Back to Journals » International Journal of Nanomedicine » Volume 15
Silencing of MEF2D by siRNA Loaded Selenium Nanoparticles for Ovarian Cancer Therapy
Authors Wang C, Xia Y, Huo S, Shou D, Mei Q, Tang W, Li Y, Liu H, Zhou Y, Zhu B
Received 2 July 2020
Accepted for publication 7 November 2020
Published 4 December 2020 Volume 2020:15 Pages 9759—9770
DOI https://doi.org/10.2147/IJN.S270441
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Thomas Webster
Changbing Wang,1,2,* Yu Xia,1,3,* Shaochuan Huo,4,5,* Diwen Shou,3 Qing Mei,3 Wenjuan Tang,3 Yinghua Li,1 Hongsheng Liu,6 Yongjian Zhou,3 Bing Zhu1
1Central Laboratory, Guangzhou Institute of Pediatrics, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou 510120, People’s Republic of China; 2State Key Laboratory of Respiratory Disease, National Clinical Research Center for Respiratory Disease, Guangzhou Institute of Respiratory Health, First Affiliated Hospital of Guangzhou Medical University, Guangzhou Medical University, Guangzhou 510230, People’s Republic of China; 3Department of Gastroenterology and Hepatology, Guangzhou Digestive Disease Center, Guangzhou First People’s Hospital, School of Medicine, South China University of Technology, Guangzhou 510180, People’s Republic of China; 4Department of Orthopedics, Shenzhen Hospital (Futian) of Guangzhou University of Chinese Medicine, Shenzhen 518048, People’s Republic of China; 5Shenzhen Research Institute of Guangzhou University of Chinese Medicine, Shenzhen 518048, People’s Republic of China; 6Department of Radiology, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou 510120, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Bing Zhu
Central Laboratory, Guangzhou Institute of Pediatrics, Guangzhou Women and Children’s Medical Center, Guangzhou Medical University, Guangzhou 510120, People’s Republic of China
Tel +86 2081322725
Email [email protected]
Yongjian Zhou
Department of Gastroenterology and Hepatology, Guangzhou Digestive Disease Center, Guangzhou First People’s Hospital, School of Medicine, South China University of Technology, Guangzhou 510180, People’s Republic of China
Tel +86 2081045106
Email [email protected]
Background: Delivery of therapeutic small interfering RNA (siRNA) via functionalized nanoparticles holds great promise for cancer therapy. However, developing a safe and efficient delivery carrier of siRNA is a challenging issue.
Methods: RGDfC peptide was used to modify the surface of selenium nanoparticles (SeNPs) to synthesize a biocompatible siRNA delivery vehicle (R-SeNPs), and MEF2D-siRNA was loaded onto R-SeNPs to prepare a functionalized selenium nanoparticle R-Se@MEF2D-siRNA. The chemical properties of R-SeNPs were characterized, and the anticancer efficacy as well as related mechanisms of R-Se@MEF2D-siRNA were further explored.
Results: R-Se@MEF2D-siRNA was significantly taken up by SKOV3 cells and could enter SKOV3 cells mainly in the clathrin-associated endocytosis way. The result of in vitro siRNA release demonstrated that R-Se@MEF2D-siRNA could release MEF2D-siRNA quicker in a microenvironment simulating a lysosomal environment in tumor cells compared to a normal physiological environment. The results of qRT-PCR assay proved that R-Se@MEF2D-siRNA could effectively silence the expression of the MEF2D gene in SKOV3 cells. R-Se@MEF2D-siRNA remarkably suppressed the proliferation of SKOV3 cells and further triggered its apoptosis. In addition, R-Se@MEF2D-siRNA had the capability to disrupt mitochondrial membrane potential (MMP) in SKOV3 cells and resulted in the overproduction of reactive oxygen species (ROS), indicating that mitochondrial dysfunction and ROS generation played an important role in the apoptosis of SKOV3 cells induced by R-Se@MEF2D-siRNA. In vivo, R-Se@MEF2D-siRNA also exhibited excellent antitumor activity mainly through decreasing tumor cells proliferation and triggering their apoptosis in tumor-bearing nude mice.
Conclusion: R-Se@MEF2D-siRNA provides an alternative strategy for ovarian cancer treatment in the clinic.
