Back to Journals » Journal of Inflammation Research » Volume 16
Significance of Pyroptosis in Immunoregulation and Prognosis of Patients with Acute Respiratory Distress Syndrome: Evidence from RNA-Seq of Alveolar Macrophages
Authors Liu B , Li Y, Xiang J, Li Y, Zhou M, Ren Y, Fu Z, Ding F
Received 13 June 2023
Accepted for publication 17 August 2023
Published 21 August 2023 Volume 2023:16 Pages 3547—3562
DOI https://doi.org/10.2147/JIR.S422585
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Bo Liu,1,2 Yan Li,2,3 Jinying Xiang,2,3 Yuehan Li,2,3 Mi Zhou,2,3 Yinying Ren,2,3 Zhou Fu,2,3 Fengxia Ding2,3
1Department of Cardiothoracic Surgery, Ministry of Education Key Laboratory of Child Development and Disorders, National Clinical Research Center for Child Health and Disorders, China International Science and Technology Cooperation Base of Child Development and Critical Disorders, Children’s Hospital of Chongqing Medical University, Chongqing, People’s Republic of China; 2Chongqing Key Laboratory of Pediatrics, Chongqing Engineering Research Center of Stem Cell Therapy, Chongqing Medical University, Chongqing, People’s Republic of China; 3Department of Respiratory Medicine, Ministry of Education Key Laboratory of Child Development and Disorders, National Clinical Research Center for Child Health and Disorders, China International Science and Technology Cooperation Base of Child Development and Critical Disorders, Children’s Hospital of Chongqing Medical University, Chongqing, People’s Republic of China
Correspondence: Fengxia Ding, Department of Respiratory Medicine, Ministry of Education Key Laboratory of Child Development and Disorder, National Clinical Research Center for Child Health and Disorders, China International Science and Technology Cooperation base of Child development and Critical Disorders, Children’s Hospital of Chongqing Medical University, No. 136, Zhongshan 2nd Road, Yuzhong Dis, Chongqing, 400014, People’s Republic of China, Tel +86-15178712548, Email [email protected]
Objective: This study aimed to investigate the role of pyroptosis in alveolar macrophages regarding the immune microenvironment of acute respiratory distress syndrome (ARDS) and its prognosis.
Methods: ARDS Microarray data were downloaded from Gene Expression Omnibus (GEO). Support vector machine (SVM) and random forest (RF) models were applied to identify hub pyroptosis-related genes (PRGs) with prognostic significance in ARDS. RT-PCR was used to detect the relative expression of PRGs mRNA in alveolar macrophages of ARDS mice. Consensus clustering analysis was conducted based on the expression of the PRGs to identify pyroptosis modification patterns. Bioinformatic algorithms were used to study the immune traits and biological functions of the pyroptosis patterns. Finally, protein-protein interaction (PPI) networks were established to identify hub regulatory proteins with implications for the pyroptosis patterns.
Results: In our study, a total of 12 PRGs with differential expression were obtained. Four hub PRGs, including GPX4, IL6, IL18 and NLRP3, were identified and proven to be predictive of ventilator-free days (VFDS) in ARDS patients. The AUC values of the 4 PRGs were 0.911 (GPX4), 0.879 (IL18), 0.851 (IL6) and 0.841 (NLRP3), respectively. In ARDS mice, GPX4 mRNA decreased significantly, while IL6, IL18, and NLRP3 mRNA increased. Functional analysis revealed that IL6 had the strongest positive correlation with the CCR pathway, while GPX4 exhibited the strongest negative correlation with the T co-inhibition pathway. Based on the expression of the 4 PRGs, three pyroptosis modification patterns representing different immune states were obtained, and pattern C might represent immune storm.
Conclusion: The results showed that pyroptosis plays an important regulatory role in the immune microenvironment of ARDS. This finding provides new insights into the pathogenesis, diagnosis, and treatment of ARDS.
Keywords: ARDS, macrophage, pyroptosis, immune, prognosis
Introduction
Acute respiratory distress syndrome (ARDS) is a multifactorial inflammatory pulmonary injury characterized by diffuse alveolar damage with hyaline membrane formation, epithelial cell hyperplasia, and interstitial edema. It clinically manifests as intractable hypoxemia and progressive dyspnea.1–3 In cases where positive end-expiratory pressure (PEEP) ≥5 cmH2O and PaO2/FiO2 ≤300 mmHg, chest imaging presents exudative lesions that involve both lungs and cannot be fully explained by heart failure. Extensive studies4–6 on ARDS have been conducted in recent years, but the pathogenesis of ARDS and the key therapeutic targets still require further exploration.
Alveolar macrophages (AMs) comprise the majority of pulmonary macrophages (approximately 95%). They can regulate the inflammation after ARDS as important innate immune cells, the first line of defense against respiratory pathogens. They recognize and engulf exogenous pathogens or endogenous damage-associated molecules via Toll-like receptors.7–9 In the early stage of ARDS, AMs release proinflammatory cytokines and chemokines to induce the migration and aggregation of neutrophils and monocytes (MONOs) into the alveolar lumen.10 The activated neutrophils further release inflammatory mediators, inducing tissue injury and barrier disruption. Meanwhile, the alveolar epithelial cells and effector T cells are activated, further promoting and maintaining inflammation and tissue injury.7,11 The exudative phase of ARDS comprises initial responses to injury, characterized by the release of pro-inflammatory cytokines and chemokines following AM activation and subsequent accumulation of MONOs and neutrophils.12,13 Under this background, the activation of AMs derived from peripheral MONOs is regarded as the leading cause of damage to the alveolar epithelium and endothelium.
