Back to Journals » OncoTargets and Therapy » Volume 12
Silencing circZFR inhibits the proliferation, migration and invasion of human renal carcinoma cells by regulating miR-206
Received 8 May 2019
Accepted for publication 27 August 2019
Published 13 September 2019 Volume 2019:12 Pages 7537—7550
DOI https://doi.org/10.2147/OTT.S215012
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjay Singh
This paper has been retracted.
Mi Wang,1 Yisheng Gao,2 Jie Liu2
1Department of Urology, The Second Affiliated Hospital, University of South China, Hengyang 421000, Hunan, People’s Republic of China; 2Department of Urology, Linyi People’s Hospital, Linyi 276003, Shandong, People’s Republic of China
Correspondence: Jie Liu
Department of Urology, Linyi People’s Hospital, No.27 East Section of Jiefang Road, Lanshan District, Linyi 276003, Shandong, People’s Republic of China
Email [email protected]
Background: Renal cell carcinoma (RCC) is the most prevalent kind of kidney cancer. At present, the most efficient treatment mean is surgery. 40% patients with clear cell RCC (ccRCC) relapse after surgery. Identifying novel therapeutic markers and spots for early detection and treatment of RCC is necessary.
Methods: qRT-PCR was utilized to quantify circZFR and miR-206 expression in CAKI-1 and ACHN cells. Cell viability was detected by CCK-8 assay. Colony formation capacity was measured by colony formation assay. Transwell assay was utilized to investigate migration and invasion capacity. Expression of migration and apoptosis-associated proteins was quantified by Western blot.
Results: As a result, circZFR was highly expressed in RCC tissues and cells. Si-circZFR suppressed cell growth, migration and invasion of experimental cells. In addition, knockdown of circZFR upregulated miR-206 expression. Moreover, the antigrowth, antimigrating and anti-invasive effects of si-circZFR were attenuated when downregulating miR-206. Furthermore, Met is the target gene of miR-206 in experimental cells. The suppression on these signaling pathways was acted by targeting miR-206/Met axis.
Conclusion: The results demonstrated si-circZFR inhibited cell growth, migration and invasion in experimental cells by up-regulating of miR-206. Furthermore, si-circZFR suppressed Wnt/β-catenin and PI3K/AKT pathways via targeting miR-206/Met axis.
Keywords: circZFR, miR-206, renal carcinoma cells
Introduction
Renal cell carcinoma (RCC) is the most common type of renal cancer, accounting for approximately 80% of the total samples.1 RCC also comprises 2–3% of all malignancies.2 So far, surgery is still the main effective treatment means for RCC. In addition, partial nephrectomy is the most efficient therapeutic measures for clear cell RCC (ccRCC). However, 40% of patients with ccRCC relapse after surgery;3 it may due to cancer cell unrelenting growth and metastasis. Moreover, other therapeutic options, such as chemotherapy, radiotherapy and immunotherapy, have not achieved satisfying therapeutic effect, because RCC is resistant to these therapies. Although some kinase inhibitors have been used in clinical practice, metastatic renal cell carcinoma is still largely incurable because of the nontargeted effects of current drugs.4 Therefore, identifying novel therapeutic markers and targets for early detection and treatment of RCC is necessary.5
Circular RNAs (circRNAs) are an innovative race of RNAs belonging to noncoding RNA (ncRNA),6 and they have been widely found in many species by high throughput sequencing in recent years.7,8 circRNAs are constituted of covalently closed-loop structures with neither 5ʹ to 3ʹ polarity nor polyadenylated tail.9 circRNAs have been widely informed to play critical roles in multifarious human cancer cells10 and regulate multiple cellular mechanisms. Moreover, compared with linear RNA, circRNAs have closed-loop structure to confer their higher stability and tolerance to RNA enzyme. There are plenty of studies which reported that circRNAs played vital roles in squamous cell carcinoma, gastric cancer and so forth.11,12 However, the mechanism of circRNAs effect on RCC is still limited. Previous studies revealed that circZFR promoted hepatocellular carcinoma,10 papillary thyroid carcinoma13 and gastric cancer.14 Nonetheless, the function of circZFR on RCC remains unclear.