Keywords: ovarian cancer, siRNA delivery, gene therapy, tumor targeting, MEF2D
Introduction
Ovarian cancer is one of the deadliest malignancies in women because of its high recurrence rate, and the 5-year survival rate is less than 50%.1 Chemotherapeutic drugs are commonly used for ovarian cancer therapy.2,3 Nevertheless, most cancers including ovarian cancer have developed inherent or acquired resistance.4 Thus, some novel therapeutic strategy should be developed to improve the treatment outcomes.5 Short interfering RNA (siRNA) technology is one of the most effective options for cancer therapy.6 Viruses were usually applied to deliver siRNA in the previous studies.7 Nevertheless, viral carriers had the risk of immunogenicity and insertional mutagenesis.8,9 Thus, non-viral vehicle holds great potential for the gene treatment of cancers owing to its better safety.10
Selenium nanoparticles (SeNPs), due to their biocompatibility, easy surface modification and low toxicity, have attracted much attention in the design of drug/gene vehicles.11–13 On the one hand, the trace element selenium (Se) plays a crucial role for preventing cancer.14 On the other hand, the presence of Se in the human body can ensure proper function of the immune system.15 The advantage of SeNPs makes it superior to organic nanoparticles or inorganic metal nanoparticles to deliver drug/gene.16 Therefore, SeNPs have gradually developed one of the most promising chemotherapeutics gene/drugs vehicles.17 However, the lack of tumor-targeted ability is the biggest obstacle to the application of nanoparticles as gene/drugs carriers.18–20 Thus, in this study, the positively charged peptide RGDfC was used to install on the surface of SeNPs to fabricate a tumor-targeted siRNA delivery vehicle (R-SeNPs). RGDfC has the ability to selectively bind with its receptor αvβ3 integrin that is highly expressed in various kinds of cancer cells, including SKVO3 human ovarian cancer cells.21 In addition, R-SeNPs with a positive charge are prone to load with negative charge nucleic acid molecule siRNA via the electrostatic interaction.22
The myocyte enhancer factor 2 (MEF2) family, containing the MEF2-A, -B, -C, and -D subtypes, belongs to one of the human transcription factors. MEF2 family members play an important role in the occurrence and development of cancers.23 Previous research showed that expression of MEF2D was elevated in ovarian cancer, pancreatic cancer, non-small cell lung cancer, colorectal cancer, and so on.24 Thus, MEF2D-siRNA was loaded onto the surface of R-SeNPs to fabricate R-Se@MEF2D-siRNA, aiming at silencing MEF2D gene for ovarian cancer therapy. The anticancer efficacy and mechanisms of R-Se@MEF2D-siRNA were studied using ovarian cancer SKVO3 cells and a nude mice model.
Materials and Methods
Materials
Lysotracker red, sodium selenite (Na2SeO3), Hoechst 33342, propidium (PI), and ascorbic acid (Vc) were obtained from Sigma (MO, USA). Annexin V/PI was purchased from Beyotime. Fetal bovine serum (FBS) was obtained from Gibco. All antibodies were purchased from CST (MA, USA). The sequence of MEF2D-siRNA was as follows: GCAACAGCCTAAACAAGGT.
Fabrication and Characterizations of R-SeNPs
Selenium nanoparticles (SeNPs) were synthesized in according with previous reported literature.10 Briefly, 0.5 mL Na2SeO3 (5 mM) and 0.5 mL vitamin C (20 mM) were mixed and the mixture solutions were gently stirred for 2 hours to prepare SeNPs. Then, 1 mL of RGDfC solution (1 mg/mL) was dripped to previous mixture solutions. The mixture solutions were stirred at 25°C for 3 hours to fabricate R-SeNPs. The pure R-SeNPs solution was acquired by dialyzing (MW cut-off=3.5 kDa). The amount of RGDfC in nanoparticles was determined by high performance liquid chromatography (HPLC). Size distribution and zeta potential of R-SeNPs was examined by a Malvern Zetasizer.25 The polydispersity index of nanoparticles in this paper was in the range of 0.1~0.25. Elemental composition analysis of R-SeNPs was carried out using energy dispersive X-Ray (EDX). The chemical structures of R-SeNPs were analyzed by Fourier transform infrared spectroscopy (FTIR). Morphology of R-SeNPs was visualized by transmission electron microscopy (TEM).26 Briefly, TEM samples were prepared by dripping the nanoparticles solution onto a holey carbon film on copper grids, and air dried for examination using TEM.
R-Se@MEF2D-siRNA was obtained via dripping MEF2D-siRNA (100 nM) to R-SeNPs solution at 15°C for 35 minutes. The N/P (nitrogen atom/phosphorus atom) ratios of R-SeNPs/MEF2D-siRNA are 1:1, 2:1, 4:1, and 8:1. The concentrations of loaded MEF2D-siRNA were tested using a nanodrop 2000 spectrophotometer, as in previous literature.5
Gel Electrophoresis Assay
A gel retardation assay was performed by incubating R-SeNPs with MEF2D-siRNA for 25 minutes at various N/P rates to determine complete complex formation. R-Se@MEF2D-siRNA was transferred to an agarose gel electrophoresis (1%) at 110 mV for 20 minutes and then the gel imagings were visualized by a gel documentation system.
Cell Culture
HUVEC and SKOV3 cells were purchased from ATCC (Manassas, VA, USA) and incubated in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% FBS at 37°C with 5% CO2.
