Back to Journals » OncoTargets and Therapy » Volume 9
Single-nucleotide polymorphism in microRNA-binding site of SULF1 target gene as a protective factor against the susceptibility to breast cancer: a case-control study
Authors Zhou Q, Jiang Y, Yin W, Wang Y, Lu J
Received 21 December 2015
Accepted for publication 15 March 2016
Published 9 May 2016 Volume 2016:9 Pages 2749—2757
DOI https://doi.org/10.2147/OTT.S102433
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Min Li
Qiong Zhou,1–3 Yiwei Jiang,1,2,4 Wenjin Yin,1,2 Yaohui Wang,1,2,4 Jinsong Lu4
1Department of Breast Surgery, Fudan University Shanghai Cancer Center, 2Department of Oncology, Shanghai Medical College, Fudan University, Shanghai, 3Department of Gynecology, Zhejiang Cancer Hospital, Hangzhou, 4Breast Cancer Center, Renji Hospital, School of Medicine, Shanghai Jiaotong University, Shanghai, People’s Republic of China
Purpose: Numerous clinical studies have suggested that chemopreventive drugs for breast cancer such as tamoxifen and exemestane can effectively reduce the incidence of estrogen receptor (ER)-positive breast cancer. However, it remains unclear how to identify those who are susceptible to ER-positive breast cancer. Accordingly, there is a great demand for a probe into the predisposing factors so as to provide precise chemoprevention. Recent evidence has indicated that ERα expression can be regulated by microRNAs (miRNAs), such as miR-206, in breast cancer. We assumed that single-nucleotide polymorphisms (SNPs) in the miR-206-binding sites of the target genes may be associated with breast cancer susceptibility with different ER statuses.
Methods: We genotyped the SNPs that reside in and around the miR-206-binding sites of two target genes – heparan sulfatase 1 (SULF1) and RPTOR-independent companion of mammalian target of rapamycin Complex 2 (RICTOR) – which were related to the progression or metastasis of breast cancer cells in 710 breast cancer patients and 294 controls by the matrix-assisted laser desorption ionization-time of flight mass spectrometry method. Modified odds ratios (ORs) with their 95% confidence intervals (CIs) were calculated by a multivariate logistic regression analysis to evaluate the potential association between the SNPs and breast cancer susceptibility.
Results: For rs3802278, which is located in the 3'-untranslated region (3'-UTR) of SULF1, the frequency of the AA genotype was less in breast cancer patients than that in the controls as compared to that of the GG + GA genotype not only for ER-positive breast cancer patients (adjusted OR =0.663, P=0.032) but also for hormone receptor-positive breast cancer patients (adjusted OR =0.610, P=0.018). Besides, the frequency of the AA genotype was less than that of the GG genotype between the ER-positive breast cancer patients and the controls (adjusted OR =0.791, P=0.038). For rs66916453, which is located in the 3'-UTR of RICTOR, no significant difference was observed between the case and the control group for the genotypes or alleles (P>0.05).
Conclusion: The SNPs in the miRNA-binding sites within the 3'-UTR of SULF1 may serve as protective factors against the susceptibility to breast cancer, especially to ER-positive breast cancer in the Chinese population. These SNPs are promising candidate biomarkers to predict the susceptibility of breast cancer and guide the administration of targeted preventive endocrine therapy.
Keywords: breast cancer susceptibility, miRNA, single-nucleotide polymorphism, SULF1
Introduction
Early in 1976, women who used exogenous estrogen were found to have a higher incidence of breast cancer.1 Subsequently, an increasing number of case-control and prospective studies reported that hormone replacement therapy increased the incidence and mortality of breast cancer.2–5 Estrogen receptor α (ERα) is crucial for the estrogen-dependent growth of breast cancer; its expression is essential for the prognosis of breast cancer patients and their responses to endocrine therapy. Numerous studies have proven that the higher the level of ERα expression in tumor cells, the greater the response to endocrine therapy.6 The human ERα gene (ESR1) is controlled at the transcriptional level by many different cofactors.7 Over the past few years, manifold genetic or epigenetic events, such as mutations of open reading frame8 and deoxyribonucleic acid (DNA) methylation of CpG islands,9 have been found to be involved in the complex mechanism that regulates ESR1 gene expression in breast cancer. An increasing amount of evidence indicates that other epigenetic changes, including microRNA (miRNA) networks, may also contribute to ESR1 regulation.
