Back to Journals » International Journal of General Medicine » Volume 14
Single-Cell Sequencing to Identify Six Heat Shock Protein (HSP) Genes-Mediated Progression Subtypes of Clear Cell Renal Cell Carcinoma
Authors Li Q, Lu M , Zhang Z, Zhang R
Received 6 May 2021
Accepted for publication 6 July 2021
Published 23 July 2021 Volume 2021:14 Pages 3761—3773
DOI https://doi.org/10.2147/IJGM.S318271
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Qinke Li, Maoqing Lu, Zhechuan Zhang, Ronggui Zhang
Department of Urology, The Second Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China
Correspondence: Ronggui Zhang
Department of Urology, The Second Affiliated Hospital, Linjiang Road, Chongqing Medical University, Chongqing, 400010, People’s Republic of China
Tel +86-23-13983790901
Fax +23-63832133
Email [email protected]
Background: Heat shock proteins (HSPs) are widely involved in tumor occurrence and development and are prognostic markers for multiple tumors. However, the role of HSPs in clear cell renal cell carcinoma (ccRCC) remains unclear.
Methods: We used Cytoscape to identify hub genes in the ccRCC single-cell sequencing data set from the Gene Expression Omnibus (GEO) data repository. We identified subtypes, C1 and C2, of The Cancer Genome Atlas (TCGA) patients based on the expression of hub genes using unsupervised consensus clustering. Principal component analysis (PCA) was used to verify the clustering differences, and Kaplan–Meier (K-M) estimate was used to verify the survival differences between C1 and C2 patients. We used TIMER 2.0 and CIBERSORT to evaluate the immune cell infiltration of HSP genes and C1 and C2 patients. The R package “pRRophetic” was used to evaluate the sensitivity in C1 and C2 patients to the four first-line treatment drugs.
Results: We identified six hub genes (HSP90AA1, HSPH1, HSPA1B, HSPA8, and HSPA1A) encoding HSP, five of which were significantly downregulated in TCGA group, and four had a protective effect on prognosis (p < 0.05). Survival analysis showed that C1 patients had a better overall survival (p < 0.001). TIMER 2.0 analysis showed that three HSP genes were significantly correlated with the infiltration of CD4+ T cells and CD4+ Th1 cells (|cor|> 0.5, p< 0.001). CIBERSORT showed significant differences in multiple infiltrating immune cells between C1 and C2 patients. Meanwhile, the expression of PD1 was significantly lower in C1 patients than in C2 patients, and the expression of PDL1 is the another way around. Drug sensitivity analysis showed that C1 patients were more sensitive to sorafenib, pazopanib, and axitinib (p < 0.001).
Conclusion: Our research revealed two molecular subtypes of ccRCC based on 6 HSP genes, and revealed significant differences between the two subtypes in terms of clinical prognosis, immune infiltration, and drug sensitivity.
Keywords: ccRCC, bioinformatics, immune, molecular subtypes, prognosis
Introduction
Kidney cancer affects nearly 300,000 individuals worldwide each year and is responsible for over 100,000 deaths annually.1,2 Clear cell renal cell carcinoma (ccRCC) is the most common RCC subtype, accounting for approximately 80% of all cases.3–5 The mortality rate of kidney cancer depends largely on the disease progression. According to the classification criteria of the eighth edition of the American Joint Committee on Cancer (AJCC), the five-year survival rate for stage I/II is approximately 93%, that for regional renal cancer (stage III) is approximately 70%, while that for metastatic renal cancer (stage IV) is only 13%.2,6,7 In terms of prognosis assessment, the five-year recurrence-free survival rate of patients with stage I disease is greater than 92%, while the risk of recurrence for patients with stage II and III diseases is as high as 40%.8,9 As such, the treatment of advanced renal cancer still faces a huge challenge.
