Back to Journals » Journal of Inflammation Research » Volume 16
Serum Protein Profiling Reveals a Decrease in Apolipoprotein A-IV During a Clinical Depressive Mood State
Authors Mun S , Lee S , Yun Y , Joo EJ , Kang HG , Lee J
Received 28 April 2023
Accepted for publication 26 August 2023
Published 5 September 2023 Volume 2023:16 Pages 3925—3936
DOI https://doi.org/10.2147/JIR.S419176
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Sora Mun,1 Seungyeon Lee,2 Yeeun Yun,3 Eun-Jeong Joo,4,5 Hee-Gyoo Kang,1– 3 Jiyeong Lee1,3
1Department of Biomedical Laboratory Science, College of Health Sciences, Eulji University, Gyeonggi, Republic of Korea; 2Department of Senior Healthcare, Graduate School, Eulji University, Gyeonggi, Republic of Korea; 3Department of Biomedical Laboratory Science, Graduate School, Eulji University, Gyeonggi, Republic of Korea; 4Department of Neuropsychiatry, School of Medicine, Eulji University, Daejeon, Republic of Korea; 5Department of Psychiatry, Uijeongbu Eulji Medical Center, Eulji University, Gyeonggi, Republic of Korea
Correspondence: Hee-Gyoo Kang, Department of Biomedical Laboratory Science, College of Health Sciences, Eulji University, 553 Sanseong-daero, Sujeong-gu, Seongnam-si, Gyeonggi-do, Republic of Korea, Tel +82-31-740-7315, Email [email protected] Jiyeong Lee, Department of Biomedical Laboratory Science, College of Health Sciences, Eulji University, 712 Dongil-ro, Uijeongbu-si, Gyeonggi-do, Republic of Korea, Tel +82-31-951-3862, Email [email protected]
Purpose: Depressive mood is a major psychiatric symptom that causes serious disturbances in daily life. Unlike physical symptoms, psychiatric symptoms are more difficult to evaluate objectively. Therefore, we aimed to discover biomarkers that reflect changes in serum protein metabolism during a clinical depressive mood.
Methods: Serum protein profiling was conducted in participants who were not experiencing a current depressive episode (healthy individuals and patients in remission). Serum proteins were identified and quantified using liquid chromatography–tandem mass spectrometry. Differentially expressed proteins with a p-value < 0.05 were selected, and candidate biomarkers were verified using multiple reaction monitoring analysis for absolute quantification.
Results: Apolipoprotein A-IV levels were lower in the group with a current episode of depression than in the remission and healthy control groups. Further, fibronectin levels were also lower in the group with a current episode of depression than in the healthy control group but not in the remission group.
Conclusion: We found that apolipoprotein A-IV-mediated inflammation is involved in clinical depressive moods, possibly by inducing neurological changes in the brain. Therefore, apolipoprotein A-IV and fibronectin levels may be explored as potentially novel biomarkers for detecting a current episode of depression.
Keywords: biomarkers, depression, liquid chromatography–tandem mass spectrometry, inflammation, neuroinflammation, serum proteomics
Introduction
The World Health Organization defines depression as a clinical symptom of sadness, irritability, emptiness, and depressive mood (like occasional suicidal ideation and behavior, a loss of pleasure or interest in activities, and anhedonia) lasting for at least two weeks.1 It causes difficulties in daily life due to various negative emotions and physical symptoms, such as sleep disorders, loss of appetite, and weight loss.1,2 The diagnosis of depression is generally based on the clinical medical examination through interviews and questionnaires about subjective mood states, such as the Beck Depression Inventory (BDI) and the Hamilton Rating Scale for Depression-17, filled out by the patient.3 According to a recent study, while environmental and other risk factors make up the majority of depression risk factors, some genetic risk factors also play a role.1 Chronic stress and stressful early life events can also cause depression.2 Alterations in several genes have been identified as potential risk factors for depression, including the genes, dopamine receptor D4, 5-hydroxytryptamine receptor 1A, monoamine oxidase A, piccolo presynaptic cytomatrix protein, solute carrier family 6 member 3, solute carrier family 6 member 4, and tryptophan hydroxylase 2. These genetic variations are involved in exchange of monoamine neuromediators. However, these genetic alterations are not the only risk factors that lead to depression. Depression is a complex and multifactorial condition influenced by various biological, psychological, and environmental factors.