MiRNAs are small noncoding RNAs that control gene expression at the translational level by targeting specific messenger RNAs (target mRNAs). Mature miRNAs become part of the RNA-induced silencing complex, which recognizes the specific sites in the 3′-untranslated region (3′-UTR) of target mRNAs and induces translational repression or mRNA cleavage.10 miRNAs are novel factors for gene regulation; their functions have not been completely recognized but are considered to serve an important role in the regulation of many biological processes, such as cellular proliferation, differentiation, and apoptosis. Studies have shown that miRNA mutations or incorrect expressions are correlated with various human cancers and that miRNAs may function as oncogenes or tumor suppressors.11 miRNA expression profiling also revealed that certain miRNAs are differentially expressed among breast cancer subtypes.12,13 Recent studies have shown that specific miRNAs may regulate ERα-mediated signaling, thereby influencing metastasis and survival in breast cancer.14–17
Several miRNAs, such as microRNA-206 (miR-206), have been reported to target ERα, repress ERα mRNA and protein expression, and inhibit estrogen-dependent growth in breast cancer cell lines. In addition, the expression of miR-206 is regulated by 17α-estradiol (E2) and ERα-selective agonist in a double-negative feedback loop.18 The expression level of miR-206 is higher in ERα-negative human breast tumor specimens than that in ERα-positive ones, which suggests that the regulation of miR-206 may have an impact on the transition between the ERα-positive phenotype and the ERα-negative phenotype.19 Thus, the interrelationship between miR-206 and ERα may be crucial to the development of breast cancer with different ERα statuses. However, the logic behind it is still open to investigation.
The majority of miRNA-binding sites are located within the 3′-UTR of mRNAs, which produces the cleavage of target mRNAs or the suppression of their translation via base pairing.20 Therefore, genetic variants that reside in the miRNA gene or its binding sites of target mRNAs may alter the binding affinities, influence the interaction between miRNAs and target mRNAs, and ultimately change the expression of target genes. As the most frequent genetic variation in the human genome, the single-nucleotide polymorphisms (SNPs) in miRNA genes and their target sites may be promising candidate biomarkers for tumor formation and development. The SNPs that are located in miRNA-binding sites have been increasingly reported to impact the regulatory loop between miRNAs and their target genes21,22 or function as a genetic marker for cancer risk.23,24 Nevertheless, it still remains ambiguous which SNPs are functional and whether the SNPs can serve as biomarkers to assess the risk and prognosis of breast cancer. As several miRNAs, including miR-206, are closely related to ERα, we hypothesize that the variation in some gene-binding sites regulated by these miRNAs may be associated with the risk of breast cancer with different ERα statuses.
On these premises, we used miR-206 as a starting point for our study to select several potential target genes of miR-206, which may be correlated with carcinogenesis, progression, or metastasis of breast cancer on the basis of recent studies. Then, we performed a case-control study to demonstrate whether the SNPs located in the miRNA-binding sites within the 3′-UTR of selected target mRNAs had an effect on the susceptibility of ER-positive breast cancers. Several clinical studies have indicated that chemoprevention for breast cancer such as tamoxifen and exemestane may effectively reduce the incidence of ER-positive breast cancer.25–27 Therefore, it may be of great value to filter the women with high susceptibility to ER-positive breast cancer from the normal population, offering the implications for predictive factors for chemoprevention in breast cancer, as well as improving the precision and cost-effectiveness of breast cancer chemoprevention as a health care intervention.