Pyroptosis is a programmed cell death that can protect cells against infection. However, it can also lead to local and systemic inflammatory responses, and even fatal infectious shock in severer cases.14 Cell pyroptosis is mainly regulated by the Caspase-1 signaling pathway and the Caspase-4, Caspase-5, and Caspase-11 signaling pathways.15 It mainly involves the activation of various Caspase family proteins by inflammasomes, leading to the cleavage and polymerization of several Gasdermin family members, including GSDMD. This process results in cell membrane perforation, ultimately causing cell death. Accumulating evidence has suggested that macrophages undergo pyroptosis in response to an exogenous infectious or non-infectious stimulus, resulting in an enhanced inflammatory response.16–18 A recently study19 reveals that NLRP3 inflammasome activation and subsequent pyroptosis in AMs are responsible for the lung injury secondary to acute pancreatitis. Inhibition of AMs pyroptosis substantially suppresses acute pancreatitis-induced pulmonary lesion. Shao et al20 found that the administration of human growth hormone-releasing peptide (Ghrelin) in mice with traumatic brain injury (TBI) led to reductions in the levels of inflammatory factors (IL-1β, IL-6, TNF-α and IL-18), pyroptosis-related proteins (NLRP3, Caspase1-P20, HMGB1 and GSDMD), and phosphorylation of NF-κB in lung tissues. At the same time, Ghrelin also decreased the pulmonary vascular permeability and reduced the number of peripheral macrophages. These findings indicated that Ghrelin inhibited TBI-induced ARDS probably by blocking the NF-κB signaling and decreasing pyroptosis. Recent research21–23 has also demonstrated that alveolar macrophage pyroptosis plays a crucial role in ARDS, and by inhibiting this process, ARDS can be effectively ameliorated. However, the comprehensive role of pyroptosis in alveolar macrophages of ARDS has been less well studied. In this study, we applied bioinformatics to systemically analyze the role of pyroptosis in the prognosis and immunoregulation of ARDS patients. Our results demonstrate that pyroptosis in alveolar macrophages plays a crucial regulatory role in the immune microenvironment of ARDS. This finding is expected to contribute to a better understanding of the pathogenesis of ARDS and provide essential targets for its prevention and treatment.
Methods
Data Preprocessing
GSE89953 and GSE116560 datasets were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/). The GSE89953 dataset included 26 pairs of AMs samples (obtained from the BALF at day 1 of onset in ARDS patients) and peripheral blood MONOs samples.24 The GSE116560 dataset documented 68 AMs samples from ARDS patients, including 30 samples obtained at day 1 of onset and used in the present study.25 Then, the total RNA from the samples was subjected to genome-wide transcriptome sequencing. Gene annotation was performed using the GPL6883 platform. Probes corresponding to multiple molecules were removed, and the probes corresponding to the same molecule were compared to preserve the probe with the maximal signal. Batch correction was performed using the ComBat function of sva package (Figure S1) to ensure there were no significant batch effects across samples. Detailed clinical characteristics of patients included in this project are shown in Supplementary File 1. Pyroptosis-related genes (PRGs) were retrieved from previous research.26–30 Ultimately, 20 annotated PRGs were involved in this study.
Identification of Hub PRGs
A protein-protein interaction (PPI) network of PRGs was constructed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://cn.string-db.org/) to explore the relationship between the PRGs. The correlations between the PRGs in the AMs samples and the total samples were analyzed using Spearman correlation. The differential expression of the 20 PRGs between the AMs and MONOs samples was analyzed using the Wilcoxon test. The Least Absolute Shrinkage and Selection Operator (LASSO) regression was applied to remove redundant genes, followed by the establishment of random forest (RF) and support vector machine (SVM) models. Residuals were calculated to evaluate the performance of the two models.
Prognostic Performance of the Hub PRGs for ARDS
Ventilator-free days (VFDs) are a common indicator of clinical outcome in ARDS patients. Here, VFDs>0 were conferred to patients who were alive within 28 days and liberated from mechanical ventilation, while VFDs=0 were conferred to patients who died or required persistent mechanical ventilation. For the 30 patients from the GSE116560 dataset, VFDs ranged from 0 to 25, and the average value of 12 was set as the threshold. VFDs≥12 were conferred to patients with a relatively good clinical outcome, while VFDs<12 was conferred to patients with a relatively poor clinical outcome. The area under the receiver operating characteristic curve (ROC-AUC) was calculated to assess the discriminative capability of the hub PRGs in patients with varying clinical outcomes.
Identification of Pyroptosis Modification Patterns
Unsupervised clustering analysis was performed to identify pyroptosis modification patterns in ARDS patients based on the expression of the hub PRGs. Consensus clustering was applied to assess the robustness of clusters using the following parameters: clustering method based on k-means, 1000 iterations, and 80% resampling of samples. The optimal number of clusters was determined from cumulative distribution function (CDF) curves. Principal component analysis (PCA) was applied to further validate the reliability of the clustering.