Cellular Uptake of R-Se@MEF2D-siRNA
SKOV3 cells at the density of 5×104 cells/mL were cultured in 6-well plate for 24 hours. Afterwards, SKOV3 cells were exposed to FAM-labeled R-Se@MEF2D-siRNAFAM (100 nM of siRNAFAM). The SKOV3 cells were washed, supplemented with fresh complete medium, and visualized under a fluorescence microscope. Uptake of R-Se@MEF2D-siRNAFAM in HUVEC was carried out by using the same method as above. The αvβ3 integrin expression in SKOV3 cells and HUVEC cells was analyzed by Western blot assay, as in a previous publication.10 In order to further verify whether RGDfC-mediated uptake occurred in SKOV3 cells, a competitive inhibition experiment has been performed. In brief, SKOV3 cells were pretreated with free RGDfC (0.5 μg/mL) for 2 hours. Subsequently, the cells were washed with cold PBS and exposed to R-Se@MEF2D-siRNAFAM for 1 hour, 2 hours, and 4 hours The uptake mechanisms of FAM-labeled R-Se@MEF2D-siRNAFAM in SKOV3 cells were studied using various endocytosis inhibitors and analyzed by flow cytometer.27
The Release of MEF2D-siRNA from R-SeNPs
For assessing the release profiles of MEF2D-siRNA from R-SeNPs, 1 mg of R-Se@MEF2D-siRNA complexes with a N/P rate (8:1) were incubated with 1 mL PBS (pH 5.4 or 7.4) and transferred into a dialysis tube. The tubes were immersed in 100 mL of PBS (pH 5.4 or 7.4) under horizontal shaking (150 rpm). The released MEF2D-siRNA was detected for 15 hours by a spectromax quickdrop. At predetermined time points, aliquots of 0.5 mL outside the dialysis tubes were withdrawn for test and replenished with the equivalent volume of fresh PBS. The released amount of payload MEF2D-siRNA was determined by comparison with an experimentally determined standard curve.28
RT-qPCR Analysis
SKOV3 cells in six-well plates were incubated for 24 hours to reach about 75% confluence. SKOV3 cells were co-incubated with 100 nM equivalent siRNA of R-Se@MEF2D-siRNA or R-Se@siNC for 24 hours. The cells were rinsed and incubated in the fresh complete medium for another 24 hours. The cells without treatment were set as the control group. Total RNA was extracted from SKVO3 cells by TRIzol reagent in accordance with manufacturer’s instruction. The RNA concentration was measured using a NanoDrop™ 1000 Spectrophotometer.29 The 2−ΔΔCT method was used to analyze the data. The primers sequences are shown in Table 1.
 |
Table 1 The Primer Sequences of MEF2D and GAPDH for Quantitative Real-Time PCR |
MTT Assay
The cytotoxicity of R-Se@MEF2D-siRNA was detected using MTT assay.30 In brief, SKOV3 cells in a 96-well plate were cultured for 12 hours to acquire about 50% confluence. Then SKOV3 cells were exposed to R-Se@MEF2D-siRNA and R-Se@siNC (various siRNA equivalent concentrations) for 48 hours. The previous media were taken out and the fresh media containing MTT (5 μg/mL) were supplemented in each well. The 96-well plate was kept in an incubator at 37°C for another 4 hours. Then the media were taken out, and 150 μL of DMSO was added to each well for shaking for 15 minutes. Finally, the cells were tested under a microplate reader at 570 nm.31
Flow Cytometry Assay
Flow cytometry was used to analyze the cell cycle distribution and apoptosis. In brief, SKOV3 cells were treated with R-Se@MEF2D-siRNA or R-Se@siNC (100 nM of equivalent siRNA) for 24 hours, and then rinsed with PBS. The collected cells were fixed with methanol overnight and then stained with 1 μg/mL of propidium (PI) for 20 minutes before testing using flow cytometry.32
Mitochondrial Membrane Potential (Δψm) Assay
For mitochondrial membrane potential (MMP) examination, the SKOV3 cells were treated with R-Se@MEF2D-siRNA or R-Se@siNC (100 nM of equivalent siRNA) for 24 hours and then were co-incubated with 1 μg/mL of JC-1 for 15 minutes. Finally, the cells were washed and tested via a FACS flow cytometer.33
Detection of Reactive Oxygen Species (ROS)
ROS level of SKOV3 cells was examined as reported.34 Briefly, SKOV3 cells were treated with R-Se@MEF2D-siRNA or R-Se@siNC (100 nM of equivalent siRNA) for 24 hours before staining with 10 μM DCFH-DA for 20 minutes. Then the imaging of cells was captured under a fluorescence microscope.
Xenograft Mouse Model
All animal experiments were approved by the Ethics Committee of Guangzhou Medical University and performed according to the protocols and guidelines of the Experimental Animal Center of Guangzhou Medical University. A BALB/c nude mice model (6-week old) was used to assess the antitumor activity of R-Se@MEF2D-siRNA. Briefly, the tumor-bearing mice were prepared by subcutaneously injecting 5×106 SKOV3 cells in the abdomen. The tumor-bearing mice were intravenously injected with saline, R-Se@siNC, or R-Se@MEF2D-siRNA (0.2 mg/kg of siRNA) once every 3 days after tumor volumes reached 100 mm3. Tumor volumes (mm3) were calculated using the following formula: (length×width2)/2.
The sections were stained with H&E, Ki67, pp53, caspase3 and TUNEL, and photographed under a microscope (Leica DMi8) for histologic analysis.