Unlike physical symptoms, the depressive mood is more difficult to judge objectively and can be judged only by subjective evaluation. A depressive mood lasting at least 2 weeks is classified as morbid depression, but it is not easy to distinguish from the routine negative emotions that occur in daily life. Therefore, biomarkers that reflect the depressive mood state in patients with major depressive disorder (MDD) and bipolar disorder (BD) could provide clues in understanding the mechanism underlying clinical depressive episodes, ultimately facilitating the prevention and treatment of depressive disorders. Given that the need for objective biomarkers for depression has been recognized, several studies have attempted to discover serum biomarkers of depression.4,5 These studies compared the expression patterns of serum proteins between patients with MDD and healthy individuals. Studies on identification of common biomarkers for depressive moods shared by multiple diseases, such as MDD, attention deficit hyperactivity disorder (ADHD), anxiety disorder, and BD have not been performed to date. Although these conditions have distinct underlying mechanisms, they share a common feature causing a negative mood. Identification of biomarkers that determine the presence or absence of depressive mood shared by different diseases may help in understanding the relation between the underlying mechanisms that contribute to depression.
To this goal, in this study serum protein profiling of patients experiencing a depressive episode was performed. Differentially expressed proteins in patients with a current depressive episode were compared to those in both healthy controls (ie, those without a history of clinical depressive mood state) and remission patients (ie, those who had previously experienced depressive episode but had been without any issues for at least 2 weeks) as controls. Although the patients in remission did not have clinical depression at that moment, they had physiological similarities to those in a current depressive episode. It has been reported that persistent inflammatory reactions are activated during a depressive episode and that the levels of inflammatory factors remain elevated even during remission.6,7 Therefore, remission patients were used to exclude biomarkers such as altered inflammatory factors in a past depressive episode. In addition, although both the remission and healthy individuals were considered asymptomatic, there was a difference in the degree of depressive mood between them. Depression levels are closer to zero in healthy individuals than in remission patients.3 Therefore, this study also aimed to discover biomarkers that reflect the extent of depressive mood and the presence of a current depressive episode.
Biomarkers that accurately reflect the presence and severity of a depressive episode would significantly improve diagnostic accuracy and provide insights into the underlying biological mechanisms involved in depression. Prior studies have identified certain biomarkers specific to individual mood disorders, but a comprehensive investigation into shared biomarkers across different depressive mood conditions, such as MDD and BD, is scarce. Thus, the present study aims to discover novel serum biomarkers that change during a current depressive episode in both MDD and BD. We hypothesize that specific serum proteins levels are associated with a current depressive episodes in both, MDD and BD patients. We anticipate that these profiles will differ from those observed in healthy individuals without any history of clinical depression and in remission patients who previously experienced depression but are currently asymptomatic. By comparing these groups, we aim to identify distinct biomarkers associated with current depressive states while excluding those influenced by past inflammatory reactions.
Materials and Methods
Participants
This study evaluated participants with and without clinical depressive episode (CDE) from Eulji University Hospital (Nowon-gu, Seoul, Republic of Korea). Patients with a current depressive episode including MDD and BD, were diagnosed by a psychiatrist based on the clinical examination through interviews and questionnaires about subjective mood states, such as the BDI and the Hamilton Rating Scale for Depression-17, filled out by the patient.8,9 There were 22 patients with a CDE in the discovery set and 49 patients with a CDE in the validation set (Table 1). Among 49 patients with a CDE in the validation set, patients with BD experiencing a current depressive episode (n = 13) and patients with MDD (n = 36) were included. As comparative controls, participants without CDE were divided into the following three groups: the first group included participants who had a history of clinical depressive episode (MDD or BD) and were treated with medication (remission controls with medication treatment [RCM]); the second group included those who had a history of clinical depressive episode (MDD or BD) but were not treated with medication (remission controls with no medication treatment [RC]); and the third group included the participants who had no history of clinical depressive episode and had a normal BDI score of <13 (healthy controls [HC] who had no history of depressive mood disorder and medication treatment). The first group (RCM) involved 19 participants in the discovery set and 48 in the validation set (MDD: n = 35 and BD: n = 13); the second group (RC) involved 5 participants in the discovery set and 7 in the validation set; and the third group (HC) involved 21 participants in the discovery set and 59 in the validation set (Table 1). Participants in the discovery set underwent serum protein profiling using liquid chromatography–tandem mass spectrometry (LC-MS/MS). The selected candidate biomarkers were verified using multiple reaction monitoring (MRM) analysis by absolutely quantifying the candidate biomarkers.