MicroRNAs (miRNAs) are small ncRNAs molecules that control gene expression level after transcription.15 Accumulating evidence shows that miRNAs represent abnormal expression in many human tumors, such as RCC, lung tumor and breast tumor.16–18 And miRNAs function as an indispensable regulation factor in initiation, development and metastasis of tumor.19 miR-206 was widely acknowledged in cancer. For instance, miR-206 reduced osteosarcoma cell malignancy in vitro.20 In addition, Cui et al elucidated that miR-206 suppressed proliferation and forecasted poor prognosis of cervical cancer cells.21 Furthermore, Met was reported to play a vital role in prompting RCC.22 And Met is the target gene of miR-206.
In the current investigation, we aim to reveal the function of circZFR on RCC and the potential mechanism of circZFR effect on RCC via regulating miR-206 and Met.
Materials And Methods
Clinical Specimens
Clinical human kidney cancer tissues and para-carcinoma tissues (n=22) were attained from Linyi People’s Hospital. All patients accepted no preoperative treatment before surgery. We informed each patient and obtained their consents. The present research was allowed by the Medical Ethics Committee Linyi People’s Hospital.
Cell Culture
Human kidney cancer CAKI-1 and ACHN cells were attained from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were developed in Roswell Park Memorial Institute-1640 (RPMI-1640) medium (Gibco BRL, Gaithersburg, MD, USA) containing 10% fetal bovine serum (FBS, HyClone Technologies, South Logan, UT, USA). The experimental cells mentioned above were inoculated in an incubator of 5% CO2 at 37°C.
Cell Transfection
circZFR small interfering RNA (si-RNA), si-negative control (NC), miR-206 inhibitor and the NC inhibitor were prefabricated (Life Technologies, Carlsbad, MD, USA) and were transfected into the cell lines used in the experiments. 48 hrs was chosen as the optimal harvest time in the consequent experiments.
Cell Viability
Cells were inoculated in a 96-well plate at the density of 5×103 cells per well. Cell viability was measured by Cell Counting Kit-8 (CCK-8, Dojido Laboratories, Tokyo, Japan). In brief, after cell were rinsed, CCK-8 solution was appended to cell culture media, and then cells were cultivated for 1 hr at 37°C in humidified 5% CO2 and 95% air atmosphere. The absorbance at 450 nm was evaluated by Microplate Reader (Bio-Rad, Hercules, CA, USA).
Colony Formation Assay
The experimental cells were inoculated in a 6-well plate. After that, cells were cultivated for 2 weeks, respectively. After incubation, cells were flushed with phosphate-buffered saline (PBS, Thermo Scientific, Waltham, MA, USA), settled with paraformaldehyde (Sigma Aldrich, St Louis, MO, USA) and stained with crystal violet (Sigma Aldrich) for 15 mins at room temperature. The number of colonies was calculated by utilizing a microscope (Olympus IX81, Tokyo, Japan).
Apoptosis Assay
Cells were inoculated in a 6-well plate for 1×105 cells per well. After incubation, treated cells were rinsed twice gently with cold PBS. Apoptotic cells were detected utilizing Annexin V-FITC/PI apoptosis detection kit (Beijing Biosea Biotechnology, Beijing, China). The apoptosis of CAKI-1 and ACHN cells was quantified by flow cytometry (Beckman Coulter, Atlanta, GA, USA).
Luciferase Reporter Assay
Cells were seeded in 12-well plates, which were divided NC mimic group and miR-206 mimic group. Each group was transfected with circZFR-wt and circZFR-mut by Lipofectamine 2000 (Invitrogen, San Diego, CA, USA) in antibiotic free medium for 48 hrs. Luciferase activity was measured with the Dual-Luciferase® Reporter Assay System (Promega, Madison, WI, USA). The relative luciferase activity was normalized.