Materials and methods
Study cohort
Between 2008 and 2011, 710 patients with breast cancer and 294 nonmalignant women were recruited as the cases and controls, respectively. The inclusion criteria were females aged between 18 years and 85 years. These breast cancer patients must be confirmed by histopathological examination of samples from core needle biopsy or open biopsy. Histological types were not limited. Participants who suffered from other malignancies or who reported a family history of other malignancies were excluded from the study. Written informed consent was obtained from each subject. For each case and control, a 5 mL of peripheral blood sample was collected and stored at −80°C. The study was approved by the ethics committee of Fudan University Shanghai Cancer Center (FDUCC).
Candidate gene loci
First, based on the predictions of TargetScan (http://www.targetscan.org/), miRanda (http://www.microrna.org/microrna/home.do), microcosm (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/), and PicTar (http://pictar.mdc-berlin.de/) networks and the review of literature, we selected the potential target genes of miR-206 that are related to malignant characteristics of breast cancer, such as proliferation, angiogenesis, invasion, metastasis, or inhibition of apoptosis. Second, we downloaded the SNPs in the 3′-UTR of the candidate genes from the National Center for Biotechnology Imformation (NCBI) database (http://www.ncbi.nlm.nih.gov/guide/all/). We screened the SNP loci that cover the extension of 2 kb at both sides of the miR-206-binding sites in the target genes with low binding free energy as candidates. Table 1 lists the characteristics of the candidate loci.
  | Table 1 Candidate target genes and the corresponding SNPs |
Polymerase chain reaction and SNP genotyping
The DNA was extracted from the peripheral blood samples of all subjects using the Qiagen DNA blood kit (Qiagen NV, Venlo, the Netherlands) according to the manufacturer’s protocols. SNP genotyping was performed at Shanghai Benegene Biotechnology Co., Ltd (Shanghai, People’s Republic of China), using the MassARRAY system (Sequenom, San Diego, CA, USA) by the matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry method. Primers for polymerase chain reaction (PCR) and single-base extension were designed by the Assay Designer software package (Sequenom). The sequences of the primers are listed in Table 2. PCR amplification was performed in a 5 μL reaction mixture that combined 5 ng DNA, 0.95 μL water, 0.625 μL of PCR buffer (containing 15 mM MgCl2), 1 μL of 2.5 mM deoxynucleotide (dNTP), 0.325 μL of 25 mM MgCl2, 1 μL of PCR primers, and 0.1 μL of 5 units/μL HotStar Taq (Qiagen NV). The reaction conditions were as follows: 94°C for 15 minutes, 45 cycles at 94°C for 20 seconds, 56°C for 30 seconds, and 72°C for 1 minute, followed by 3 minutes at 72°C. After the amplification, the remaining dNTPs were dephosphorylated by adding 1.53 μL of water, 0.17 μL of shrimp alkaline phosphatase (SAP) buffer, and 0.3 units of SAP (Sequenom). The reaction was placed at 37°C for 40 minutes, and the enzyme was deactivated by incubating at 85°C for 5 minutes. After SAP treatment, the single primer extension over the SNP was combined with 0.755 μL water, 0.2 μL of 10× iPLEX buffer, 0.2 μL of termination mix, 0.041 μL of iPLEX enzyme (Sequenom), and 0.804 μL of 10 μM extension primer. The single-base extension reaction was performed at 94°C for 30 seconds, 40 cycles at 94°C for 5 seconds, 52°C for 5 seconds, and 80°C for 5 seconds, followed by 72°C for 3 minutes. The reaction mix was desalted by adding 6 mg of cation exchange resin (Sequenom), mixed, and resuspended in 25 μL water. The completed genotyping reactions were spotted onto a 384-well spectroCHIP (Sequenom) using the MassARRAY Nanodispenser (Sequenom) and determined by the MALDI-TOF mass spectrometer. Genotype calling was performed in real time with the MassARRAY RT software Version 3.0.0.4 and analyzed using the MassARRAY Typer software Version 3.4 (Sequenom).