Statistical Analysis
Data are presented as the means±SD. A one-way ANOVA test was used to determine significance among groups. *P<0.05 and **P<0.01 were considered significant and highly significant, respectively.
Results and Discussion
Characterization of R-SeNPs
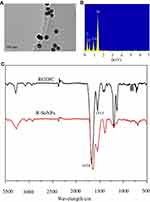
The morphology and size of the prepared selenium nanoparticles R-SeNPs were visualized by transmission electron microscopy (TEM), which indicated the uniform spherical particles of R-SeNPs (~80 nm) (Figure 1A). Elemental compositions of R-SeNPs was analyzed by EDX. From Figure 1B, oxygen and carbon atom signal derived from RGDfC and a typical Se atom signal derived from SeNPs were detected in the spectrum of R-SeNPs, indicating the successful loading of RGDfC onto SeNPs. Moreover, FTIR spectrum of R-SeNPs (Figure 1C) demonstrated the similar FTIR spectrum of RGDfC with the characteristic peak at 1,658 and 1,533 cm−1 of amide derived from RGDfC, indicating the effectual preparation of R-SeNPs. The schematic illustration of the formation of RGDfC-Se@MEF2D-siRNA was presented in Scheme 1. From Figure 2, size distributions of R-SeNPs in deionized water and PBS observed for 15 days, and the result showed that R-SeNPs could keep a stable size of particles (<150 nm) for within 15 days. The favorable stability of R-SeNPs in water and PBS supports its biological application. From Figure 3A, zeta potential of SeNPs transformed from −30.7 mV to 13.7 mV by loading with RGDfC, and the zeta potential of R-Se@MEF2D-siRNA was about 7.3 mV.
 |
Figure 2 Particle size survey of R-SeNPs in deionized water (A) and PBS (B). Abbreviations: PBS, phosphate buffer saline; R-SeNPs, RGDfC-modified selenium nanoparticles. |
Study on siRNA Loading Ability
An agarose gel assay was applied to assess siRNA loading ability of R-SeNPs. Seen from Figure 3B, R-SeNPs could significantly bind MEF2D-siRNA and MEF2D-siRNA migration started to be retarded at a N/P ratio of 1:1. When a N/P ratio of R-SeNPs/MEF2D-siRNA reached 8:1, R-SeNPs could completely retard the migration of MEF2D-siRNA. These results showed that R-SeNPs had the ability to bind MEF2D-siRNA and protect its degradation.
Selective Uptakes of R-Se@MEF2D-siRNA
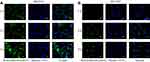
High uptake of therapeutic siRNA makes a great contribution to good treatment efficacy. Many studies showed that αvβ3 integrin, a receptor of RGD, was overexpressed in most cancer cells, including SKOV3 cells.35 The expression level of the integrin receptor on the cell membrane should be examined first in order to analyze whether the receptor contributes to the cellular uptake of R-Se@MEF2D-siRNA. The results indicated that the expression level of integrin receptor was higher in SKOV3 cells compared to HUVEC cells (Supplementary Figure S1), suggesting that RGDfC had a superior guided selectivity for SKOV3 cells to HUVEC cells. From Figure 4, the selective uptake of R-Se@MEF2D-siRNA in two cells was visualized under a fluorescence microscope, and the results showed that R-Se@MEF2D-siRNA exhibited greater cellular uptake in SKOV3 cells than that in HUVEC, suggesting RGDfC-modified functionalized selenium nanoparticles could enhance the uptake of MEF2D-siRNA in SKOV3 ovarian cancer cells. In order to further verify whether RGDfC-mediated uptake occurred in SKOV3 cells, a competitive inhibition experiment was carried out. Free RGDfC was added to SKOV3 cells first to block the interaction between RGDfC and its receptor integrin, and then R-Se@MEF2D-siRNA was added to the medium to study whether less R-Se@MEF2D-siRNA were taken by SKOV3 cells. As shown in Supplementary Figure S2, the pretreatment with free RGDfC in SKOV3 cells resulted in less uptake of R-Se@MEF2D-siRNA compared to SKOV3 cells without pretreatment, suggesting RGDfC-mediated targeting played an important role in the uptake of R-Se@MEF2D-siRNA in SKOV3 cells. In addition, fluorescence signal of SKOV3 cells increased from 1 hour to 4 hours of co-incubated with R-Se@MEF2D-siRNA, suggesting R-Se@MEF2D-siRNA entered SKOV3 cells in a time-dependent way.