 |
Table 1 Participant Characteristics |
Serum Sample Collection and Processing for Mass Spectrometry Analysis
The serum of patients with clinical depression and asymptomatic controls, including patients in remission, was collected from Eulji University Hospital. Blood samples were obtained in an anticoagulant-free vacutainer. After 2 h at 24°C, blood samples were centrifuged at 4000 × g for 5 min to separate the serum. To separate six proteins highly abundant in human serum, a multiple-affinity removal system comprising a liquid chromatography (LC) column (human 6-HC, 4.6 × 50 mm; Agilent Technologies, Santa Clara, CA, USA) was used before tryptic digestion, as previously described.10 Briefly, samples were loaded onto the column, and the eluted high- and low-abundance proteins were collected separately. The low-abundance proteins were used for analysis. Proteins in the samples were concentrated using a Nanosep Centrifugal Device with an Omega™ Membrane 3 K (Pall Corporation, Port Washington, NY, USA).
Determination of Protein Concentration and Tryptic Digestion
To measure the levels of serum protein, a bicinchoninic acid (BCA) Protein Assay Kit (Thermo Fisher Scientific, Cleveland, OH, USA) was used in accordance with the manufacturer’s instructions. All samples were prepared to a concentration of 100 μg of protein in a total volume of 50 μL. For reduction, samples were prepared as peptides via treatment with 5 mM Tris (2-carboxyethyl) phosphine (Pierce, Rockford, IL, USA) at 37°C, centrifuged at 400 rpm for 30 min, and then treated with 15 mM iodoacetamide (Sigma-Aldrich, St. Louis, MO, USA) for alkylation by incubating at 25°C for 1 h in the dark. Finally, serum proteins were cleaved into peptides by mass spectrometry (MS)-grade trypsin gold (Promega, Madison, WI, USA) at 37°C overnight. The cleavage products were desalted using a C18 cartridge (Waters, Milford, MA, USA). Cleaned samples were dried using a vacuum dryer (Scan Vac, LaboGene, Lynge, Denmark).
Sample Protein Fractionation and LC-MS/MS Analysis
All individual samples were pooled, and the pooled samples were analyzed by the data-dependent acquisition (DDA, also known as information-dependent acquisition, or IDA) method, and this data was used to create a protein library. Pooled serum samples were separated into 12 fractions using a 3100 OFFGEL Low Res Kit (pH 3–10; Agilent Technologies) in accordance with the manufacturer’s instructions. The cleavage products were cleaned using a C18 Macro spin column (Harvard Apparatus, Holliston, MA, USA). Cleaned samples were dried using a vacuum dryer (Scan Vac, LaboGene). The obtained twelve fractions were loaded onto an Eksigent nanoLC 400 system and the cHiPLC® (AB Sciex, Concord, ON, Canada). Apart from the pooled sample, the individual samples were analyzed for relative analysis using a TripleTOF 5600 mass spectrometer (AB Sciex) by sequential window acquisition of all theoretical mass spectra (SWATH) acquisition which is based on the data-independent acquisition (DIA) method. In each run, samples containing 1 µg/µL were injected onto an Eksigent ChromXP nanoLC trap column (350 µm i.d. × 0.5 mm, ChromXP C18 3 µm) at a flow rate of 5000 nL/min. Thereafter, samples were eluted from the Eksigent ChromXP nanoLC column (75 µm i.d. × 15 cm) at a flow rate of 300 nL/min for 95 min. Mobile phase B buffer was added gradually into the column (5–90%) over a 95-min total run time. The gradient of the mobile phase B buffer was as follows: (time/% B) 0 min/mobile phase B 5%, 10.5 min/40%, 80 min/90%, and 95 min/5%.