  | Table 2 Sequences of primers |
Statistical analysis
Both cases and controls were analyzed with a chi-squared test to determine whether they were in Hardy–Weinberg equilibrium (HWE) in order to exclude the possibility of experimental artifacts. Clinical parameters were compared between the two groups using a Student’s t-test for continuous variables and a chi-squared test for unordered categorical variables. The data for the age of disease onset were presented as a mean value ± SD. A one-way analysis of variance and a Student’s t-test for differences in age of disease onset were used among the three genotypes and between the two genotypes, respectively, for each SNP, and a general linear model was applied for the trend test of age. The modified odds ratios (ORs) with their 95% confidence intervals (CIs) were calculated by a multivariate logistic regression analysis to evaluate the potential association between the SNPs and breast cancer susceptibility. All statistical analyses were processed by Stata 12.0 (StataCorp LP, College Station, TX, USA).
Results
Characteristics of the participants
Between 2008 and 2011, 710 patients with breast cancer (mean age 49.68±9.27 years) and 294 non-malignant women (mean age 49.69±9.86 years) were recruited as the cases and controls, respectively. No statistically significant distribution difference was observed between the two groups in terms of age and menopause status. Table 3 lists the characteristics of the enrolled patients.
  | Table 3 Main characteristics of the enrolled patients |
Hardy–Weinberg equilibrium
The results of the HWE analysis for both cases and controls are shown in Table 4. When the P-value was >0.01, the cohort was considered in HWE. We discovered that both cases and controls of each SNP were in HWE (all P>0.01, Table 4), which suggests that the results of the study were reliable.
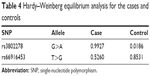  | Table 4 Hardy–Weinberg equilibrium analysis for the cases and controls |
Association between rs3802278 and breast cancer susceptibility
For the SNP rs3802278 located within the 3′-UTR of heparan sulfatase 1 (SULF1), the frequency of the AA genotype was less in the breast cancer patients than the controls (adjusted OR =0.663, P=0.032, Table 5) compared with that of the GG + GA genotype, which implies that the AA genotype shows a protective effect on breast cancer risk. Similar results were achieved not only in ER-positive breast cancer patients (adjusted OR =0.610, P=0.018, Table 6) but also in hormone receptor (HR)-positive (ER and/or progesterone receptor positive) breast cancer patients (adjusted OR =0.642, P=0.030, Table 7), which suggest that the AA genotype reveals a protective impact against the susceptibility to ER-positive and HR-positive breast cancers compared with the GG + GA genotype. Besides, the frequency of the AA genotype was less than that of the GG genotype between ER-positive breast cancer patients and controls (adjusted OR =0.791, P=0.038, Table 6), which indicates that the AA genotype presents a protective association against the risk of ER-positive breast cancer compared with the GG genotype.
Association between rs66916453 and breast cancer susceptibility
For rs66916453 located within the 3′-UTR of RPTOR-independent companion of mammalian target of rapamycin (mTOR) Complex 2 (RICTOR), no significant difference was observed between the case and the control group for genotypes or alleles (P>0.05, Table 8).
Association between SNP genotype and age of disease onset
For rs3802278, the AA genotype delayed the onset of disease as compared to the GG + GA genotype for breast cancer overall (P=0.046), ER-positive breast cancer (P=0.013), and HR-positive breast cancer (P=0.032). The trend test also showed an increasing age with the AA genotype in a dose-dependent manner (Table 9). However, we found no association with the age of breast cancer onset for rs66916453 (all P>0.05, Table 10).
Discussion
To the best of our knowledge, this study first focused on the relationship of rs3802278 (located in the 3′-UTR of SULF1 gene) with the susceptibility of breast cancer overall as well as ER-positive or HR-positive breast cancer, which may be regulated by miR-206 or other miRNAs.