Endocytosis is a key cellular process that leads to the internalization of selenium nanoparticles in cancer cells.36 The uptake pathway of R-Se@MEF2D-siRNA was studied using an endocytosis inhibition experiment. As shown in Figure 5A, the incubation of R-Se@MEF2D-siRNA in SKOV3 cells at low temperature (4°C) or pretreatment with NaN3/DOG (a cell energy metabolism inhibitor) at 37°C remarkably reduced uptake of R-Se@MEF2D-siRNA, indicating an energy-dependent endocytosis way of R-Se@MEF2D-siRNA. Different endocytosis inhibitors, chlorpromazine (clathrin-associated endocytosis), amiloride (macropinocytosis), and nystatin (caveolae-mediated endocytosis) were used to study endocytosis mechanisms of R-Se@MEF2D-siRNA. From Figure 5A, uptake of R-Se@MEF2D-siRNA by SKOV3 cells was decreased by 24% or 27% in the presence of nystatin or amiloride. Nevertheless, chlorpromazine-treatment made a great effect on the uptake of R-Se@MEF2D-siRNA, and the uptake was decreased by 48%, suggesting that SKOV3 cells take up the R-Se@MEF2D-siRNA mainly by the clathrin-associated endocytosis pathway.
The pH-Sensitive Release of MEF2D-siRNA
The releases of MEF2D-siRNA from R-SeNPs were detected in PBS of pH 5.4 and pH 7.4 simulating an acidic endosomes/lysosomes microenvironment and normal physiological environment, respectively.37 From Figure 5B, the obvious burst releases of MEF2D-siRNA in both pH values were observed within the initial 2 hours. Furthermore, R-Se@MEF2D-siRNA in PBS of pH 5.4 exhibited quicker release of MEF2D-siRNA and the release rate was about 83% at 15 hours. Nevertheless, the release rate of MEF2D-siRNA in pH 7.4 (61%) was lower than that in pH5.4 (83%) at 15 hours. This phenomenon could explain that acidic conditions increased the protonation of R-SeNPs, and thus weakened electrostatic interactions between R-SeNPs and MEF2D-siRNA, which promoted the release of MEF2D-siRNA from R-SeNPs. The acid-sensitive siRNA release feature of R-Se@MEF2D-siRNA exhibited unique advantages in cancer therapy.
R-Se@MEF2D-siRNA Silences the Expression of the MEF2D Gene
R-SeNPs was applied to deliver MEF2D-siRNA to SKOV3 cells, aiming at silencing the gene expression of MEF2D. A quantitative real time polymerase chain reaction (qRT-PCR) was applied to assess the mRNA level of MEF2D in SKOV3 cells. Seen from Figure 5C, R-Se@MEF2D-siRNA remarkably decreased the MEF2D mRNA level of SKOV3 cells. However, R-Se@siNC hardly influenced the expression of the MEF2D gene. The above results revealed that R-Se@MEF2D-siRNA could significantly silence the MEF2D gene in SKOV3 cells.
R-Se@MEF2D-siRNA Inhibits the Proliferation of SKOV3 Cells
The proliferation inhibition of SKOV3 cells was assessed using MTT assay. From Figure 6A, the proliferation of SKOV3 cells was remarkably suppressed by R-Se@MEF2D-siRNA and the cell viability rate was 39.6% after 48 hours. Nevertheless, no obvious effect on viability of cells treated with R-Se@siNC was observed, indicating that R-Se@MEF2D-siRNA inhibits the proliferation of SKOV3 ovarian cancer cells by silencing gene expression of MEF2D. The carrier itself (R-Se@siNC) did not exhibit obvious cytotoxicity against SKOV3 cells at such low concentrations as used in this study. Besides, R-Se@MEF2D-siRNA exhibited weak cellular uptake in HUVEC and had slight cytotoxicity against HUVEC (Figure 6B), further confirming the good biocompatibility of R-Se@MEF2D-siRNA. This finding was consistent with previous publications.13,29
Apoptosis of SKOV3 cells induced by R-Se@MEF2D-siRNA was assessed using flow cytometry. From Figure 6C, a sub-G1 apoptosis peak in the R-Se@MEF2D-siRNA group was more obvious (20.15%) compared to the R-Se@siNC group (6.42%) or untreated control group (6.19%), proving that R-Se@MEF2D-siRNA obviously induced the apoptosis of SKOV3 cells. In addition, R-Se@MEF2D-siRNA did not have a significant effect on cell cycle distribution of SKOV3 cells.
R-Se@MEF2D-siRNA Induces Mitochondria Dysfunction and ROS Overproduction
The loss of mitochondrial membrane potential (MMP, ΔΨm) can initiate apoptosis of tumor cells. Thus, we used flow cytometry to examine whether R-Se@MEF2D-siRNA triggered SKOV3 cells apoptosis by damaging the MMP.38 The change of ΔΨm of the cells were detected by staining with JC-1 to study the initiation of cell apoptosis, in which the red fluorescence indicates normal cells and the green fluorescence signal indicates apoptotic cells with mitochondrial dysfunction. Seen from Figure 7A, a part of red fluorescence transformed into green fluorescence, indicating R-Se@MEF2D-siRNA elevated the mitochondria depolarization of SKOV3 cells. The mitochondrial dysfunctions of SKOV3 cells exposed to R-Se@MEF2D-siRNA remarkably increased from 1.7% (untreated cells) to 21.32%. The above result indicates that the mitochondrial dysfunction induced by R-Se@MEF2D-siRNA makes an important contribution to the apoptosis of SKOV3 ovarian cancer cells.