Heat shock proteins (HSPs) are highly conserved chaperone molecules that are synthesized in large quantities to protect cells from damage when the body is in various stress states, such as high temperature, cold, mechanical damage, ischemia, or radiation.10,11 According to their relative molecular weights, HSPs are divided into six families-HSP27, HSP40, HSP60, HSP70, HSP90, and the large HSP family (HSP110 and glucose regulatory protein 170, (GRP170)).12–14 HSPs are involved in various cellular processes, including protein assembly, secretion, transportation, protein degradation, and transcription factor regulation, mainly to prevent protein misfolding and accelerate protein refolding.12,13,15–22 Previous studies have shown that the expression of HSPs is elevated in a variety of cancers and is associated with the proliferation, metastasis, apoptosis, and invasion of tumor cells and can also be a biomarker for certain cancers.23 In terms of immune regulation, HSPs can regulate the production of a series of cytokines, including TNF-α, IL-6, IL-10, and IL-12, which can further mediate the immune regulation of tumors.24 Extracellular HSPs can bind to the characteristic receptors on dendritic cells, affecting the delicate balance of immune regulation in the tumor microenvironment.25 HSPs can bind to antigen peptides on the surface of cancer cells, making them viable targets in cancer immunotherapy.25,26
Next-generation sequencing (NGS) has promoted in-depth research in the fields of tumor Intratumor heterogeneity (ITH) and tumor somatic mutations in the past ten years.27 However, traditional RNA sequencing only provides the average gene expression of various types of cells in the tissue, so it is hard to peek the transcriptome characteristics of the cell subsets in the tumor.28 Single-cell sequencing (scRNA-seq) is a high-throughput experimental technology for quantifying gene expression profiles of cell populations at the single-cell level.29 Therefore, scRNA-seq can analyze the gene expression or epigenetic modification of the cells that make up the tumor microenvironment with higher sensitivity, and provides a higher resolution scan for the description of ITH.30,31
In this study, we obtained scRNA-seq data related to metastatic ccRCC from the Gene Expression Omnibus (GEO) data repository to determine more precise molecular changes that mediate the progression of ccRCC. Besides, we used an independent patient cohort from The Cancer Genome Atlas (TCGA) to identify the potential functions, clinical prognosis, and therapeutic potential of these molecules. Our research provides new ideas for the mechanism of HSP molecules in ccRCC.
Materials and Methods
Sample Collection
Single-cell sequencing samples of 46 primary ccRCC (PDX-pRCC) and 36 lung metastatic ccRCC (PDX-mRCC) cases were obtained from the GSE73121 dataset of the GEO (https://www.ncbi.nlm.nih.gov/geo/) data repository.32 The R “hclust” algorithm was used to cluster the samples hierarchically, and outliers were removed. The principal component analysis (PCA) was used to test the clustering effect. Finally, 44 pRCC and 27 mRCC samples were included for further analysis (Figure S1 shows the PCA results of pRCC and mRCC). The transcriptome expression data (Count) and sample annotation files of 539 patients were obtained from TCGA database (https://portal.gdc.cancer.gov/). We manually screened out the patients who were officially recommended not to be included in the analysis (n = 19, “Do not use” = ‘‘TURE”) as well as those without follow-up information (n = 11). Finally, 509 patients with ccRCC were included in the follow-up analysis. Meanwhile, we converted all “Count” expression data into “TPM” to correct the impact of transcription sequencing depth and gene length and used “log2 (TPM + 1)” for clustering and correlation analyses to reduce the bias caused by excessive expression values.
Difference Analysis and HUB Gene Identification
The R package “DESeq2”33 was used to analyze the differentially expressed genes (DEGs) between mRCC and pRCC (screening criteria were |LogFC| >2 and FDR <0.01). The R package “enrichplot”34 was used to perform GO and KEGG enrichment analyses of the selected DEGs. The online website “STRING”35 was used to construct a protein-protein interaction (PPI) network for differential protein-coding genes, and the CytoHubba36 plug-in of Cytoscape 3.7.237 was used to screen important nodes and identify hub genes in the network.
Identification of Molecular Subtypes
Through the R package “ConsensusClusterPlus”,38 based on the expression of six HUB genes, unsupervised consensus clustering (kmax=9) was performed for 509 patients in TCGA cohort, and the k=2 was taken as the best cluster (Figure S2 shows the clustering results and Table 1 shows the clinical information associated with the C1 and C2 cohorts). PCA analysis was performed on the whole transcriptome data of 509 patients (Number of gene = 55,268) to clarify the validity of clustering C1 and C2.
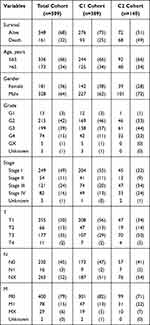 |
Table 1 Characteristics of TCGA Cohort with Clear Cell Renal Cell Carcinoma |
Prognostic Evaluation
According to the median value of HSP gene expression as the intercept, the patients were divided into a high expression group and a low expression group. The R package “survival”39 was used to assess the overall survival difference between the two groups and betwixt C1 and C2 patients.
Immune Correlation Assessment
TIMER 2.040 was used to evaluate the correlation between the expression level of six HSP genes and the abundance immune cell infiltration in ccRCC patients. CIBERSOFT,41 a deconvolution algorithm based on gene expression, which can infer 22 types of human immune cell, was used to analyze immune cell infiltration in C1 and C2 patients (p<0.05 is considered a valid result). The T test was used to evaluate the differences in the expression of immune checkpoint inhibitors PD1, PDL1 and CTLA4 in C1 and C2 patients.