Western Blot Assay
Proteins of experimental cells were separated by RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China) fortified with protease inhibitor (Roche, Basel, Switzerland). An equal amount of extracted protein was evaluated utilizing the BCATM Protein Assay Kit (Pierce, Rockford, IL, USA). Each protein band were detached by 10% SDS-PAGE and then transferred onto a nitrocellulose membrane (Millipore, Boston, MA, USA). Afterward, the appropriate primary antibodies were diluted and incubated with the membranes at 4°C overnight. Primary antibodies were listed as shown below: cleaved-caspase-3 (No. ab2302, Abcam, Cambridge, MA, USA), cleaved-caspase-9 (No. ab2324, Abcam), MMP-9 (No. ab38898, Abcam), vimentin (No. ab8978, Abcam), Wnt3a (No. ab28472, Abcam), PI3K (No. ab151549, Abcam), p-PI3K (No. ab138364, Abcam), β-catenin (No. 9562, Cell Signaling, Boston, MA, USA), AKT (No. 4685, Cell Signaling) and p-AKT (No. sc-271966, Santa Cruz Biotechnology, Santa Cruz, CA, USA). β-actin (No. sc-47778, Santa Cruz Biotechnology) was utilized as an endogenous protein for normalization. After the treated membranes were rinsed, they were conjunct with appropriate secondary antibodies labeled with horseradish peroxidase for 1 hr at room temperature. Ultimately, the membranes with protein bands were removed into the Bio-Rad ChemiDoc™ XRS system. Finally, each area of the protein band was detected and assessed by Image Lab™ Software (Bio-Rad). Each test was performed in triplicate experiments.
Migration And Invasion Assay
8 μM transwell chambers (Millipore) were placed in 24-well plates in the transwell assay. To assess the migration capacity, experimental cells were inoculated in the upper chambers. As for invasion assay, cells were inoculated in the upper chamber preprocessing with Matrigel (Corning, Corning, NY, USA). Cells in the upper chamber were suspended in 200 μL serum-free culture media. The lower chamber was added with 600 μL complete media containing 10% FBS. After cultivation for 48 hrs, the experimental cells which were not able to migrate or invade would maintain in the upper chamber and they were transferred gently with a cotton swab, and the migrated or invaded experimental cells were immobilized with 4% paraformaldehyde (Beyotime) for 30 mins and then dyed utilizing crystal violet solution for 15 mins. The cells were calculated using a microscope. And statistics were illustrated as the mean value of experimental cells adhered to the underside of the chamber of the five stochastic selected fields for each chamber.
Quantitative Real-Time PCR
Total RNA was separated from transfected cells utilizing tizol reagent (Invitrogen) as stated in the manufacturer’s protocols. RNA concentration and purity were measured by UV spectrophotometer at 260 nm and 280 nm. miRNA reverse transcription was worked utilizing the MultiscribeRTkit (Biosystems, Barcelona, Spain). The PCR was utilized of the SYBR® Green Master Mix (TaKaRa, Tokyo, Japan) following the protocols. The relative expression levels were calculated by the 2−△△Ct method, all experiments were repeated 3 times.
Statistical Analysis
Each analysis was repeated at least triple times. The evidence of various experiments was performed as the mean ± SD and measured utilizing SPSS 19.0 statistical software (SPSS, Chicago, IL, USA). The p-values were determined by a one-way analysis of variance (ANOVA) or Student t-test. A p-value of <0.05 was considered statistically significant.
Results
circZFR Is Remarkably Expressed In Renal Carcinoma Tissues And Cells
To investigate whether circZFR expression was changed in RCC tissues and cells, qRT-PCR was utilized to assess circZFR expression. As shown in Figure 1A, circZFR expression was remarkably escalated in renal carcinoma tissues as compared with para-carcinoma tissues (p<0.001). Moreover, the circZFR expression in renal carcinoma cells and normal tubular epithelial cell line was detected. And the expression was notably raised in CAKI-1, ACHN, A498 and KTCTL-26 cells (p<0.05 or p<0.01, Figure 1B).
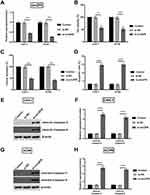
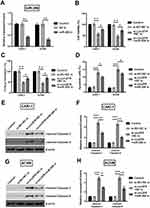
Knockingdown circZFR Inhibits Cell Proliferation And Induces Apoptosis Of CAKI-1 And ACHN Cells
To investigate whether si-circZFR was transfected into the experimental cells successfully, qRT-PCR was carried out to access the expression level of experimental cells. Both in CAKI-1 and in ACHN cells, circZFR expression was significantly decreased under the condition of knocking down circ-ZFR (p<0.01, Figure 2A). The consequences of CCK-8 assay suggested that cell viability was inhibited by knocking down the expression of circZFR (p<0.05 or p<0.01, Figure 2B). Sequentially, colony formation assay was utilized to measure the colony formation capacity of experimental cells; knockdown of circZFR strongly suppressed the proliferation (p<0.01, Figure 2C). Furthermore, apoptosis of cells was estimated by flow cytometry, and knockdown of circZFR notably developed the apoptosis rate (p<0.001, Figure 2D). As indicated in Figure 2E–H, the cleaved-caspae-3 and cleaved-caspase-9 expression was all notably raised in CAKI-1 and ACHN cells (p<0.001). All data indicated that silencing circZFR could inhibit the proliferation and induce apoptosis and the cleavage of caspases in CAKI-1and ACHN cells.