Immune Analysis and Biological Functions in Different Pyroptosis Modification Patterns
Single-sample Gene Set Enrichment Analysis (ssGSEA) was applied to assess immune activity.31 Kruskal–Wallis test was used to compare the enrichment scores of immune pathways between MONOs and AMs samples. Spearman correlation was performed to assess the associations between the hub PRGs and immune reactivity as well as HLA expression. The immune differences across different pyroptosis modification patterns were explored using the Kruskal–Wallis test. The gene set “c2.cp.kegg.v7.4.symbols” was used as the reference gene set to reflect the changes in biological pathways. The clusterProfiler package was applied to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The GSVA algorithm was adopted to convert the expression matrix to a score matrix, and the limma package was used to perform differential analysis in the scores of biological pathways across different patterns (P<0.05). Additionally, the LIMMA package was also applied to obtain differentially expressed genes (DEGs) across different pyroptosis modification patterns (P<0.05).
Co-Expression Network Analysis
Weighted gene coexpression network analysis (WGCNA) was used to assess the expression matrix extracted from the DEGs across different pyroptosis modification patterns.32 Network construction and module detection were performed using a scale-free topological overlap matrix (TOM). The optimal soft threshold was set to 21. Modules were merged at a cut height of 0.2, and the minimum module size was set to 20. Spearman correlation was used to evaluate the association between the modules and the pyroptosis modification patterns.
Establishment of the Mouse ARDS Model and Histopathological Evaluation
Animal experimental procedures were approved by the Ethics Committee of Children’s Hospital of Chongqing Medical University. C57BL/6 mice (8–12 weeks old), weighing 20–25g, were purchased from the Animal Experiment Center of Chongqing Medical University and housed under standard conditions at the Animal Experimental Center of Children’s Hospital of Chongqing Medical University. The mice were divided into two groups: the Control group (normal mice) and the ARDS group, with each group containing 10 mice. After intraperitoneal injection of pentobarbital (50mg/kg) for anesthesia, the ARDS model in mice was established by intratracheal instillation of lipopolysaccharide (LPS) (5mg/kg, Solarbio Technology, China). After 24 hours, the mice were euthanized.33 The lower left lung lobe was fixed in 4% paraformaldehyde for 24 hours and then embedded in paraffin. Subsequently, the sections were deparaffinized with xylene, washed in different gradients of alcohol, incubated in distilled water, stained with hematoxylin and eosin, and mounted with neutral resin. Two observers independently examined the lung tissue sections under an optical microscope and recorded the pathological changes according to the previously described.34 Briefly, the histological index of lung injury was assessed by grading the following features on a five-point scale from 0 to 4: lung edema, infiltration of inflammatory cells, alveolar haemorrhage, hyaline membrane, and atelectasis. The scoring system was as follows: 0 = normal, 1 = injured area ≤25%, 2 = injured area 25–50%, 3 = injured area 51–70%, and 4 = injured area >70%. Ten fields were randomly selected for evaluation in each animal, and the scores were averaged for analysis.
Extraction of Primary Alveolar Macrophages from Mice
The procedure for extracting primary alveolar macrophages from mice is as follows:35 Perform alveolar lavage by endotracheal intubation three times to collect the bronchoalveolar lavage fluid. Centrifuge the collected bronchoalveolar lavage fluid at 4°C and 1500rpm for 10 minutes. Discard the supernatant and resuspend the pellet in RPMI 1640 complete medium containing 20% fetal bovine serum and 1% antibiotics. Place the suspension in a cell culture incubator at 37°C with 5% CO2 for 2 hours. Non-adherent cells are removed by gently washing the culture dish, and the adherent cells that remain attached to the dish are considered alveolar macrophages.
Quantitative Real-Time Polymerase Chain Reaction
We isolated total mRNA from alveolar macrophages using an RNA extraction kit (BioTeke, RP1202, China) following the manufacturer’s instructions. The mRNA was then reverse transcribed into cDNA using a reverse transcription kit (TAKARA-RR037A, Japan). RT-PCR was performed using SYBR Q-PCR Premix (TAKARA-RR820A, Japan) according to the provided protocol. GAPDH was used as the internal reference, and the relative expression levels of four hub PRGs (GPX4, IL6, IL18, and NLRP3) were determined using the 2−ΔΔCt method. The primer sequences used were as follows (5’-3’): GPX4: Forward: GTAACCAGTTCGGGAAGCAG, Reverse: TGTCGATGAGGAACTGTGGA; IL-6: Forward: GCCTTCTCTGATGCT, Reverse: TGTGACTCCAGCTTATCTCTTGG; IL-18: Forward: GCCTCTATTTGAAGATATGACTGA, Reverse: GAGATAGTACAGCCATACCTCTA; NLRP3: Forward: TCTTTGCGGCTATGTACTATCT, Reverse: TTCTAATAGGACCTTCACGT; GAPDH: Forward: TGATGACATCAAGAAGGTGGTGAAG, Reverse: TCCTTGGAGGCCATGTAGGCCAT. The data were expressed as mean ± SD. Statistical analysis was conducted using GraphPad software version 9.5.1. The comparison between two groups was performed using an unpaired, two-tailed Student’s t-test. Statistical significance was considered when the p-value was less than 0.05.