Drug Susceptibility Prediction
The R package “pRRophetic”42 was used to evaluate the sensitivity of C1 and C2 patients to four advanced ccRCC first-line drugs, and the algorithm evaluated the maximum inhibitory concentration (IC50) and prediction accuracy through ridge regression and 10-fold cross-validation of the training set of the Dependent Cancer Drug Sensitivity Genomics (GDSC) database (https://www.cancerrxgene.org/).
Results
Gene Enrichment Analysis and HUB Gene Identification
We used the the R package “DEseq2” to evaluate the differences in single-cell transcriptome levels between 46 cases of PDX-pRCC and 36 cases of lung metastasis PDX-mRCC, and finally identified 494 DEGs, of which 173 were downregulated and 321 were upregulated (The specific results are shown in Table S1). Subsequently, we evaluated the possible pathways involved in the expression of the DEGs through GO and KEGG enrichment analyses. Figure 1A shows the results of GO enrichment analysis (adj. p <0.05). Interestingly, multiple pathways related to protein folding were significantly enriched. Among them, biological processes (BP) are mainly enriched in protein folding, response to unfolded protein and chaperone−mediated protein folding pathways. The cellular component (CC) is mainly enriched in inclusion body, ficolin−1−rich granule, U2−type precatalytic spliceosome pathways. Molecular function (MF) is mainly enriched in protein folding chaperone, MHC protein complex binding, unfolded protein binding and misfolded protein binding pathways. This suggests that in metastatic ccRCC cells, protein folding-related pathways may be widely activated or silenced. Figure 1B shows the association of DEGs in the five most significant enrichment pathways (lysosomal transport, positive regulation of cytokine production, protein folding, response to unfolded protein and vacuolar transport).
Subsequently, we conducted PPI analysis on the protein-coding genes of DEGs using STRING, and we used the various algorithms built into Cytoscape to identify the most closely linked key genes (hub genes). Interestingly, the enrichment results of multiple algorithms showed that the 6 genes (HSPA1A, HSPA1B, HSPA4L, HSPA8, HSP90AA1 and HSPH1) of the HPS family all showed significant connections with other DEGs. These genes are widely involved in the biological processes related to protein folding. As such, we speculate that these HPS genes may play an essential role in ccRCC. Figure 1C shows the interaction network between six HPS family genes and DEGs.
Prognosis Analysis of the Six HSP Genes
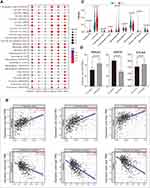
To further confirm the role of these six HSP genes in ccRCC, we analyzed the expression data of 509 ccRCC patients in the TCGA cohort. Compared with those in the normal group, HSP90AA1, HSPH1, HSPA1B, HSPA8 and HSPA1A genes were significantly downregulated in the tumor group (p<0.01, Figure 2A). Furthermore, the expression of HSP90AA1, HSPA4L and HSPA8 were significantly downregulated in patients at an advanced stage (Stage III and IV) than that in patients at an early stage (stage I and II) (p <0.01, Figure 2B). Meanwhile, the K-M survival analysis showed that low expression of HSP90AA1, HSPA8, HSPA1A and HSPA4L were associated with poor prognosis (p<0.05, Figure 2C).
Identification of ccRCC Molecular Subtypes
To identify the presence of molecular subtypes related to HSP in ccRCC patients, we performed unsupervised consensus clustering of 509 ccRCC patients based on the expression of the six HSP genes using the R package “ConsensusClusterPlus”, and successfully divided the patients into two Subtypes-C1 and C2 (k = 2, Figure 3A). Significantly, the expression of HSP90AA1, HSPA8, HSPA1A, and HSPA4L genes in C2 patients were remarkably downregulated, while HSPA1A and HSPA1B seemed to have a low contribution to clustering (Figure 3B). Besides, we conducted PCA on patients with expression information at the whole gene level (n = 55,268) to further evaluate the effectiveness of clustering. The results revealed that the transcriptome expression patterns of patients with C1 and C2 showed significant differences, indicating that clustering based on the six genes was feasible and effective (Figure 3C). We used a heat map to evaluate the difference between the C1 and C2 patients with the expression levels of the top 5000 genes, and found that the transcription profiles of the two types of patients were significantly different (Figure S3). Subsequently, we conducted a survival analysis of the patients with C1 and C2. The prognosis of the two subtypes was significantly different (p<0.0001), with a risk ratio of 1.909 (95% CI = 1.351–2.697) for C2/C1 (Figure 3D).