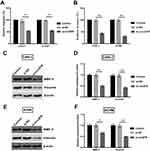
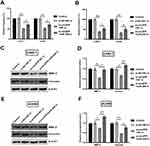
Knockingdown circZFR Inhibits The Migration And Invasion Of CAKI-1 And ACHN Cells
To further investigate whether circZFR affects the experimental cell migration and invasion, transwell assays were utilized. Ability of cell migration and invasion was notably declined by silencing circZFR (p<0.01, Figure 3A and B). MMP-9 and Vimentin expression was determined by Western blot. The protein expression was markedly declined when silencing circZFR in CAK-1 and ACHN cells (p<0.05 or p<0.01, Figure 3C and F). Thus, this evidence indicated that knockdown of circZFR restrained invasion and migration capacity in experimental cells.
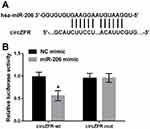
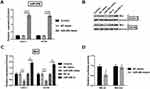
miR-206 Is A Target Of circZFR
Statistical analysis indicated that miR-200 was a candidate circZFR target. The circZFR sequences were cloned to form reporter plasmids. Luciferase reporter assay results showed that upregulating miR-206 reduced the luciferase activity of circZFR-wt (p<0.05). However, the luciferase activity showed no significance on circZFR-mut when up-regulating miR-206 (Figure 4).
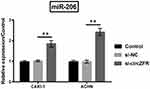
Knockingdown circZFR Elevates miR-206 Expression
In order to determine the relation between circZFR and miR-206, miR-206 expression was stimulated by silencing circZFR. miR-206 expression level was strongly raised by knockdown of circZFR (p<0.01, Figure 5).
Knockingdown circZFR Inhibits Proliferation And Induces Apoptosis Via Upregulating miR-206
To identify whether miR-206 was transfected into experimental cells successfully, qRT-PCR was utilized to identify miR-206 expression. miR-206 expression was remarkably enhanced after transfected with miR-206 inhibitor (p<0.01, Figure 6A). As shown in Figure 6B, viability was attenuated when downregulating miR-206 (p<0.05). Similarly, the decline of colony formation capacity was alleviated by downregulating miR-206 (p<0.05, Figure 6C). Flow cytometry was utilized to measure the apoptosis of experimental cells. The apoptotic cells were partly lessened by when downregulating miR-206 both in CAKI-1 and in ACHN group (p<0.05, Figure 6D). Changes of cleaved-caspase-3 and cleaved-caspase-9 expression in CAKI-1 cells were identified by Western blot. This protein expression was notably declined by downregulating miR-206 in CAKI-1 cells (p<0.01, Figure 6E and F). The expression of proteins mentioned above in ACHN cells was detected by Western blot. As expected, this protein expression was notably declined by downregulating miR-206 in ACHN cells (p<0.05 or p<0.01, Figure 6G and H).
Knockingdown The circZFR Suppresses Migration And Invasion Capacity By Upregulation Of miR-206 In CAKI-1 And ACHN Cells
In the transwell assay, cell migration and invasion were notably raised in a certain degree in both CAKI-1 and ACHN cells (p<0.05, Figure 7A and B). As the data suggested in Figure 7C and D, MMP-9 and vimentin expression was declined by knockdown of circZFR in CAKI-1 cells (p<0.05 or p<0.01). Meanwhile, this protein expression mentioned above was notably raised by downregulating miR-206 expression in ACHN cells (p<0.05 or p<0.01, Figure 7E and F).