Results
Expression of PRGs in Different Clinical Samples
The chromosomal positions and the PPI network of the 20 PRGs are displayed in Figure 1A and B. It was found that the majority of the PRGs, except PRKACA, might play their roles in ARDS as a complex. The associations between different PRGs in the total samples (Figure 1C) and AM samples (Figure 1D) were analyzed. The Wilcox test resulted in 12 PRGs with differential expression in AMs and MONOs samples (Figure 1E and F). All the results demonstrated that pyroptosis may play an important role in the occurrence of ARDS.
PRGs as Potential Prognostic Markers for ARDS
To explore the prognostic significance of the 20 PRGs for ARDS, LASSO regression was applied to perform feature selection for dimension reduction to exclude redundant genes (Figure 2A and B). Hub PRGs were further identified by SVM (Figure 2C) and RF (Figure 2D) models. The SVM model was regarded as the optimal model with a smaller value of residuals (Figure 2E), and the genes involved in the model, including GPX4, IL6, IL18 and NLRP3, were considered as hub PRGs. ROC analysis revealed that the AUC values of the 4 PRGs were 0.911 (GPX4), 0.879 (IL18), 0.851 (IL6) and 0.841 (NLRP3), respectively, suggesting good prognostic performance for ARDS (Figure 2F).
Hub PRGs are Potentially Involved in Immunoregulation in ARDS
To investigate the significance of the hub PRGs (GPX4, IL6, IL18 and NLRP3) in the immunity of ARDS, correlations between PRGs expression and the activity of immune-related pathways, as well as HLA expression, were analyzed. Differential analysis revealed differences in immune function between the MONOs and AMs samples. As compared to the MONOs sample, AMs samples exhibited significant activation of some immune pathways, such as APC co-inhibition and CCR (Figure 3A). Additionally, the expression of HLA between the two types of samples was also remarkably different (Figure 3B). Correlational analysis indicated that the expression of the 4 PRGs was closely associated with multiple immune functions in AMs samples (Figure 4A). IL6 exhibited the strongest positive correlation with the CCR pathway (r=0.9), while GPX4 had the strongest negative correlation with the T co-inhibition (r= −0.55). Moreover, the expression of HLA-DMA exhibited the strongest negative correlation with NLRP3 (r= −0.66), but had the strongest positive correlation with GPX4 (r=0.59) (Figure 4B). Collectively, the 4 PRGs may participate in immunoregulation in ARDS.
Pyroptosis Modification Patterns in ARDS
Unsupervised consensus clustering analysis was performed on AMs samples based on the expression of the hub PRGs (Figure 5A–C). Three PRG-Clusters (A, B and C) were identified and could be well distinguished, as validated by further PCA (Figure 5D). Most of the 20 PRGs showed significant differential expression between different clusters. Among them, the pro-inflammatory cytokine IL-1β and inflammasome NLRP3 exhibited the highest expression in PRG-Cluster C, while the lowest expression in PRG-Cluster B (Figure 5E and F). The results demonstrate that ARDS may present multiple pyroptosis modification patterns, while PRG-Cluster C may represent an immune storm.
Immune Functions in the Three PRG-Clusters
Immune function and HLA expression in the three PRG Clusters were assessed. PRG-Cluster C exhibited the highest activity in immune responses (Figure 6A), and PRG-Cluster B had the highest mRNA expression of HLA (Figure 6B). These results show that pyroptosis may play an important regulatory role in the immune microenvironment of ARDS.
Biological Characteristics in the Three PRG-Clusters
Enrichment analysis was conducted to assess the biological functions in different pyroptosis modifications patterns. Compared to PRG-Cluster B, PRG-Cluster A exhibited higher activity in KEGG pathways, such as PROTEIN_EXPORT and N_GLYCAN_BIOSYNTHESIS (Figure 7A, Supplementary File 2). When compared with PRG-Cluster C, PRG-Cluster A had significantly up-regulated CITRATE_CYCLE_TCA_CYCLE and PYRIMIDINE_METABOLISM (Figure 7B, Supplementary File 3). Comparisons between PRG-Cluster B and C demonstrated significant up-regulation of pathways such as NOD_LIKE_RECEPTOR_SIGNALING_PATHWAY and CYTOKINE_CYTOKINE_RECEPTOR_INTERACTION in PRG-Cluster C (Figure 7C, Supplementary File 4). To gain more insight into the molecular mechanism, we obtained 597 DEGs among the three PRG-Clusters (Figure 8A). The most enriched GO terms of the DEGs included positive regulation of DNA-binding transcription factor activity, among others. (Figure 8B), while the most activated KEGG pathways included Toll-like receptor signaling pathway and TNF signaling pathway, among others. (Figure 8C). WGCNA was applied to obtain three gene modules associated with the three PRG-Clusters (Figure 8D–G). PRG-Cluster B exhibited the strongest positive correlation with the grey module (r=0.77), and PRG-Cluster C had the strongest positive correlation with the blue module (r=0.8). The above results also clarified the gene expression regulatory network and the changes in relevant pathways mediated by pyroptosis in ARDS.