Evaluation of Immune Response of Six HSP Genes and ccRCC Subtypes
Given the close relationship between HSPs and immune responses, we further evaluated the correlation between the six HSP genes and immune cell infiltration. Using TIMER 2.0, we found that the expression of the six HSP genes in ccRCC was significantly related to CD4+ T cells, CD8+ T cells, B cells, and macrophages (Figure 4A). Remarkably, HSP90AA1, HSPA4L, and HSPA8 were significantly positively correlated with CD4+ T cells (Pho >0.4, p <0.0001), and significantly negatively correlated with CD4+ Th1 cells (Pho <-0.4, p <0.0001) (Figure 4B). We used CIBERSORT to evaluate the differences in immune cell infiltration between C1 and C2 patients. Plasma cells, CD8+ T cells, follicular helper T cells, Tregs, and M0 macrophages showed lower infiltration levels in C1 patients than in C2 patients (p <0.05), while M1 and M2 macrophages, resting dendritic cells, and resting mast cells showed higher infiltration levels (p <0.05) (Figure 4C). Furthermore, we evaluated the differences in the expression of immune checkpoint protein programmed cell death 1 (PD1/PDCD1), programmed death-ligand 1 (PD_L1/CD274), and cytotoxic T-lymphocyte associated protein 4 (CTLA4) in C1 and C2 patients. The expression of PD1 in C1 patients was significantly lower than that in C1 patients, while the content of PD_L1 was significantly higher than that in C2 patients (p <0.001) (Figure 4D). In summary, the subtypes of kidney cancer mediated by six HSP genes showed significant differences in tumor immune response.
Prediction of Drug Sensitivity for ccRCC Subtypes
Given that members of the HSP family are widely involved in resistance to chemotherapeutics, we evaluated the sensitivity of C1 and C2 patients to the first-line treatment drugs sorafenib, sunitinib, pazopanib, and axitinib, using the R package “pRRophetic”. Interestingly, C2 patients were more sensitive to sunitinib (Figure 5A), and C1 patients were more sensitive to sorafenib, pazopanib, and axitinib (Figure 5B–D). These results further indicated that patients of different subtypes may have more significant differences in sensitivity to different chemotherapeutic drugs.
 |
Figure 5 Differences in response sensitivity of C1 and C2 patients to sunitinib (A), sorafenib (B), pazopanib (C), and axitinib (D). |
Discussion
The accurate definition of tumor subtypes based on sequencing results is very important for precise treatment of patients. Previous studies have identified multiple tumor subtypes of ccRCC, such as 3 subtypes based on tumor suppressor genes, or 4 stratification based on transcription factor activity.43,44 These subtypes reveal part of the transcriptome characteristics of ccRCC and have guiding significance for the treatment of patients. It is worth noting that the tumor tissues derived from sequencing include many different types of cells, including immune cells. Therefore, traditional transcriptome sequencing is difficult to describe the evolutionary characteristics of different subgroups of tumor cells.27 In addition, ITH will change with the progress of treatment and time, which is a huge challenge for tumor treatment.45,46 While single-cell sequencing (scRNA-seq), as the highest resolution method currently used to describe cell genomes and epigenetic modifications, has promoted our understanding of cancer progression and ITH47 Gene expression profiles at the single-cell level can more sensitively show the transcriptional characteristics of different subgroups of tumor cells at different reaction stages.30
In this study, we used published scRNA-seq data to obtain a higher-resolution understanding of molecular changes inside metastatic ccRCC cells. Through differential and enrichment analyses, we identified six hub genes encoding heat shock proteins. Among them, HSPA1A, HSPA1B, and HSPA8 encode HSP70. HSP90AA1 encodes HSP90, and HSPA4L and HSPH1 encode HSP110.13
Specifically, HSPA1A and HSPA1B are paralogous genes located in the major histocompatibility complex (MHC) class III region on the short arm of chromosome 6, and code for the classical HSP70 protein.48,49 Previous studies have shown that HSP70 is overexpressed in liver, prostate, colorectal, lung, and cervical cancers.50 In human acute leukemia cells, Hsp70 can bind to death receptors 4 and 5 (DR4 and DR5), inhibit Apo-2L/TRAIL-induced assembly and death-inducing signaling complex (DISC) activity, further inhibiting apoptosis.51 In addition, Gabai et al demonstrated that HSP70 (HSPA1) could inhibit oncogene induction through two separate signal transduction pathways (the PI3K-mediated p53-dependent pathway or RAS/ERK-mediated p53-independent pathway) and cell senescence.52 However, HSP70 is downregulated in kidney cancer cells.53 Ramp et al analyzed the expression of HSP70 in 145 patients with renal cell carcinoma. In contrast to healthy cells, HSP70 in renal cancer cells was significantly downregulated in both the cytoplasm and nucleus (p = 0.0176), both in well-differentiated (G1) and poorly differentiated cells.53
The HSPA8 gene, located on chromosome 11q23.3–25, encodes the structural HSP70 protein HSC70, which has 85% homology with HSPA1A/B.54,55 HSC70 regulates cell signal transduction in normal cells and can mediate the migration and angiogenesis of endothelial cells induced by VEGF-induced Akt phosphorylation.56,57 Previous studies have shown that HSC70 is overexpressed in human colon cancer and glioma, and may be a poor prognostic indicator.58,59 Azuma et al found that HSC70 can induce cytotoxic T lymphocyte (CTL) responses by binding and carrying anti-cancer peptides.60 In contrast, Yehiely et al showed that HSC70 could bind to mutants of the tumor suppressor protein p53 and inhibit oncogene-mediated transformation.61
HSP90AA1 is located on the complement chain of chromosome 14q32.33, and encodes two different mRNA transcripts, Hsp90α TV1 and TV2.62,63 Elevated Hsp90α levels have previously been observed in leukemia, breast cancer, and pancreatic cancer.64,65 Teng et al found that the proto-oncogene MYC can bind to the transcription start site (TSS) at the proximal end of the HSP90AA1 gene and induce its expression.66 Perotti et al observed that in breast cancer cells, the growth hormone prolactin induces the expression of HSP90AA1 through STAT5.