HSULF1/SULF1 is a sulfatase that can selectively desulfate heparin sulfate proteoglycans. Heparin sulfate proteoglycans mediate the activation of tyrosine kinase receptors in numerous cell signaling pathways that are regulated by the heparin-binding growth factors and cytokines.28–30 Therefore, SULF1 serves an important role in the activation of many cell signaling pathways. Previous studies31–33 suggested that SULF1 was stably expressed in various normal tissues, whereas the expression level of SULF1 was downregulated in several tumor cells, including breast cancer, pancreatic cancer, renal cell carcinoma, and hepatocellular carcinoma cells. The reexpression of the SULF1 gene can inhibit the proliferation and migration of tumor cells as well as promote chemotherapy-induced apoptosis.34 Additionally, SULF1 suppressed the proliferation of MDA-MB-468 breast cancer cells and tumor angiogenesis of transplanted tumors in nude mice.31 A recent study showed that the lack of SULF1 expression enhanced the migration and invasion ability of MCF10DCIS breast cancer cells, and the elevated expression of SULF1 was associated with improved disease-free survival and overall survival of breast cancer patients.35
When it comes to the studies concerning the association between the SNP of SULF1 and breast cancer susceptibility, only one study has reported such findings for rs262347 to date.36 In that study, Okolicsanyi et al observed that rs2623047 in SULF1 was significantly associated with breast cancer risk. Despite different SNPs, both their study and ours focused on the same gene SULF1 with a similar conclusion, which indicates SULF1 as a potentially important gene in the development of breast cancer. However, disparities existed between their study and ours. Okolicsanyi et al conducted the genotyping by using the restriction fragment length polymorphism analysis instead of the MALDI-TOF applied in our study. On the other hand, their study failed to evaluate the association of various genotypes with the risk of developing both breast cancer and different subtypes.
Our data implied that the subjects with the GG + GA genotype of rs3802278 had a higher risk of breast cancer, which had never been published before. The SNP rs3802278 is located within the 3′-UTR of the SULF1 gene, which may be the target gene of miR-206 according to the prediction by several methods, such as TargetScan and miRanda. Meanwhile, it is of note that we screened SNP loci that cover the extension of 2 kb at both sides of the miR-206-binding sites in the target genes (rs3802278 located ~350 bp upstream of the miR-206 target sites in the SULF1 gene), which implies that SULF1 is likely the target gene of miR-206 and may also be regulated by other miRNAs. A portion of studies has revealed that SNPs in and around the miRNA-binding sites within 3′-UTR of the target mRNAs can affect miRNA-mediated regulatory function by changes in both the sequence and the secondary structure of mRNA induced by the SNPs, which may cause changes in the binding affinity and interaction between the miR-206 and the SULF1 genes.37–39 These changes may accordingly cause an altered regulatory effect of miR-206 or other miRNAs on the SULF1 mRNA level, then the protein expression, and finally the role of SULF1 in breast cancer risk. However, this issue merits further study, regarding whether and how miR-206 or other miRNAs function with SULF1.
For rs3802278, our data have also shown a protective effect of the AA genotype against the susceptibility to ER-positive and HR-positive breast cancers, which indicates that SULF1 may be involved in the regulation of ERα signaling pathway and has a potential role in the carcinogenesis of the ER-positive or HR-positive breast cancer. Meanwhile, miR-206 is reportedly associated with the ERα signaling pathway. However, evidence is lacking as to the network among miR-206, SULF1, and ERα.
Our findings also revealed that rs3802278 was associated with the age of breast cancer onset. Han et al40 observed a marginally significant association of rs3802278 with the age of ovarian cancer onset and a significant trend for a decreasing age with the A allele of rs3802278 in a dose-dependent manner, which was more or less consistent with our data. Unfortunately, there is little evidence for the potential mechanism underlying this effect, which hints at a demand for additional studies.
mTOR is a downstream protein of the phosphatidylinositol-3-kinase signaling pathway. When mTOR combines with the RICTOR, the mTOR–RICTOR complex (mTORC2) is formed. Several studies confirmed the importance of mTORC2 to the development of breast cancer;41–45 its blockade in breast cancer cells would significantly promote apoptosis and inhibit migration.46 However, our study failed to identify the association between genotypes and alleles of the SNP within RICTOR and breast cancer risk. Considering the complexity of the regulation process, the binding ability of certain miRNAs or their regulation effects on the target gene may not change even with different variants or a relationship with breast carcinogenesis may not exist.