Met Is The Target Of miR-206 In CAKI-1 And ACHN Cells
To confirm the effect of miR-206 on Met, gene expression and protein expression of Met was quantified utilizing qRT-PCR and Western blot. First, miR-206 gene expression was dramatically elevated both in CAKI-1 and in ACHN cells (p<0.001, Figure 8A). As indicated in Figure 8B and C, Met expression was remarkably declined by upregulating miR-206 in CAKI-1 and ACHN cells (p<0.01). Oppositely, Met expression was markedly raised by downregulating miR-206 (p<0.05). Furthermore, the luciferase activities of Met-wt and Met-mut in HEK293 cells were determined. Met-wt luciferase activity was notably reduced by upregulating miR-206 (p<0.05). However, Met-mut luciferase activity showed no significance when upregulating miR-206 (Figure 8D).
Knockingdown circZFR Suppressed Wnt/β-catenin And PI3K/AKT Pathways Through Upregulating miR-206
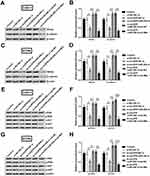
As a next step, Met expression was silenced in CAKI-1 and ACHN cells. Met expression was remarkably declined in both cells (p<0.01, Figure 9A and B).
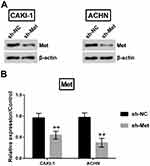 |
Figure 9 Met expression was dow-regulated in CAKI-1 and ACHN cells. (A-B) Met expression was estimated utilizing Western blot. **p<0.01 compared with marked group in the graph. |
To determine the potential mechanism of circZFR, Western blot was carried out to assess the associated protein expression. Wnt3a and β-catenin expression was obviously reduced when silencing circ-ZFR (p<0.05). Conversely, Wnt3a and β-catenin expression was remarkably elevated by downregulating miR-206 in CAKI-1 cells (p<0.01). Subsequently, the expression was markedly declined when downregulating Met (p<0.01, Figure 10A and B). Similarly, Wnt3a and β-catenin expression showed the same changes in ACHN cells (p<0.05 or p<0.01, Figure 10C and D). In addition, the rates of p/t-PI3K and p/t-AKT were strongly reduced when silencing circ-ZFR (p<0.05). Moreover, the rates were escalated when downregulating miR-206 (p<0.01). Furthermore, the escalation was reversed by downregulating Met in CAKI-1 cells (p<0.01 or p<0.001, Figure 10E and F). Sequentially, PI3K and AKT expression was detected in ACHN cells. The rates of p/t-PI3K and p/t-AKT were similarly reduced by silencing circZFR (p<0.05). And the rates then elevated by downregulating miR-206 (p<0.05 or p<0.01). Finally, the rates were dramatically declined when downregulating Met (p<0.01 or p<0.001, Figure 10G and H).
Discussion
circRNAs have been found for more than 40 years. However, the functions of circRNAs are recognized in recent years. Accumulating researches illustrated that circRNAs played vital roles in human cancer pathogenesis.13 For instance, circPPP1R12A promoted the growth, proliferation, invasion and metastasis of colon cancer via Hippo-YAP signaling pathway.23 Wu et al demonstrated circTADA2A increased the migration, invasion and proliferation of osteosarcoma cells and functioned as a tumor promoter in osteosarcoma.24 Hsa_circ_0000337 was proved to conduct proliferation, migration and invasion capacity of esophageal squamous cancer, indicating hsa_circ_0000337 as a lardaceous diagnostic biological marker and possible therapeutic spot.25 According to previous research, Wei et al investigated the function of circZFR in papillary thyroid cancer cells and the relationship between circZFR and miR-126.13 However, the relationship between circZFR and RCC remains widely unknown. We examined the effect of circZFR on experimental cells for the first time in the current investigation. The data suggested that circZFR was exceedingly expressed in tumor tissues. We revealed that knockdown of circZFR suppressed viability, migration and invasion capacity of the experimental cells. Moreover, we speculated the potential pathways and the relation between circZFR and miR-206. The results indicated that circZFR promoted RCC progression via Wnt/β-catenin and PI3K/AKT pathways.