Although HSPA4L (heat shock protein family A (Hsp70) member 4 like) is homologous to HSPA4, it is a member of HSP110. HSPA4L was found to be expressed mainly in human testicular germ cells and sperm.67 HSPA4L was found to be highly expressed in leukemia cells. Hosaka et al showed that HSPA4L might be involved in the resistance of cancer cells to apoptosis.68
HSPH1 is a member of the HSP110 family, and the encoded protein acts as the nucleotide exchange factor for HSPA1A/B, promoting the release of ADP from HSPA1A/B.69 Zappasodi et al showed that HSPH1 expression is elevated in aggressive human B-cell non-Hodgkin lymphoma (B-NHL) and positively correlated with c-Myc or Bcl-6 expression.70 Zhang et al confirmed that elevated HSPH1 levels might mediate tamoxifen resistance.71
Although these six HSP genes and their encoded proteins have been shown to promote tumor development in many studies, their role in ccRCC is unknown, and their direct impact on ccRCC is still unclear.
In this study, besides HSPA4L, the other five genes were seen to be downregulated in ccRCC tissues. In addition, HSPA4L, HSP90AA1, and HSPA8 were downregulated in patients with stages III and IV, suggesting that they may be involved in tumor progression. Based on the expression of HSP genes, obtaining molecular subtypes through consistent clustering seems to be effective. The expression of HSPA8, HSP90AA1, HSPA4L, and HSPH1 in C1 patients was much higher than that in C2 patients, and their survival was also better. As such, our research seems to strengthen the evidence that HSP is downregulated in kidney cancer. In addition, 6 HSPs are not only significantly associated with CD4+ T cells and CD4+ Th1 cells, but the expression of immune checkpoints (ICPs, PD1 and PDL1) in C1 and C2 subtypes is also significantly different, suggesting that HSPs may be potential immunotherapy maker. Furthermore, we used drug sensitivity prediction and found that C2 patients were less sensitive to a wider range of drugs. This may be the underlying reason for the poor prognosis. However, it is worthwhile to further study the specific mechanisms of HSPs directly or indirectly responding to ccRCC immunotherapy and drug sensitivity. Although our study obtained a considerable amount of positive results, it still has limitations, including the lack of in vitro and in vivo studies and external verification of larger sample sizes. Our work requires independent, clinical prospective studies.
Conclusion
In summary, we identified six heat shock protein-encoding genes that may have protective functions in ccRCC using single-cell sequencing data. We identified two ccRCC subtypes based on the expression of six genes, and there were significant differences in the prognosis, immune cell infiltration, and drug sensitivity between the two subtypes. Our research provides a potential direction for studying the mechanism of these six HSP molecules in mediating immunity and drug sensitivity in ccRCC.
Abbreviations
AJCC, American Joint Committee on Cancer; B-NHL, B-cell non-Hodgkin lymphoma; BP, biological processes; CC, cellular component; ccRCC, clear cell renal cell carcinoma; CTL, cytotoxic T lymphocyte; CTLA4, cytotoxic T-lymphocyte associated protein 4; DISC, death-inducing signaling complex; DR4, death receptors 4; DR5, death receptors 5; GEO, Gene Expression Omnibus; HSP, Heat shock protein; MF, Molecular function; MHC, major histocompatibility complex; PCA, Principal component analysis; PD1, programmed cell death 1; PD_L1, programmed death-ligand 1; TCGA, The Cancer Genome Atlas; TSS, transcription start site.
Data Sharing Statement
All data can be obtained from the corresponding author in a reasonable reasons.
Ethics
The original data required for this research were collected from public databases, so no medical ethics review is required.
Consent for Publication
All authors approved publication.