Overproduction of reactive oxygen species (ROS) caused by antitumor drugs plays an essential role in the apoptosis and death of cancer cells.39 Therefore, in this study, the ROS level of SKOV3 cells was detected via fluorescence method. Figure 7B shows that R-Se@MEF2D-siRNA-treatment resulted in an obvious increase of DCF green fluorescence signal in SKOV3 cells, suggesting the ROS overproduction. Nevertheless, R-Se@siNC did not affect the MMP and the ROS level in comparison with the control group. The above results indicate that mitochondrial dysfunction and ROS overproduction induced by R-Se@MEF2D-siRNA play an important role in apoptosis of SKOV3 cells.
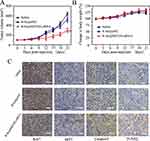
In vivo Anti-Tumor Efficacy and Toxicity Studies
The SKOV3 tumor xenograft was applied to assess the in vivo anticancer efficacy of R-Se@MEF2D-siRNA. From Figure 8A, as expected, R-Se@MEF2D-siRNA significantly delayed the tumor growth compared to the saline group (control) and R-Se@siNC group (negative control), verifying good antitumor efficacy of R-Se@MEF2D-siRNA. As seen from Figure 8B, the body weights of mice during the treatment period showed a slight increase, indicating that R-Se@MEF2D-siRNA was biocompatible materials in mice at the used dose. The anticancer mechanism of R-Se@MEF2D-siRNA was further elaborated by histological studies. Ki67-staining was used to examine the proliferation of tumor cells. The caspase-3, pp53, and TUNEL staining were applied to study tumor cells apoptosis (Figure 8C). Compared to the saline-treatment group, the treatment of R-Se@MEF2D-siRNA observably reduced the Ki67-positive tumor cells, suggesting that the proliferation of tumor cells was suppressed by R-Se@MEF2D-siRNA. As expected, R-Se@MEF2D-siRNA could obviously induce tumor cell apoptosis. These results indicated that R-Se@MEF2D-siRNA presented a significant antitumor efficacy in ovarian cancer treatments by suppressing the proliferation of SKOV3 ovarian cancer cells and inducing its apoptosis.
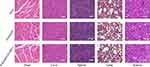
To assess in vivo toxicity of R-Se@MEF2D-siRNA, heart, kidneys, liver, lung and spleen of mice were collected and stained with H&E. As shown in Figure 9, the H&E stained heart, kidneys, liver, lung, and spleen tissue sections in the R-Se@MEF2D-siRNA-treatment group showed no obvious abnormality compared to saline or R-Se@siNC negative groups, further proving the good in vivo biocompatibility of R-Se@MEF2D-siRNA. Thus, R-Se@MEF2D-siRNA exhibited promising potential to be a prodrug for ovarian cancer therapy.
Conclusions
In this study, a tumor-targeted siRNA delivery carrier R-SeNPs was fabricated to deliver MEF2D-siRNA to SKOV3 cells for ovarian cancer therapy. R-Se@MEF2D-siRNA exhibited significant uptake in SKOV3 cells by clathrin-associated endocytosis and release of MEF2D-siRNA from R-SeNPs in acidic condition was quicker than that in normal physiological condition. R-Se@MEF2D-siRNA could efficaciously silence gene expression of MEF2D in SKOV3 cells and suppress proliferations of SKOV3 cells, and trigger SKOV3 cells apoptosis. More importantly, R-Se@MEF2D-siRNA exerted significant antitumor activity with low toxic side-effects in tumor-bearing nude mice. Taken together, R-Se@MEF2D-siRNA holds promising potential for ovarian cancer gene therapy.
Acknowledgments
This work was supported by the Science and Technology Planning Project of Guangdong Province (2014A020212024, 2015A020211002), the National Natural Science Foundation of China (81970507, 82004395), the Natural Science Foundation of Guangdong Province (2020A1515010100), the Technology Planning Project of Guangzhou City (201904010132; 201804010142), Innovative Clinical Technique of Guangzhou (2019GX05), China Postdoctoral Science Foundation (2018M633088), Scientific Research Project of Guangdong Provincial Bureau of Traditional Chinese Medicine (20192054).
Disclosure
The authors declare no conflicts of interest.