Hub Regulatory Proteins in Different Pyroptosis Modification Patterns
PPI networks corresponding to the grey and blue modules were established using the STRING database. The top 10 proteins with the highest MCC value calculated by Cytoscape were selected as hub regulatory proteins. PRG-Cluster C might be regulated by IL6, IL1B, CXCL8, CCL3, MMP9, IL1RN, CXCL10, TLR2, PTGS2, CCL20 (Figure 9A), while PRG-Cluster B might be under the control of LRTOMT, GSTM2 and ALDH3B2 (Figure 9B).
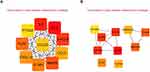 |
Figure 9 PPI networks. (A) Hub regulatory proteins of PRG-Cluster C based on blue module genes; (B) Hub regulatory proteins of PRG-Cluster B based on grey module genes. |
Expression of Hub PRGs in Alveolar Macrophages of ARDS Mice
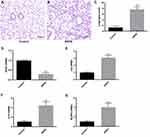
Finally, we validated the expression of Hub PRGs in alveolar macrophages of ARDS mice. Firstly, we established a mouse model of ARDS induced by LPS. In the lung tissues of ARDS mice, there was a significant infiltration of inflammatory cells, thickening of alveolar septa, and severe interstitial edema (Figure 10A and B). Compared to the control group, the lung tissue injury score in the ARDS group was significantly higher (P < 0.001) (Figure 10C). qRT-PCR was performed to measure the expression of Hub PRGs (GPX4, IL6, IL18, and NLRP3) in alveolar macrophages of mice. Compared to the control group, the expression of three hub PRGs (IL6, IL18, and NLRP3) was significantly upregulated in the ARDS group (P < 0.001). In contrast, the expression of one hub PRG, GPX4, showed a significant decrease (P < 0.001) (Figure 10D–G).
Discussion
ARDS is an inflammatory pulmonary disease characterized by acute onset and high mortality. In response to inflammatory stimulation, severe hypoxemia is induced by diffuse injury of the alveolar–capillary barrier, immune infiltration, flooding of alveolar airspaces with protein-rich edema fluid, and severe gas exchange abnormalities.8,36 Increasing evidence37–39 has suggested that the pyroptosis of AMs is involved in the occurrence and development of ARDS, and its suppression may be a new treatment strategy. The present study, for the first time, analyzed the mRNA level of 20 PRGs in AMs and MONOs samples from ARDS patients and revealed 12 PRGs that exhibited differential expression between the two types of samples. Further analysis found that GPX4, IL6, IL18 and NLRP3 were hub PRGs with prognostic significance for ARDS and implications for immunoregulation. Furthermore, we also discovered significant changes in the mRNA expression of four hub PRGs in AMs of ARDS mice. Three pyroptosis modification patterns in AMs samples were identified based on the expression of the four PRGs. According to the analyses of the pattern-mediated gene expression regulatory network and changes in relevant pathways, we proved that pyroptosis may play an important regulatory role in immune microenvironment in ARDS.
Currently, several studies have focused on the four hub PRGs. GPX4 is an inhibitory protein of lipid peroxidation that can degrade the lipid peroxides. It was recently reported as a key regulatory protein of pyroptosis in macrophages, as it could decrease the activity of Caspase-1, Caspase-11, and PLCG1 to prevent GSDMD-N production and pyroptosis.40 IL-6 is a multi-functional cytokine involved in multiple physiological functions and essential for maintaining immune homeostasis. It can inhibit the inflammatory injury associated with pyroptosis in AMs, which might be achieved via the suppression of Caspase-3-GSDME-mediated apoptosis-to-pyroptosis transformation or typical pyroptosis.41 IL-18 is a member of the IL-1 cytokine family and stimulates innate lymphocytes and antigen-experienced, but not naive, T cells. Both Caspase-1-dependent and independent pyroptosis pathways can induce the activation of Caspase-1, which then cleaves the precursors of IL-18. The bioactive IL-18 is then released into the extracellular environment and induces inflammatory responses.42 NLRP3, composed of 1016 amino acids, is encoded by NLRP3 gene mapped to chromosome 1q44. The carboxy-terminal LRR domain is responsible for ligand recognition; the intermediate NOD domain hydrolyzes ATP to GTP; the amino-terminal PYD domain participates in cellular inflammatory response by binding to molecules sharing an identical DNA-binding domain.43 Activated NLRP3 exposes the NACHT domain, which induces ATP-dependent oligomerization, resulting in the formation of an inflammasome containing Caspase and ASC (Apoptosis-associated speck-like protein containing a CARD). The NLRP3 inflammasome then activates Caspase-1 to induce pyroptosis in cells.