Acknowledgments
We are grateful to the uploader of the original data and the public database.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
The study was sponsored by Research on the Frontiers and Application of Chongqing Science and Technology, Commission No. cstc2015jcyjA10030.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Turajlic S, Swanton C, Boshoff C. Kidney cancer: the next decade. J Exp Med. 2018;215(10):2477–2479. doi:10.1084/jem.20181617
2. Barata PC, Rini BI. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. 2017;67(6):507–524. doi:10.3322/caac.21411
3. Störkel S, van den Berg E. Morphological classification of renal cancer. World J Urol. 1995;13(3):153–158. doi:10.1007/bf00184870
4. Srigley JR, Delahunt B, Eble JN, et al. The International Society of Urological Pathology (ISUP) vancouver classification of renal neoplasia. Am J Surg Pathol. 2013;37(10):1469–1489. doi:10.1097/PAS.0b013e318299f2d1
5. Nickerson ML, Jaeger E, Shi Y, et al. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14(15):4726–4734. doi:10.1158/1078-0432.Ccr-07-4921
6. Team TACSmaec. American Cancer Society. Survival rates for kidney cancer. Available from: https://www.cancer.org/cancer/kidney-cancer/detection-diagnosis-staging/survival-rates.html#references.
7. Tsukimi Y, Okabe S. Recent advances in gastrointestinal pathophysiology: role of heat shock proteins in mucosal defense and ulcer healing. Biol Pharm Bull. 2001;24(1):1–9. doi:10.1248/bpb.24.1
8. Janowitz T, Welsh SJ, Zaki K, Mulders P, Eisen T. Adjuvant therapy in renal cell carcinoma-past, present, and future. Semin Oncol. 2013;40(4):482–491. doi:10.1053/j.seminoncol.2013.05.004
9. Stephenson AJ, Chetner MP, Rourke K, et al. Guidelines for the surveillance of localized renal cell carcinoma based on the patterns of relapse after nephrectomy. J Urol. 2004;172(1):58–62. doi:10.1097/01.ju.0000132126.85812.7d
10. Jäättelä M. Heat shock proteins as cellular lifeguards. Ann Med. 1999;31(4):261–271. doi:10.3109/07853899908995889
11. Jego G, Hazoumé A, Seigneuric R, Garrido C. Targeting heat shock proteins in cancer. Cancer Lett. 2013;332(2):275–285. doi:10.1016/j.canlet.2010.10.014
12. Wang X, Chen M, Zhou J, Zhang X. HSP27, 70 and 90, anti-apoptotic proteins, in clinical cancer therapy (Review). Int J Oncol. 2014;45(1):18–30. doi:10.3892/ijo.2014.2399
13. Wu J, Liu T, Rios Z, Mei Q, Lin X, Cao S. Heat shock proteins and cancer. Trends Pharmacol Sci. 2017;38(3):226–256. doi:10.1016/j.tips.2016.11.009
14. Ciocca DR, Calderwood SK. Heat shock proteins in cancer: diagnostic, prognostic, predictive, and treatment implications. Cell Stress Chaperones. 2005;10(2):86–103. doi:10.1379/csc-99r.1
15. Beere HM. The stress of dying”: the role of heat shock proteins in the regulation of apoptosis. J Cell Sci. 2004;117(Pt13):2641–2651. doi:10.1242/jcs.01284
16. Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–677. doi:10.1146/annurev.ge.22.120188.003215
17. Liu T, Daniels CK, Cao S. Comprehensive review on the HSC70 functions, interactions with related molecules and involvement in clinical diseases and therapeutic potential. Pharmacol Ther. 2012;136(3):354–374. doi:10.1016/j.pharmthera.2012.08.014
18. Macario AJ, Conway de macario E. Molecular chaperones: multiple functions, pathologies, and potential applications. Front Biosci. 2007;12:2588–2600. doi:10.2741/2257
19. Jolly C, Morimoto RI. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J Natl Cancer Inst. 2000;92(19):1564–1572. doi:10.1093/jnci/92.19.1564
20. Lianos GD, Alexiou GA, Mangano A, et al. The role of heat shock proteins in cancer. Cancer Lett. 2015;360(2):114–118. doi:10.1016/j.canlet.2015.02.026
21. Gething MJ, Sambrook J. Protein folding in the cell. Nature. 1992;355(6355):33–45. doi:10.1038/355033a0
22. Nollen EA, Morimoto RI. Chaperoning signaling pathways: molecular chaperones as stress-sensing ‘heat shock’ proteins. J Cell Sci. 2002;115(Pt 14):2809–2816.
