Mol Sci. 2020;21(20):7624. doi:10.3390/ijms21207624
41. Gonzalez-Avila G, Sommer B, Mendoza-Posada DA, Ramos C, Garcia-Hernandez AA, Falfan-Valencia R. Matrix metalloproteinases participation in the metastatic process and their diagnostic and therapeutic applications in cancer. Crit Rev Oncol Hematol. 2019;137:57–83. doi:10.1016/j.critrevonc.2019.02.010
42. Prasad AS. Effects of zinc deficiency on Th1 and Th2 cytokine shifts. J Infect Dis. 2000;182(Suppl 1):S62–68. doi:10.1086/315916
43. Skrajnowska D, Bobrowska-Korczak B. Role of zinc in immune system and anti-cancer defense mechanisms. Nutrients. 2019;11(10):2273. doi:10.3390/nu11102273
44. Lee HL, Jang JW, Lee SW, et al. Inflammatory cytokines and change of Th1/Th2 balance as prognostic indicators for hepatocellular carcinoma in patients treated with transarterial chemoembolization. Sci Rep. 2019;9(1):3260. doi:10.1038/s41598-019-40078-8
45. Kogame M, Nagai H, Shinohara M, Igarashi Y, Sumino Y, Ishii K. Th2 dominance might induce carcinogenesis in patients with HCV-related liver cirrhosis. Anticancer Res. 2016;36(9):4529–4536. doi:10.21873/anticanres.11000
46. Saxena R, Kaur J. Th1/Th2 cytokines and their genotypes as predictors of hepatitis B virus related hepatocellular carcinoma. World J Hepatol. 2015;7(11):1572–1580. doi:10.4254/wjh.v7.i11.1572
47. Wu MY, Yiang GT, Cheng PW, Chu PY, Li CJ. Molecular targets in hepatocarcinogenesis and implications for therapy. J Clin Med. 2018;7(8). doi:10.3390/jcm7080213
48. Mao JX, Guo WY, Guo M, Liu C, Teng F, Ding GS. Acute rejection after liver transplantation is less common, but predicts better prognosis in HBV-related hepatocellular carcinoma patients. Hepatol Int. 2020;14(3):347–361. doi:10.1007/s12072-020-10022-4
49. Zhuang H, Zhou Z, Zhang Z, et al. B3GNT3 overexpression promotes tumor progression and inhibits infiltration of CD8(+) T cells in pancreatic cancer. Aging (Albany NY). 2020;13(2):2310–2329. doi:10.18632/aging.202255
50. Yi Y, He HW, Wang JX, et al. The functional impairment of HCC-infiltrating gammadelta T cells, partially mediated by regulatory T cells in a TGFbeta- and IL-10-dependent manner. J Hepatol. 2013;58(5):977–983. doi:10.1016/j.jhep.2012.12.015
51. Wang WC, Zhang ZQ, Li PP, et al. Anti-tumor activity and mechanism of oligoclonal hepatocellular carcinoma tumor-infiltrating lymphocytes in vivo and in vitro. Cancer Biol Ther. 2019;20(9):1187–1194. doi:10.1080/15384047.2019.1599663
52. Yang Y, Ye YC, Chen Y, et al. Crosstalk between hepatic tumor cells and macrophages via Wnt/beta-catenin signaling promotes M2-like macrophage polarization and reinforces tumor malignant behaviors. Cell Death Dis. 2018;9(8):793. doi:10.1038/s41419-018-0818-0
53. Kinosada H, Okada-Iwasaki R, Kunieda K, et al. The dual pocket binding novel tankyrase inhibitor K-476 enhances the efficacy of immune checkpoint inhibitor by attracting CD8(+) T cells to tumors. Am J Cancer Res. 2021;11(1):264–276.
54. Spranger S, Bao R, Gajewski TF. Melanoma-intrinsic beta-catenin signalling prevents anti-tumour immunity. Nature. 2015;523(7559):231–235. doi:10.1038/nature14404
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.