All the four hub PRGs are associated with immune pathways and may play a role in immunoregulation. Our results demonstrated that IL6 had the strongest positive correlation with the CCR pathway. It has been established that activated M1 macrophages induce the expression of the pro-inflammatory cytokine IL-6 and chemokine receptors CCR1/CCR5 to promote T-helper type 1 (Th1) and Th17 responses, which are essential in pathogen killing.44 A study on colitis-related colorectal cancer revealed that IL-6 induces macrophages to become tumor-promoting macrophages by producing CCL20 in the tumor microenvironment. CCL-20 recruits B cells and γδ T cells expressing CCR6 to advance the tumor progression.45 Additionally, our study also found the strongest negative correlation between GPX4 and T co-inhibition. It is noteworthy that the activity of GPX4 is essential for T cell survival. Experiments in genetically engineered mouse models have shown that deficiency of GPX4 in myeloid cells increases the rate of lethality in sepsis. Furthermore, GPX4 knockout in myeloid cells also facilitates the occurrence of intestinal tumors by increasing the genetic instability in intestinal epithelial cells. Further research has demonstrated that GPX4-deficient antigen-specific CD8 + and CD4 + T cells fail to proliferate and protect host mice from infection by lymphocytic choriomeningitis virus and Leishmania.8 Another study reported that enhanced expression of GPX4 in T cells increases the number of follicular helper T cells and facilitates antibody responses in vaccinated mice and young people.46 The studies mentioned above collectively indicate that GPX4 is crucial for T cell immunity. Additionally, we found that HLA-DMA expression had the strongest negative correlation with NLRP3 (r= −0.66) and the strongest positive correlation with GPX4 (r= 0.59), which also implies the potential role of the four hub PRGs in immunoregulation of ARDS.
The present study identified three pyroptosis modification patterns using unsupervised clustering analysis and PCA, and demonstrated differential expression in the majority of the PRGs across different patterns. It inferred that ARDS has diverse pyroptosis modification patterns represented by different immune characteristics and microenvironments. The results of KEGG pathway enrichment analysis also indicated the activation of different signaling pathways in different patterns, demonstrating the complexity of immune responses in ARDS. To gain more insight into the molecular mechanism, 597 DEGs among the three pyroptosis modification patterns were obtained and found to enrich in GO terms such as positive regulation of DNA-binding transcription factor activity, as well as KEGG pathways such as Toll-like receptor signaling pathway and TNF signaling pathway. Further WGCNA obtained three gene modules associated with the pyroptosis modification patterns. Based on the result of correlation analysis, the grey and blue modules with significant correlation with cluster B and C were selected for further analysis. PPI networks based on the genes involved in the two modules were generated, and the top 10 proteins with the highest MCC value were selected by Cytoscape and considered as hub regulatory proteins for the corresponding pyroptosis modification patterns. This analysis helps unravel the pyroptosis-mediated gene expression regulatory network in ARDS.
There are still some limitations to the study. For instance, the study results should be interpreted with caution due to the small size and heterogeneity of the samples included. Validation of the results is required in further large-scale studies, and both in vivo and in vitro experiments are needed to explore the specific mechanisms. Moreover, all the data in this study were obtained at the mRNA level, which may lead to less satisfactory results regarding the assessment for protein-based immune signaling pathways. Regardless of the limitations, our study also proved that pyroptosis modification is of vital significance in the immune microenvironment in ARDS, which deepens our understanding of the potential pathogenesis of ARDS.
Conclusion
In conclusion, the present study conducted bioinformatic analysis on previously published human sample datasets, GSE89953 and GSE116560 datasets, and identified the important roles of the PRGs and pathways associated with AMs pyroptosis in the immunoregulation of ARDS. Additionally, we identified four hub PRGs (GPX4, IL6, IL18 and NLRP3) associated with AMs pyroptosis in ARDS. The differential expression of these four PRGs was confirmed in both normal and ARDS mice. The findings of this study may provide new targets and insights for the diagnosis and treatment of ARDS.
Abbreviations
ARDS, Acute respiratory distress syndrome; BALF, Bronchoalveolar lavage fluids; CDF, Cumulative distribution function; DEGs, Differentially expressed genes; GO, Gene ontology; GEO, Gene Expression omnibus; KEGG, Kyoto Encyclopedia of genes and genomes; PCA, Principal component analysis; PPI, Protein-protein interaction; PRGs, Pyroptosis-related genes; RF, Random forest; TBI, Traumatic brain injury; SVM, Support vector machine; VFDS, Ventilator-free days.
Data Sharing Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Statement
Animal experimental procedures were approved by the Ethics Committee of Children’s Hospital of Chongqing Medical University (2022-083) and followed the standards of the National Institutes of Health Guide for the Care and Use of Laboratory Animals.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by Scientific Natural Science Foundation of Chongqing (CSTB2022NSCQ-MSX0930), Project of China Postdoctoral Science Foundation (2022MD713713), Specially Funded Project of Chongqing Postdoctoral Science Foundation (2022CQBSHTB1009), Chongqing Municipal Education Natural Science Foundation (KJQN202200419) and Children’s Hospital of Chongqing Medical University, National Clinical Research Center for Child Health and Disorders (NCRCCHD-2022-YP-05).
Disclosure
The authors have declared that no competing interest exists in this work.
References
1. Matthay MA, McAuley DF, Ware LB. Clinical trials in acute respiratory distress syndrome: challenges and opportunities. Lancet Respir Med. 2017;5(6):524–534. doi:10.1016/S2213-2600(17)30188-1
2. Meyer NJ, Calfee CS. Novel translational approaches to the search for precision therapies for acute respiratory distress syndrome. Lancet Respir Med. 2017;5(6):512–523. doi:10.1016/S2213-2600(17)30187-X
3. Maiolo G, Collino F, Vasques F, et al. Reclassifying acute respiratory distress syndrome. Am J Respir Crit Care Med. 2018;197(12):1586–1595.