23. Calderwood SK, Khaleque MA, Sawyer DB, Ciocca DR. Heat shock proteins in cancer: chaperones of tumorigenesis. Trends Biochem Sci. 2006;31(3):164–172. doi:10.1016/j.tibs.2006.01.006
24. Tsan MF, Gao B. Cytokine function of heat shock proteins. Am J Physiol Cell Physiol. 2004;286(4):C739–44. doi:10.1152/ajpcell.00364.2003
25. Das JK, Xiong X, Ren X, Yang JM, Song J. Heat shock proteins in cancer immunotherapy. J Oncol. 2019;2019:3267207. doi:10.1155/2019/3267207
26. Murshid A, Theriault J, Gong J, Calderwood SK. Investigating receptors for extracellular heat shock proteins. Methods Mol Biol. 2011;787:289–302. doi:10.1007/978-1-61779-295-3_22
27. González-Silva L, Quevedo L, Varela I. Tumor functional heterogeneity unraveled by scRNA-seq technologies. Trends Cancer. 2020;6(1):13–19. doi:10.1016/j.trecan.2019.11.010
28. Wu H, Humphreys BD. The promise of single-cell RNA sequencing for kidney disease investigation. Kidney Int. 2017;92(6):1334–1342. doi:10.1016/j.kint.2017.06.033
29. Mereu E, Lafzi A, Moutinho C, et al. Benchmarking single-cell RNA-sequencing protocols for cell atlas projects. Nat Biotechnol. 2020;38(6):747–755. doi:10.1038/s41587-020-0469-4
30. Navin NE. Cancer genomics: one cell at a time. Genome Biol. 2014;15(8):452. doi:10.1186/s13059-014-0452-9
31. Zheng GX, Terry JM, Belgrader P, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. 2017;8:14049. doi:10.1038/ncomms14049
32. Kim KT, Lee HW, Lee HO, et al. Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol. 2016;17:80. doi:10.1186/s13059-016-0945-9
33. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi:10.1186/s13059-014-0550-8
34. Yu G. Enrichplot: visualization of functional enrichment result. Available from: https://github.com/GuangchuangYu/enrichplot.
35. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–d613. doi:10.1093/nar/gky1131
36. Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol. 2014;8(Suppl 4):S11. doi:10.1186/1752-0509-8-s4-s11
37. Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi:10.1101/gr.1239303
38. Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics (Oxford, England). 2010;26(12):1572–1573. doi:10.1093/bioinformatics/btq170
39. Therneau TM. A Package for Survival Analysis in R; 2020. Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Springer (New York).2000; ISBN:0-387-98784-3.
40. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–w514. doi:10.1093/nar/gkaa407
41. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337
42. Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9):e107468. doi:10.1371/journal.pone.0107468
43. Zhu Y, Cang S, Chen B, et al. Patient stratification of clear cell renal cell carcinoma using the global transcription factor activity landscape derived from RNA-Seq data. Front Oncol. 2020;10:526577. doi:10.3389/fonc.2020.526577
44. Zhong W, Zhang F, Huang C, Lin Y, Huang J. Classification of clear cell renal cell carcinoma based on tumor suppressor genomic profiling. J Cancer. 2021;12(8):2359–2370. doi:10.7150/jca.50462
45. Kosari F, Parker AS, Kube DM, et al. Clear cell renal cell carcinoma: gene expression analyses identify a potential signature for tumor aggressiveness. Clin Cancer Res. 2005;11(14):5128–5139. doi:10.1158/1078-0432.Ccr-05-0073
46. Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi:10.1056/NEJMoa1113205
47. Potter SS. Single-cell RNA sequencing for the study of development, physiology and disease. Nat Rev Nephrol. 2018;14(8):479–492. doi:10.1038/s41581-018-0021-7
48. Wu B, Hunt C, Morimoto R. Structure and expression of the human gene encoding major heat shock protein HSP70. Mol Cell Biol. 1985;5(2):330–341. doi:10.1128/mcb.5.2.330
49. Milner CM, Campbell RD. Structure and expression of the three MHC-linked HSP70 genes. Immunogenetics. 1990;32(4):242–251. doi:10.1007/bf00187095
50. Murphy ME. The HSP70 family and cancer. Carcinogenesis. 2013;34(6):1181–1188. doi:10.1093/carcin/bgt111
51. Guo F, Sigua C, Bali P, et al. Mechanistic role of heat shock protein 70 in Bcr-Abl-mediated resistance to apoptosis in human acute leukemia cells. Blood. 2005;105(3):1246–1255. doi:10.1182/blood-2004-05-2041