miRNAs participated in numerous biological and pathological proceedings, such as proliferation, metastasis, apoptosis and so on.3,14 According to Wei’s research, miR-206 expression in RCC cells and tissues was both notably downregulated; miR-206 suppressed the growth, proliferation and migration in RCC cell lines.26 Similarly, Cai et al investigated that downregulating miR-206 expression had an effect on suppressing proliferation and invasion in ccRCC.27 Those research indicated miR-206 was a critical factor in tumorgenesis and development. Besides, miR-206 inhibited proliferation and migration on hepatocellular carcinoma cells.28 In the current research, we also suggested that miR-206 played a crucial role as a suppression factor in RCC. Furthermore, miR-206 could act as a downstream effector of circZFR, as its expression was negatively regulated by circZFR, and the effects of circZFR silence on RCC cell lines were attenuated when miR-206 was silenced. Nowadays, accumulating studies indicated circRNAs could exert their function through binding with miRNAs.29,30 For instance, circZFR was proved to promote hepatocellular carcinoma development via regulation of miR-3619-5p/CTNNB1 axis.10 circZFR promoted cell proliferation as well as invasion capacity on papillary thyroid tumor by regulating miR-1261.13 In our study, we investigated the correlation of circZFR and miR-206, knockdown of circZFR remarkably upregulated miR-206 expression both in experimental cells. Furthermore, circZFR could effect viability, proliferation, apoptosis, migration and invasion capacity by upregulating miR-206. In addition, further studies are still required to confirm this hypothesis.
Met is a receptor tyrosine kinase that is dysregulated by gene mutation, gene amplification, protein overexpression and ligand-dependent autocrine or paracrine loops.31 In both clear and nonclear cell RCC, the increase of both total Met and phospho Met expression is associated with poor prognosis.32 Besides, Met was the target gene according to a previous study. Moreover, Met was studied to be functioned in RCC.33 Another study reported that Wnt/β-catenin pathway was the downstream effector of Met in glioblastoma stem cells.34 In the current study, Met expression was remarkably declined by upregulating miR-206 and then notably elevated when downregulating miR-206 in CAKI-1 and ACHN cells. Combined with luciferase report assay, we confirmed that Met could be targeted by miR-206.
Wnts function in encoding a wide family secreted proteins which have various roles in cell proliferation, migration and differentiation.35 Wnt family genes play a vital part in human organogenesis and tumorgenesis. Additionally, they are associated with renal progress and initiation of several kinds of renal diseases including renal malignancy.36,37 Wnts also regulate comprehensive cellular behavior including differentiation, proliferation, migration and survival.38 A previous report showed that Wnt signaling pathway promoted proliferation in early kidney development.37 β-catenin is the ultimate effector of Wnt signaling pathway and is emerging as a decisive molecule in the pathogenesis of renal cancer. Accumulating research reported that the overexpression of β-catenin in RCC is correlated with high incidence.39 Thus, we detected Wnt3a and β-catenin expression, which was decreased by knocking-down circZFR, and the decrease was reversed by silencing the expression of miR-206. Subsequently, the expression was dramatically declined by downregulating Met. The data indicated knockdown of circZFR suppressed Wnt/β-catenin signaling pathway via upregulation of miR-206 and Met expression.
PI3K/AKT pathway is genetically spotted in more pathway ingredients and in more tumor sorts than other growth factor signaling pathways. PI3K/AKT signaling pathway is moderately muted but exceedingly triggered in RCC and represented a bright treatment target. As a matter of fact, PI3K pathway inhibitors are authorized for use in RCC.40 In our study, we proved that PI3K/AKT signaling pathway was suppressed by circZFR. On the other hand, silencing miR-206 and Met reversed the suppression. These findings indicated the mechanism of circZFR had an affect on RCC through these two pathways mentioned above by targeting miR-206/Met axis.
Conclusion
On the whole, the present study is the first to indicate that si-circZFR could affect growth, migration and invasion in RCC. Moreover, the suppression of si-circZFR on cell growth, migration and invasion capacity was attenuated via upregulation of miR-206 expression. And si-circZFR may, therefore, act as a regulator through targeting miR-206/Met axis in RCC progression. These findings uncovered a novel molecular mechanism of circZFR and miR-206 effect on RCC and provided an innovative target for clinical treatment.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. We informed each patient, and written informed consents were obtained from patients before surgery. The present research was allowed by the Medical Ethics Committee Linyi People’s Hospital.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgment
This research was supported by Shandong Provincial Natural Science Foundation, China (No. ZR2017PH021).
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that there are no conflicts of interest in this work.