4. Spengler D, Winoto-Morbach S, Kupsch S, et al. Novel therapeutic roles for surfactant-inositols and -phosphatidylglycerols in a neonatal piglet ARDS model: a translational study. Am J Physiol Lung Cell Mol Physiol. 2018;314(1):L32–L53. doi:10.1152/ajplung.00128.2017
5. Liang Q, Lin Q, Li Y, et al. Effect of SIS3 on extracellular matrix remodeling and repair in a lipopolysaccharide-induced ARDS rat model. J Immunol Res. 2020;2020:6644687. doi:10.1155/2020/6644687
6. De Luca D, Cogo P, Kneyber MC, et al. Surfactant therapies for pediatric and neonatal ARDS: ESPNIC expert consensus opinion for future research steps. Crit Care. 2021;25(1):75. doi:10.1186/s13054-021-03489-6
7. Wen Z, Fan L, Li Y, et al. Neutrophils counteract autophagy-mediated anti-inflammatory mechanisms in alveolar macrophage: role in posthemorrhagic shock acute lung inflammation. J Immunol. 2014;193(9):4623–4633. doi:10.4049/jimmunol.1400899
8. Matsushita M, Freigang S, Schneider C, Conrad M, Bornkamm GW, Kopf M. T cell lipid peroxidation induces ferroptosis and prevents immunity to infection. J Exp Med. 2015;212(4):555–568. doi:10.1084/jem.20140857
9. Huang X, Xiu H, Zhang S, Zhang G. The role of macrophages in the pathogenesis of ALI/ARDS. Mediators Inflamm. 2018;2018:1264913. doi:10.1155/2018/1264913
10. Mei H-X, Ye Y, Xu H-R, et al. LXA4 inhibits lipopolysaccharide-induced inflammatory cell accumulation by resident macrophages in mice. J Inflamm Res. 2021;14:1375–1385. doi:10.2147/JIR.S301292
11. Tan W, Zhang C, Liu J, Miao Q. Regulatory T-cells promote pulmonary repair by modulating T helper cell immune responses in lipopolysaccharide-induced acute respiratory distress syndrome. Immunology. 2019;157(2):151–162. doi:10.1111/imm.13060
12. Potera RM, Cao M, Jordan LF, Hogg RT, Hook JS, Moreland JG. Alveolar macrophage chemokine secretion mediates neutrophilic lung injury in Nox2-deficient mice. Inflammation. 2019;42(1):185–198. doi:10.1007/s10753-018-0883-7
13. Zhang H-W, Wang Q, Mei H-X, et al. RvD1 ameliorates LPS-induced acute lung injury via the suppression of neutrophil infiltration by reducing CXCL2 expression and release from resident alveolar macrophages. Int Immunopharmacol. 2019;76:105877. doi:10.1016/j.intimp.2019.105877
14. Bergsbaken T, Fink SL, Cookson BT. Pyroptosis: host cell death and inflammation. Nat Rev Microbiol. 2009;7(2):99–109. doi:10.1038/nrmicro2070
15. Man SM, Karki R, Kanneganti T-D. Molecular mechanisms and functions of pyroptosis, inflammatory caspases and inflammasomes in infectious diseases. Immunol Rev. 2017;277(1):61–75. doi:10.1111/imr.12534
16. Fan E, Brodie D, Slutsky AS. Acute respiratory distress syndrome: advances in diagnosis and treatment. JAMA. 2018;319(7):698–710. doi:10.1001/jama.2017.21907
17. Shen Y, Liu -W-W, Zhang X, et al. TRAF3 promotes ROS production and pyroptosis by targeting ULK1 ubiquitination in macrophages. FASEB J. 2020;34(5):7144–7159. doi:10.1096/fj.201903073R
18. Li -Z-Z, Wang H, Wang H, et al. The effect of erythrocyte transfusion on macrophage pyroptosis and inflammation in a sepsis model. Adv Clin Exp Med. 2021;30(6):617–622. doi:10.17219/acem/133490
19. Wu X-B, Sun H-Y, Luo Z-L, Cheng L, Duan X-M, Ren J-D. Plasma-derived exosomes contribute to pancreatitis-associated lung injury by triggering NLRP3-dependent pyroptosis in alveolar macrophages. Biochim Biophys Acta Mol Basis Dis. 2020;1866(5):165685. doi:10.1016/j.bbadis.2020.165685
20. Shao X-F, Li B, Shen J, et al. Ghrelin alleviates traumatic brain injury-induced acute lung injury through pyroptosis/NF-κB pathway. Int Immunopharmacol. 2020;79:106175. doi:10.1016/j.intimp.2019.106175
21. Tang Y, Yu Y, Li R, et al. Phenylalanine promotes alveolar macrophage pyroptosis via the activation of CaSR in ARDS. Front Immunol. 2023;14:1114129. doi:10.3389/fimmu.2023.1114129
22. Wu YT, Xu WT, Zheng L, et al. 4-octyl itaconate ameliorates alveolar macrophage pyroptosis against ARDS via rescuing mitochondrial dysfunction and suppressing the cGAS/STING pathway. Int Immunopharmacol. 2023;118:110104. doi:10.1016/j.intimp.2023.110104
23. Liu C, Zhou Y, Tu Q, Yao L, Li J, Yang Z. Alpha-linolenic acid pretreatment alleviates NETs-induced alveolar macrophage pyroptosis by inhibiting pyrin inflammasome activation in a mouse model of sepsis-induced ALI/ARDS. Front Immunol. 2023;14:1146612. doi:10.3389/fimmu.2023.1146612
24. Morrell ED, Radella F, Manicone AM, et al. Peripheral and alveolar cell transcriptional programs are distinct in acute respiratory distress syndrome. Am J Respir Crit Care Med. 2018;197(4):528–532. doi:10.1164/rccm.201703-0614LE
25. Morrell ED, Bhatraju PK, Mikacenic CR, et al. Alveolar macrophage transcriptional programs are associated with outcomes in acute respiratory distress syndrome. Am J Respir Crit Care Med. 2019;200(6):732–741. doi:10.1164/rccm.201807-1381OC
26. Wang K, Lv Z, Fang C, et al. Pyroptosis patterns are involved in immune microenvironment regulation of dilated cardiomyopathy. Dis Markers. 2022;2022:4627845. doi:10.1155/2022/4627845
27. Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunol Rev. 2015;265(1):6–21.