52. Gabai VL, Yaglom JA, Waldman T, Sherman MY. Heat shock protein Hsp72 controls oncogene-induced senescence pathways in cancer cells. Mol Cell Biol. 2009;29(2):559–569. doi:10.1128/mcb.01041-08
53. Ramp U, Mahotka C, Heikaus S, et al. Expression of heat shock protein 70 in renal cell carcinoma and its relation to tumor progression and prognosis. Histol Histopathol. 2007;22(10):1099–1107. doi:10.14670/hh-22.1099
54. Sonna LA, Fujita J, Gaffin SL, Lilly CM. Invited review: effects of heat and cold stress on mammalian gene expression. J Appl Physiol. 2002;92(4):1725–1742. doi:10.1152/japplphysiol.01143.2001
55. Dworniczak B, Mirault ME. Structure and expression of a human gene coding for a 71 kd heat shock ‘cognate’ protein. Nucleic Acids Res. 1987;15(13):5181–5197. doi:10.1093/nar/15.13.5181
56. Rutherford SL, Zuker CS. Protein folding and the regulation of signaling pathways. Cell. 1994;79(7):1129–1132. doi:10.1016/0092-8674(94)90003-5
57. Shiota M, Kusakabe H, Izumi Y, et al. Heat shock cognate protein 70 is essential for Akt signaling in endothelial function. Arterioscler Thromb Vasc Biol. 2010;30(3):491–497. doi:10.1161/atvbaha.109.193631
58. Kubota H, Yamamoto S, Itoh E, et al. Increased expression of co-chaperone HOP with HSP90 and HSC70 and complex formation in human colonic carcinoma. Cell Stress Chaperones. 2010;15(6):1003–1011. doi:10.1007/s12192-010-0211-0
59. Helmbrecht K, Rensing L. Different constitutive heat shock protein 70 expression during proliferation and differentiation of rat C6 glioma cells. Neurochem Res. 1999;24(10):1293–1299. doi:10.1023/a:1020933308947
60. Azuma K, Shichijo S, Takedatsu H, Komatsu N, Sawamizu H, Itoh K. Heat shock cognate protein 70 encodes antigenic epitopes recognised by HLA-B4601-restricted cytotoxic T lymphocytes from cancer patients. Br J Cancer. 2003;89(6):1079–1085. doi:10.1038/sj.bjc.6601203
61. Yehiely F, Oren M. The gene for the rat heat-shock cognate, hsc70, can suppress oncogene-mediated transformation. Cell Growth Differentiation. 1992;3(11):803–809.
62. Ozawa K, Murakami Y, Eki T, Soeda E, Yokoyama K. Mapping of the gene family for human heat-shock protein 90 alpha to chromosomes 1, 4, 11, and 14. Genomics. 1992;12(2):214–220. doi:10.1016/0888-7543(92)90368-3
63. Zuehlke AD, Beebe K, Neckers L, Prince T. Regulation and function of the human HSP90AA1 gene. Gene. 2015;570(1):8–16. doi:10.1016/j.gene.2015.06.018
64. Jameel A, Skilton RA, Campbell TA, Chander SK, Coombes RC, Luqmani YA. Clinical and biological significance of HSP89 alpha in human breast cancer. Int J Cancer. 1992;50(3):409–415. doi:10.1002/ijc.2910500315
65. Gress TM, Müller-Pillasch F, Weber C, et al. Differential expression of heat shock proteins in pancreatic carcinoma. Cancer Res. 1994;54(2):547–551.
66. Teng SC, Chen YY, Su YN, et al. Direct activation of HSP90A transcription by c-Myc contributes to c-Myc-induced transformation. J Biol Chem. 2004;279(15):14649–14655. doi:10.1074/jbc.M308842200
67. Nonoguchi K, Tokuchi H, Okuno H, et al. Expression of Apg-1, a member of the Hsp110 family, in the human testis and sperm. Int J Urol. 2001;8(6):308–314. doi:10.1046/j.1442-2042.2001.00304.x
68. Hosaka S, Nakatsura T, Tsukamoto H, Hatayama T, Baba H, Nishimura Y. Synthetic small interfering RNA targeting heat shock protein 105 induces apoptosis of various cancer cells both in vitro and in vivo. Cancer Sci. 2006;97(7):623–632. doi:10.1111/j.1349-7006.2006.00217.x
69. Rauch JN, Gestwicki JE. Binding of human nucleotide exchange factors to heat shock protein 70 (Hsp70) generates functionally distinct complexes in vitro. J Biol Chem. 2014;289(3):1402–1414. doi:10.1074/jbc.M113.521997
70. Zappasodi R, Ruggiero G, Guarnotta C, et al. HSPH1 inhibition downregulates Bcl-6 and c-Myc and hampers the growth of human aggressive B-cell non-Hodgkin lymphoma. Blood. 2015;125(11):1768–1771. doi:10.1182/blood-2014-07-590034
71. Zhang K, Jiang K, Hong R, et al. Identification and characterization of critical genes associated with tamoxifen resistance in breast cancer. Peer J. 2020;8:e10468. doi:10.7717/peerj.10468
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.