References
1. Zhang Q, Kuang G, He S, et al. Photoactivatable prodrug-backboned polymeric nanoparticles for efficient light-controlled gene delivery and synergistic treatment of platinum-resistant ovarian cancer. Nano Lett. 2020;20:3039–3049. doi:10.1021/acs.nanolett.9b04981
2. Karimi S, Fouani MH, Moshaii A, Nikkhah M, Hosseinkhani S, Sheikhnejad R. Development of dual functional nucleic acid delivery nanosystem for DNA induced silencing of Bcl-2 oncogene. Int J Nanomedicine. 2020;15:1693–1708. doi:10.2147/IJN.S236217
3. Paro AD, Hossain M, Webster TJ, Su M. Monte Carlo and analytic simulations in nanoparticle-enhanced radiation therapy. Int J Nanomedicine. 2016;11:4735–4741. doi:10.2147/IJN.S114025
4. Mahmoodi Chalbatani G, Dana H, Gharagouzloo E, et al. Small interfering RNAs (siRNAs) in cancer therapy: a nano-based approach. Int J Nanomedicine. 2019;14:3111–3128. doi:10.2147/IJN.S200253
5. Zhang F, Zhao L, Wang S, et al. Construction of a biomimetic magnetosome and its application as a SiRNA carrier for high-performance anticancer therapy. Adv Funct Mater. 2018;28:1703326. doi:10.1002/adfm.201703326
6. Guo Z, Li S, Liu Z, Xue W. Tumor-penetrating peptide-functionalized redox-responsive hyperbranched poly(amido amine) delivering siRNA for lung cancer therapy. ACS Biomater Sci Eng. 2018;4:988–996. doi:10.1021/acsbiomaterials.7b00971
7. Vaidya AM, Sun Z, Ayat N, et al. Systemic delivery of tumor-targeting siRNA nanoparticles against an oncogenic LncRNA facilitates effective triple-negative breast cancer therapy. Bioconjugate Chem. 2019;30:907–919. doi:10.1021/acs.bioconjchem.9b00028
8. Sakurai Y, Hada T, Kato A, Hagino Y, Mizumura W, Harashima H. Effective therapy using a liposomal siRNA that targets the tumor vasculature in a model murine breast cancer with lung metastasis. Mol Ther Oncolytics. 2018;11:102–108. doi:10.1016/j.omto.2018.10.004
9. Rehman M, Ihsan A, Madni A, et al. Solid lipid nanoparticles for thermoresponsive targeting: evidence from spectrophotometry, electrochemical, and cytotoxicity studies. Int J Nanomedicine. 2017;12:8325–8336. doi:10.2147/IJN.S147506
10. Xia Y, Tang G, Guo M, et al. Silencing KLK12 expression via RGDfC-decorated selenium nanoparticles for the treatment of colorectal cancer in vitro and in vivo. Mater Sci Eng C-Mater. 2020;110:110594. doi:10.1016/j.msec.2019.110594
11. Guo M, Li Y, Lin Z, et al. Surface decoration of selenium nanoparticles with curcumin induced HepG2 cell apoptosis through ROS mediated p53 and AKT signaling pathways. RSC Adv. 2017;7(83):52456–52464. doi:10.1039/C7RA08796A
12. Xia Y, Xu T, Zhao M, et al. Delivery of doxorubicin for human cervical carcinoma targeting therapy by folic acid-modified selenium nanoparticles. Int J Mol Sci. 2018;19:3582. doi:10.3390/ijms19113582
13. Li Y, Lin Z, Gong G, et al. Inhibition of H1N1 influenza virus-induced apoptosis by selenium nanoparticles functionalized with arbidol through ROS-mediated signaling pathways. J Mater Chem B. 2019;7(27):4252–4262. doi:10.1039/C9TB00531E
14. Xia Y, Xu T, Wang C, et al. Novel functionalized nanoparticles for tumor-targeting co-delivery of doxorubicin and siRNA to enhance cancer therapy. Int J Nanomedicine. 2017;13:143–159. doi:10.2147/IJN.S148960
15. Lin Z, Li Y, Xu T, et al. Inhibition of Enterovirus 71 by selenium nanoparticles loaded with siRNA through bax signaling pathways. ACS Omega. 2020;5(21):12495–12500. doi:10.1021/acsomega.0c01382
16. Gong G, Li Y, He K, et al. The inhibition of H1N1 influenza induced apoptosis by sodium selenite through ROS-mediated signaling pathways. RSC Adv. 2020;10:8002–8007. doi:10.1039/C9RA09524A
17. Xia Y, Tang G, Chen Y, et al. Tumor-targeted delivery of siRNA to silence Sox2 gene expression enhances therapeutic response in hepatocellular carcinoma. Bioact Mater. 2021;6:1330–1340. doi:10.1016/j.bioactmat.2020.10.019
18. Yang X, Zhang C, Li A, Wang J, Cai X. Red fluorescent ZnO nanoparticle grafted with polyglycerol and conjugated RGD peptide as drug delivery vehicles for efficient target cancer therapy. Mater Sci Eng C-Mater. 2019;95:104–113. doi:10.1016/j.msec.2018.10.066
19. Zhu P, Gao S, Lin H, et al. Inorganic nanoshell-stabilized liquid metal for targeted photonanomedicine in NIR-II biowindow. Nano Lett. 2019;19(3):2128–2137. doi:10.1021/acs.nanolett.9b00364
20. Tao S-C, Guo S-C, Zhang C-Q. Modularized extracellular vesicles: the dawn of prospective personalized and precision medicine. Adv Sci. 2018;5:1700449. doi:10.1002/advs.201700449