References
1. Znaor A, Lortet-Tieulent J, Laversanne M, Jemal A, Bray F. International variations and trends in renal cell carcinoma incidence and mortality. Eur Urol. 2015;67(3):519–530. doi:10.1016/j.eururo.2014.10.002
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
3. Wang W, Hu W, Wang Y, Yang J, Yue Z. MicroRNA-508 is downregulated in clear cell renal cell carcinoma and targets ZEB1 to suppress cell proliferation and invasion. Exp Ther Med. 2019;17(5):3814–3822. doi:10.3892/etm.2019.7332
4. Das M. Cabozantinib improves clinical outcomes in renal cancer. Lancet Oncol. 2017;18(1):e2. doi:10.1016/S1470-2045(17)30072-4
5. Xia L, Li S, Liu Y, et al. NDNF inhibits the migration and invasion of human renal cancer cells through epithelial-mesenchymal transition. Oncol Lett. 2019;17(3):2969–2975. doi:10.3892/ol.2019.9937
6. Xu Z, Li P, Fan L, Wu M. The potential role of circRNA in tumor immunity regulation and immunotherapy. Front Immunol. 2018;9:9. doi:10.3389/fimmu.2018.00009
7. Feng J, Chen K, Dong X, et al. Genome-wide identification of cancer-specific alternative splicing in circRNA. Mol Cancer. 2019;18(1):35. doi:10.1186/s12943-019-1010-6
8. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
9. Liu J, Liu T, Wang X, He A. Circles reshaping the RNA world: from waste to treasure. Mol Cancer. 2017;16(1):58. doi:10.1186/s12943-017-0630-y
10. Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/beta-catenin pathway. Arch Biochem Biophys. 2019;661:196–202. doi:10.1016/j.abb.2018.11.020
11. Tian L, Cao J, Jiao H, et al. circRASSF2 promotes laryngeal squamous cell carcinoma progression by regulating the miR-302b-3p/IGF-1R axis. Clin Sci (Lond). 2019;133:1053–1066. doi:10.1042/CS20190110
12. Hao S, Lv J, Yang Q, et al. Identification of key genes and circular RNAs in human gastric cancer. Med Sci Monit. 2019;25:2488–2504. doi:10.12659/MSM.915382
13. Wei H, Pan L, Tao D, Li R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochem Biophys Res Commun. 2018;503(1):56–61. doi:10.1016/j.bbrc.2018.05.174
14. Liu T, Liu S, Xu Y, et al. Circular RNA-ZFR inhibited cell proliferation and promoted apoptosis in gastric cancer by sponging miR-130a/miR-107 and modulating PTEN. Cancer Res Treat. 2018;50(4):1396–1417. doi:10.4143/crt.2017.537
15. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. doi:10.1016/0092-8674(93)90529-y
16. Garcia-Vazquez R, Marchat LA, Ruiz-Garcia E, et al. MicroRNA-143 is associated with pathological complete response and regulates multiple signaling proteins in breast cancer. Technol Cancer Res Treat. 2019;18:1533033819827309. doi:10.1177/1533033819827309
17. Chen G, Ye B. The key microRNAs regulated the development of non-small cell lung cancer by targeting TGF-beta-induced epithelial-mesenchymal transition. Comb Chem High Throughput Screen. 2019;22:238–244. doi:10.2174/1386207322666190410151945
18. Carlsson J, Christiansen J, Davidsson S, Giunchi F, Fiorentino M, Sundqvist P. The potential role of miR-126, miR-21 and miR-10b as prognostic biomarkers in renal cell carcinoma. Oncol Lett. 2019;17(5):4566–4574. doi:10.3892/ol.2019.10142
19. Hidaka H, Seki N, Yoshino H, et al. Tumor suppressive microRNA-1285 regulates novel molecular targets: aberrant expression and functional significance in renal cell carcinoma. Oncotarget. 2012;3(1):44–57. doi:10.18632/oncotarget.417
20. Zhan FB, Zhang XW, Feng SL, et al. MicroRNA-206 reduces osteosarcoma cell malignancy in vitro by targeting the PAX3-MET axis. Yonsei Med J. 2019;60(2):163–173. doi:10.3349/ymj.2019.60.2.163