The results of a randomized clinical trial (Breast Cancer Prevention Trial P-1) for breast cancer prevention, which was implemented by the National Surgical Adjuvant Breast and Bowel Project (NSABP), indicated that tamoxifen reduced the occurrence of ER-positive tumors by 69% when compared with the placebo.47 Similarly, a randomized, placebo-controlled clinical trial, which was named the National Cancer Institute of Canada Clinical Trials Group Mammary Prevention.3 trial (NCIC CTG MAP.3), demonstrated that exemestane can significantly reduce invasive breast cancers, particularly for ER-positive or progesterone receptor-positive breast cancers, in postmenopausal women with risk factors for breast cancer.25 Furthermore, similar results were also achieved in the International Breast Cancer Intervention Study-II (IBIS-II) for anastrozole.28 However, tamoxifen, exemestane, and anastrozole failed to exert a significant preventive effect on HR-negative breast cancer.25,47 Women who are susceptible to HR-negative breast cancers will not only fail to benefit from preventive endocrine therapy but also suffer from the side effect of these drugs. Our study may provide new candidate biomarkers for the selection of women with a high susceptibility of HR-positive breast cancer from the normal population and the administration of specific drugs as chemoprevention to reduce their breast cancer risks. Furthermore, these findings might pave the way for precision medicine, especially in precision prevention.
However, this study has some limitations. The number of subjects is relatively limited. Therefore, the susceptibility of breast cancer is not identical to that in the general population, and additional studies are expected.
Conclusion
Our study first provided the evidence that the SNP rs3802278 located in the miRNA-binding site within the 3′-UTR of the target gene SULF1 exhibited a protective effect against the susceptibility to breast cancer, especially to ER-positive or HR-positive breast cancer, which suggests that a portion of miRNAs may be involved in the carcinogenesis of ER-positive or HR-positive breast cancer via its target genes. The results of this study also provided promising candidate biomarkers to predict the susceptibility of breast cancer and guide the administration of targeted preventive endocrine therapy.
Acknowledgments
The research is supported by grants from the National Natural Science Foundation of China (81172505 and 81302302), the Doctoral Programs Foundation of the Ministry of Education of China (20120071120105), and the Shanghai Natural Science Foundation (13ZR1452800). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Hoover R, Gray LA Sr, Cole P, et al. Menopausal estrogens and breast cancer. N Engl J Med. 1976;295(8):401–405. | ||
Ross RK, Paganini-Hill A, Gerkins VR, et al. A case-control study of menopausal estrogen therapy and breast cancer. JAMA. 1980;243(16):1635–1639. | ||
Bergkvist L, Adami HO, Persson I, Hoover R, Schairer C. The risk of breast cancer after estrogen and estrogen-progestin replacement. N Engl J Med. 1989;321(5):293–297. | ||
Colditz GA, Hankinson SE, Hunter DJ, et al. The use of estrogens and progestins and the risk of breast cancer in postmenopausal women. N Engl J Med. 1995;332(24):1589–1593. | ||
[No authors listed.] Breast cancer and hormone replacement therapy: collaborative reanalysis of data from 51 epidemiological studies of 52,705 women with breast cancer and 108,411 women without breast cancer. Collaborative Group on Hormonal Factors in Breast Cancer. Lancet. 1997;350(9084):1047–1059. | ||