21. Sun L, Zheng C, Webster TJ. Self-assembled peptide nanomaterials for biomedical applications: promises and pitfalls. Int J Nanomedicine. 2017;12:73–86. doi:10.2147/IJN.S117501
22. Xia Y, Lin Z, Li Y, et al. Targeted delivery of siRNA using RGDfC-conjugated functionalized selenium nanoparticles for anticancer therapy. J Mater Chem B. 2017;5:6941–6952. doi:10.1039/C7TB01315A
23. Chow R, Wessels JM, Foster WG. Brain-derived neurotrophic factor (BDNF) expression and function in the mammalian reproductive Tract. Hum Reprod Update. 2020;26:545–564. doi:10.1093/humupd/dmaa008
24. Xiang J, Sun H, Su L, et al. Myocyte enhancer factor 2D promotes colorectal cancer angiogenesis downstream of hypoxia-inducible factor 1alpha. Cancer Lett. 2017;400:117–126. doi:10.1016/j.canlet.2017.04.037
25. Li K, Lu X, Zhang Y, Liu K, Huang Y, Liu H. Bi3TaO7/Ti3C2 heterojunctions for enhanced photocatalytic removal of water-borne contaminants. Environ Res. 2020;185:109409. doi:10.1016/j.envres.2020.109409
26. Huang Y, Long B, Tang M, et al. Bifunctional catalytic material: an ultrastable and high-performance surface defect CeO2 nanosheets for formaldehyde thermal oxidation and photocatalytic oxidation. Appl Catal B Environ. 2016;181:779–787. doi:10.1016/j.apcatb.2015.08.047
27. Xia Y, Zhong J, Zhao M, et al. Galactose-modified selenium nanoparticles for targeted delivery of doxorubicin to hepatocellular carcinoma. Drug Deliv. 2019;26:1–11. doi:10.1080/10717544.2018.1556359
28. Zhou Y, Yu Q, Qin X, et al. Improving the anticancer efficacy of laminin receptor-specific therapeutic ruthenium nanoparticles (RuBB-Loaded EGCG-RuNPs) via ROS-dependent apoptosis in SMMC-7721 cells. ACS Appl Mater Interfaces. 2015;8:15000–15012. doi:10.1021/acsami.5b02261
29. Xia Y, Tang G, Wang C, et al. Functionalized selenium nanoparticles for targeted siRNA delivery silence Derlin1 and promote antitumor efficacy against cervical cancer. Drug Deliv. 2019;27:15–25. doi:10.1080/10717544.2019.1667452
30. Yin T, Yang L, Liu Y, Zhou X, Sun J, Liu J. Sialic acid (SA)-modified selenium nanoparticles coated with a high blood–brain barrier permeability peptide-B6 peptide for potential use in Alzheimer’s disease. Acta Biomater. 2015;25:172–183. doi:10.1016/j.actbio.2015.06.035
31. Li Y, Xia Y, Liu K, et al. Constructing Fe-MOF-derived Z-scheme photocatalysts with enhanced charge transport: nanointerface and carbon sheath synergistic effect. ACS Appl Mater Interfaces. 2020;12:25494–25502. doi:10.1021/acsami.0c06601
32. Zheng W, Yin T, Chen Q, et al. Co-delivery of Se nanoparticles and pooled SiRNAs for overcoming drug resistance mediated by P-glycoprotein and class III beta-tubulin in drug-resistant breast cancers. Acta Biomater. 2016;31:197–210. doi:10.1016/j.actbio.2015.11.041
33. Xia Y, Xiao M, Zhao M, et al. Doxorubicin-loaded functionalized selenium nanoparticles for enhanced antitumor efficacy in cervical carcinoma therapy. Mater Sci Eng C. 2020;106:110100. doi:10.1016/j.msec.2019.110100
34. Gurunathan S, Qasim M, Park CH, et al. Cytotoxicity and transcriptomic analyses of biogenic palladium nanoparticles in human ovarian cancer cells (SKOV3). Nanomaterials. 2019;9(5):787. doi:10.3390/nano9050787
35. Xia Y, Guo M, Xu T, et al. siRNA-loaded selenium nanoparticle modified with hyaluronic acid for enhanced hepatocellular carcinoma therapy. Int J Nanomedicine. 2018;13:1539–1552. doi:10.2147/IJN.S157519
36. Zeng P, Xu Y, Zeng C, Ren H, Peng M. Chitosan-modified poly(D,L-lactide-co-glycolide) nanospheres for plasmid DNA delivery and HBV gene-silencing. Int J Pharm. 2011;415:259–266. doi:10.1016/j.ijpharm.2011.05.053
37. Xia Y, Chen Y, Hua L, et al. Functionalized selenium nanoparticles for targeted delivery of doxorubicin to improve non-small-cell lung cancer therapy. Int J Nanomedicine. 2018;13:6929–6939. doi:10.2147/IJN.S174909
38. Zhang J, Wang Q, Guo Z, et al. High‐fidelity trapping of spatial–temporal mitochondria with rational design of aggregation‐induced emission probes. Adv Funct Mater. 2019;29(16):1808153. doi:10.1002/adfm.201808153
39. Gupta PK, Pappuru S, Gupta S, Patra B, Chakraborty D, Verma RS. Self-assembled dual-drug loaded core-shell nanoparticles based on metal-free fully alternating polyester for cancer theranostics. Mater Sci Eng C-Mater. 2019;101:448–463. doi:10.1016/j.msec.2019.03.041
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.