28. Wang B, Yin Q. AIM2 inflammasome activation and regulation: a structural perspective. J Struct Biol. 2017;200(3):279–282.
29. Xia X, Wang X, Cheng Z, et al. The role of pyroptosis in cancer: pro-cancer or pro-“host”. Cell Death Dis. 2019;10(9):650.
30. Karki R, Kanneganti TD. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat Rev Cancer. 2019;19(4):197–214.
31. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
32. Ge X. iDEP web application for RNA-Seq data analysis. Methods Mol Biol. 2021;2284417–2284443. doi:10.1007/978-1-0716-1307-8_22
33. Jiao Y, Yong C, Zhang R, Qi D, Wang D. Hepcidin alleviates LPS-induced ARDS by regulating the ferritin-mediated suppression of ferroptosis. Shock. 2022;57(6):274–281. doi:10.1097/SHK.0000000000001941
34. Zhou F, Zhang Y, Chen J, et al. Liraglutide attenuates lipopolysaccharide-induced acute lung injury in mice. Eur J Pharmacol. 2016;791:735–740. doi:10.1016/j.ejphar.2016.10.016
35. Xia L, Zhang C, Lv N, et al. AdMSC-derived exosomes alleviate acute lung injury via transferring mitochondrial component to improve homeostasis of alveolar macrophages. Theranostics. 2022;12(6):2928–2947. doi:10.7150/thno.69533
36. Liu C, Xiao K, Xie L. Advances in the regulation of macrophage polarization by mesenchymal stem cells and implications for ALI/ARDS treatment. Front Immunol. 2022;13:928134. doi:10.3389/fimmu.2022.928134
37. Jiang P, Jin Y, Sun M, et al. Extracellular histones aggravate inflammation in ARDS by promoting alveolar macrophage pyroptosis. Mol Immunol. 2021;135:53–61. doi:10.1016/j.molimm.2021.04.002
38. Li H, Li Y, Song C, et al. Neutrophil extracellular traps augmented alveolar macrophage pyroptosis via AIM2 inflammasome activation in LPS-induced ALI/ARDS. J Inflamm Res. 2021;14:4839–4858. doi:10.2147/JIR.S321513
39. Wu D, Zhang H, Wu Q, et al. Sestrin 2 protects against LPS-induced acute lung injury by inducing mitophagy in alveolar macrophages. Life Sci. 2021;267:118941. doi:10.1016/j.lfs.2020.118941
40. Kang R, Zeng L, Zhu S, et al. Lipid peroxidation drives gasdermin D-mediated pyroptosis in lethal polymicrobial sepsis. Cell Host Microbe. 2018;24(1):97–108.e4. doi:10.1016/j.chom.2018.05.009
41. Gou X, Xu W, Liu Y, et al. IL-6 prevents lung macrophage death and lung inflammation injury by inhibiting GSDME- and GSDMD-mediated pyroptosis during pneumococcal pneumosepsis. Microbiol Spectr. 2022;10(2):e0204921. doi:10.1128/spectrum.02049-21
42. Wang J, Sahoo M, Lantier L, et al. Caspase-11-dependent pyroptosis of lung epithelial cells protects from melioidosis while caspase-1 mediates macrophage pyroptosis and production of IL-18. PLoS Pathog. 2018;14(5):e1007105. doi:10.1371/journal.ppat.1007105
43. Zaki MH, Lamkanfi M, Kanneganti TD. The Nlrp3 inflammasome: contributions to intestinal homeostasis. Trends Immunol. 2011;32(4):171–179. doi:10.1016/j.it.2011.02.002
44. Shapouri-Moghaddam A, Mohammadian S, Vazini H, et al. Macrophage plasticity, polarization, and function in health and disease. J Cell Physiol. 2018;233(9):6425–6440. doi:10.1002/jcp.26429
45. Wunderlich CM, Ackermann PJ, Ostermann AL, et al. Obesity exacerbates colitis-associated cancer via IL-6-regulated macrophage polarisation and CCL-20/CCR-6-mediated lymphocyte recruitment. Nat Commun. 2018;9(1):1646. doi:10.1038/s41467-018-03773-0
46. Yao Y, Chen Z, Zhang H, et al. Selenium-GPX4 axis protects follicular helper T cells from ferroptosis. Nat Immunol. 2021;22(9):1127–1139. doi:10.1038/s41590-021-00996-0
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.