Yamashita H, Yando Y, Nishio M, et al. Immunohistochemical evaluation of hormone receptor status for predicting response to endocrine therapy in metastatic breast cancer. Breast Cancer. 2006;13(1):74–83. | ||
Reid G, Denger S, Kos M, Gannon F. Human estrogen receptor-alpha: regulation by synthesis, modification and degradation. Cell Mol Life Sci. 2002;59(5):821–831. | ||
Herynk MH, Fuqua SA. Estrogen receptor mutations in human disease. Endocr Rev. 2004;25(6):869–898. | ||
Giacinti L, Claudio PP, Lopez M, Giordano A. Epigenetic information and estrogen receptor alpha expression in breast cancer. Oncologist. 2006;11(1):1–8. | ||
Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8(2):93–103. | ||
Melo SA, Esteller M. Dysregulation of microRNAs in cancer: playing with fire. FEBS Lett. 2011;585(13):2087–2099. | ||
Mattie MD, Benz CC, Bowers J, et al. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol Cancer. 2006;5:24. | ||
Blenkiron C, Goldstein LD, Thorne NP, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8(10):R214. | ||
Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302(1):1–12. | ||
Iorio MV, Casalini P, Tagliabue E, Ménard S, Croce CM. MicroRNA profiling as a tool to understand prognosis, therapy response and resistance in breast cancer. Eur J Cancer. 2008;44(18):2753–2759. | ||
Adams BD, Guttilla IK, White BA. Involvement of microRNAs in breast cancer. Semin Reprod Med. 2008;26(6):522–536. | ||
Ma L, Weinberg RA. Micromanagers of malignancy: role of microRNAs in regulating metastasis. Trends Genet. 2008;24(9):448–456. | ||
Adams BD, Furneaux H, White BA. The micro-ribonucleic acid (miRNA) miR-206 targets the human estrogen receptor-alpha (ERalpha) and represses ERalpha messenger RNA and protein expression in breast cancer cell lines. Mol Endocrinol. 2007;21(5):1132–1147. | ||
Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. miR-206 Expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68(13):5004–5008. | ||
Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431(7006):343–349. | ||
Mishra PJ, Humeniuk R, Mishra PJ, Longo-Sorbello GS, Banerjee D, Bertino JR. A miR-24 microRNA binding-site polymorphism in dihydrofolate reductase gene leads to methotrexate resistance. Proc Natl Acad Sci U S A. 2007;104(33):13513–13518. | ||
Morley M, Molony CM, Weber TM, et al. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430(7001):743–747. | ||
Song F, Zheng H, Liu B, et al. An miR-502-binding site single-nucleotide polymorphism in the 3′-untranslated region of the SET8 gene is associated with early age of breast cancer onset. Clin Cancer Res. 2009;15(19):6292–6300. | ||
Ratner E, Lu L, Boeke M, et al. A KRAS-variant in ovarian cancer acts as a genetic marker of cancer risk. Cancer Res. 2010;70(16):6509–6515. | ||
Goss PE, Ingle JN, Ales-Martinez JE, et al. Exemestane for breast-cancer prevention in postmenopausal women. N Engl J Med. 2011;364(25):2381–2391. | ||
Land SR, Wickerham DL, Costantino JP, et al. Patient-reported symptoms and quality of life during treatment with tamoxifen or raloxifene for breast cancer prevention: the NSABP Study of Tamoxifen and Raloxifene (STAR) P-2 trial. JAMA. 2006;295(23):2742–2751. | ||
Fisher B, Costantino JP, Wickerham DL, et al. Tamoxifen for the prevention of breast cancer: current status of the National Surgical Adjuvant Breast and Bowel Project P-1 study. J Natl Cancer Inst. 2005;97(22):1652–1662. | ||