21. Cui J, Pan Y, Wang J, Liu Y, Wang H, Li H. MicroRNA-206 suppresses proliferation and predicts poor prognosis of HR-HPV-positive cervical cancer cells by targeting G6PD. Oncol Lett. 2018;16(5):5946–5952. doi:10.3892/ol.2018.9326
22. Wang M, Sun Y, Xu J, et al. Preclinical studies using miR-32-5p to suppress clear cell renal cell carcinoma metastasis via altering the miR-32-5p/TR4/HGF/Met signaling. Int J Cancer. 2018;143(1):100–112. doi:10.1002/ijc.31289
23. Zheng X, Chen L, Zhou Y, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18(1):47. doi:10.1186/s12943-019-1010-6
24. Wu Y, Xie Z, Chen J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression. Mol Cancer. 2019;18(1):73. doi:10.1186/s12943-019-1010-6
25. Song H, Xu D, Shi P, et al. Upregulated circ RNA hsa_circ_0000337 promotes cell proliferation, migration, and invasion of esophageal squamous cell carcinoma. Cancer Manag Res. 2019;11:1997–2006. doi:10.2147/CMAR.S195546
26. Wei C, Wang S, Ye ZQ, Chen ZQ. miR-206 inhibits renal cell cancer growth by targeting GAK. J Huazhong Univ Sci Technolog Med Sci. 2016;36(6):852–858. doi:10.1007/s11596-016-1674-8
27. Cai Y, Li H, Zhang Y. Downregulation of microRNA-206 suppresses clear cell renal carcinoma proliferation and invasion by targeting vascular endothelial growth factor A. Oncol Rep. 2016;35(3):1778–1786. doi:10.3892/or.2015.4538
28. Wang Y, Tai Q, Zhang J, et al. MiRNA-206 inhibits hepatocellular carcinoma cell proliferation and migration but promotes apoptosis by modulating cMET expression. Acta Biochim Biophys Sin (Shanghai). 2019;51(3):243–253. doi:10.1093/abbs/gmy119
29. Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi:10.1186/s12943-018-0827-8
30. Wang L, Wei Y, Yan Y, et al. CircDOCK1 suppresses cell apoptosis via inhibition of miR196a5p by targeting BIRC3 in OSCC. Oncol Rep. 2018;39(3):951–966. doi:10.3892/or.2017.6174
31. Peters S, Adjei AA. MET: a promising anticancer therapeutic target. Nat Rev Clin Oncol. 2012;9(6):314–326. doi:10.1038/nrclinonc.2012.71
32. Gibney GT, Aziz SA, Camp RL, et al. c-Met is a prognostic marker and potential therapeutic target in clear cell renal cell carcinoma. Ann Oncol. 2013;24(2):343–349. doi:10.1093/annonc/mds463
33. Zhou L, Liu XD, Sun M, et al. Targeting MET and AXL overcomes resistance to sunitinib therapy in renal cell carcinoma. Oncogene. 2016;35(21):2687–2697. doi:10.1038/onc.2015.343
34. Kim KH, Seol HJ, Kim EH, et al. Wnt/beta-catenin signaling is a key downstream mediator of MET signaling in glioblastoma stem cells. Neuro-oncology. 2013;15(2):161–171. doi:10.1093/neuonc/nos299
35. Miller JR. The Wnts. Genome Biol. 2001;3(1):1–15. doi:10.1186/gb-2001-3-1-reviews3001
36. Banumathy G, Cairns P. Signaling pathways in renal cell carcinoma. Cancer Biol Ther. 2010;10(7):658–664. doi:10.4161/cbt.10.7.13247
37. McCoy KE, Zhou X, Vize PD. Non-canonical wnt signals antagonize and canonical wnt signals promote cell proliferation in early kidney development. Dev Dyn. 2011;240(6):1558–1566. doi:10.1002/dvdy.22626
38. Xavier CP, Melikova M, Chuman Y, Uren A, Baljinnyam B, Rubin JS. Secreted Frizzled-related protein potentiation versus inhibition of Wnt3a/beta-catenin signaling. Cell Signal. 2014;26(1):94–101. doi:10.1016/j.cellsig.2013.09.016
39. Wen JL, Wen XF, Li RB, et al. UBE3C promotes growth and metastasis of renal cell carcinoma via activating Wnt/beta-catenin pathway. PLoS One. 2015;10(2):e0115622. doi:10.1371/journal.pone.0115622
40. Guo H, German P, Bai S, et al. The PI3K/AKT pathway and renal cell carcinoma. J Genet Genomics. 2015;42(7):343–353. doi:10.1016/j.jgg.2015.03.003
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.