Fernandes VM, Pradhan-Sundd T, Blaquiere JA, Verheyen EM. Ras/MEK/MAPK-mediated regulation of heparin sulphate proteoglycans promotes retinal fate in the Drosophila eye-antennal disc. Dev Biol. 2015;402(1):109–118. | ||
Ma Y, Wang J, Gao J, et al. Antithrombin up-regulates AMP-activated protein kinase signalling during myocardial ischaemia/reperfusion injury. Thromb Haemost. 2015;113(2):338–349. | ||
Quarto N, Amalric F. Heparan sulfate proteoglycans as transducers of FGF-2 signalling. J Cell Sci. 1994;107(Pt 11):3201–3212. | ||
Narita K, Staub J, Chien J, et al. HSulf-1 inhibits angiogenesis and tumorigenesis in vivo. Cancer Res. 2006;66(12):6025–6032. | ||
Narita K, Chien J, Mullany SA, et al. Loss of HSulf-1 expression enhances autocrine signaling mediated by amphiregulin in breast cancer. J Biol Chem. 2007;282(19):14413–14420. | ||
Li J, Kleeff J, Abiatari I, et al. Enhanced levels of Hsulf-1 interfere with heparin-binding growth factor signaling in pancreatic cancer. Mol Cancer. 2005;4(1):14. | ||
Lai J, Chien J, Staub J, et al. Loss of HSulf-1 up-regulates heparin-binding growth factor signaling in cancer. J Biol Chem. 2003;278(25):23107–23117. | ||
Khurana A, Liu P, Mellone P, et al. HSulf-1 modulates FGF2- and hypoxia-mediated migration and invasion of breast cancer cells. Cancer Res. 2011;71(6):2152–2161. | ||
Okolicsanyi RK, Faure M, Jacinto JM, et al. Association of the SNP rs2623047 in the HSPG modification enzyme SULF1 with an Australian Caucasian breast cancer cohort. Gene. 2014;547(1):50–54. | ||
Sabarinathan R, Tafer H, Seemann SE, Hofacker IL, Stadler PF, Gorodkin J. The RNAsnp web server: predicting SNP effects on local RNA secondary structure. Nucleic Acids Res. 2013;41(Web Server issue):W475–W479. | ||
Sabarinathan R, Wenzel A, Novotny P, Tang X, Kalari KR, Gorodkin J. Transcriptome-wide analysis of UTRs in non-small cell lung cancer reveals cancer-related genes with SNV-induced changes on RNA secondary structure and miRNA target sites. PLoS One. 2014;9(1):e82699. | ||
Haas U, Sczakiel G, Laufer SD. MicroRNA-mediated regulation of gene expression is affected by disease-associated SNPs within the 3′-UTR via altered RNA structure. RNA Biol. 2012;9(6):924–937. | ||
Han CH, Huang YJ, Lu KH, et al. Polymorphisms in the SULF1 gene are associated with early age of onset and survival of ovarian cancer. J Exp Clin Cancer Res. 2011;30:5. | ||
Qiao M, Iglehart JD, Pardee AB. Metastatic potential of 21T human breast cancer cells depends on Akt/protein kinase B activation. Cancer Res. 2007;67(11):5293–5299. | ||
Hietakangas V, Cohen SM. TOR complex 2 is needed for cell cycle progression and anchorage-independent growth of MCF7 and PC3 tumor cells. BMC Cancer. 2008;8:282. | ||
McDonald PC, Oloumi A, Mills J, et al. Rictor and integrin-linked kinase interact and regulate Akt phosphorylation and cancer cell survival. Cancer Res. 2008;68(6):1618–1624. | ||
Zhang F, Zhang X, Li M, et al. mTOR complex component Rictor interacts with PKCzeta and regulates cancer cell metastasis. Cancer Res. 2010;70(22):9360–9370. | ||
Wen ZH, Su YC, Lai PL, et al. Critical role of arachidonic acid-activated mTOR signaling in breast carcinogenesis and angiogenesis. Oncogene. 2013;32(2):160–170. | ||
Li H, Lin J, Wang X, et al. Targeting of mTORC2 prevents cell migration and promotes apoptosis in breast cancer. Breast Cancer Res Treat. 2012;134(3):1057–1066. | ||
Fisher B, Costantino JP, Wickerham DL, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90(18):1371–1388. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.