Synthesis and Purification of Label-Free Standard Peptides
Two candidate proteins were selected as potential novel biomarkers that reflected the current active depressive mood. Peptides for absolute quantification were synthesized by Peptron Co. (Yousung, Korea). The following criteria were set for peptide selection: peptides that did not involve miscleaved sites, unmodified peptides, peptides without Met, within 7–15 residues, and with a low false discovery rate (<1). After peptide standards were synthesized, two-fold serial dilutions were conducted from 1 μM peptide in high-performance liquid chromatography (HPLC)-grade water in accordance with the manufacturer’s protocol. Particular peptide(s) for study as candidate biomarker was selected based on the results of SWATH acquisition. The best-performing peptide in the SWATH acquisition was selected as a final candidate peptide to reflect the protein expression pattern. We confirmed that the selected peptide had similar patterns to its respective protein expression patterns. The final goal of MRM analysis was applying these proteins as biomarkers in the clinical field for rapid, easy, and efficient diagnosis. Therefore, a peptide with powerful and strong expression was needed.
Label-Free Quantification Through Multiple Reaction Monitoring Analysis
MRM Q1/Q3 ion pairs of peptides were selected using the Skyline library for absolute quantification. Voltage parameters for each transition, including collision energy, declustering potential, and collision cell exit potential, were determined using the Skyline library or compound optimization. An SCIEX Exion LC was used to segregate samples by using a C18 column (ACQUITY UPLC BEH C18 Column [130 Å, 1.7 µm, 2.1 mm × 150 mm] with an ACQUITY UPLC BEH C18 VanGuard Pre-column [130 Å, 1.7 µm, 2.1 mm × 5 mm]). Thereafter, samples were analyzed using a QTRAP 5500 (AB Sciex) in symptomatic patients with a depressive mood and asymptomatic controls. Each sample was loaded onto an LC column with a gradient of 5–90% mobile phase B and a total run time of 30 min. The mobile phase B buffer was gradually introduced into the LC column: (time/% B) 1 min/mobile phase B 5%, 50 min/40%, 21–25 min/90%, 25.5–30 min/5%. Mobile phase B comprised 0.1% formic acid in HPLC-grade acetonitrile, while mobile phase A comprised 0.1% formic acid in HPLC-grade water. The source parameters for MRM analysis were as follows: curtain gas, 30 psi; low collision gas; ion spray voltage, 5500 V; temperature, 400°C; ion source gas 1, 40 psi; and ion source gas 2, 60 psi.
Statistical Analysis
Data processing was conducted using PeakView for peak detection and MarkerView software for total sum normalization and t-tests. Proteins with a p-value <0.05 were further analyzed. Results of MRM analysis were processed using MultiQuant software to calculate the absolute concentration of the analyte. Groups were compared using the Mann–Whitney test. Statistical analysis was performed using GraphPad Prism software version 5.0 (La Jolla, CA, USA).
Results
Sample Characteristics
The top five symptoms of depressive mood, as evaluated using the BDI score, were anxiety, nervousness, a decline in cognitive function, physical symptoms, and increased dreams, in that order (Table 1). Moreover, 86.36% of patients with depressive mood disorder had anxiety and nervousness, 63.64% had declined cognitive function, 54.54% had physical symptoms, and 50% had more frequent dreams. In addition, some of those patients had attempted suicide, were volatile, had nausea, demonstrated weight increase, experienced increased sleepiness, showed psychotic symptoms, had a history of panic disorder, had increased appetite, and were dysphoric or had mixed manic features (Table 1).
Sample Distribution
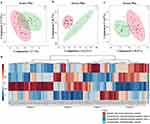
Sparse partial least squares discriminant analysis (sPLS-DA) was conducted to analyze the expression pattern of identified proteins in sample groups to distinguish CDE from three control subjects without CDE (RCM, RC, and HC). sPLS-DA analysis is based on the sample protein profile that is at least identified in one sample group. In the protein profiling list, if the expression of some proteins cannot be confirmed in a certain sample, a relative intensity value of 0 is displayed. CDE and RCM were identified as component 1 (7.7%) and component 2 (7.3%), respectively (Figure 1a). CDE and RC were identified as component 1 (13.2%) and component 2 (8.7%), respectively (Figure 1b). CDE and HC were identified as component 1 (8.4%) and component 2 (6.5%), respectively (Figure 1c). A total of 22 proteins were found to be differentially expressed between CDE and the three control samples (RCM, RC, and HC). These were divided into four clusters according to their expression patterns by heatmap analysis (Figure 1d). Each group’s average protein relative intensity was used for heatmap clustering analysis. The expressions of proteins in Cluster 1 were markedly upregulated in RC compared to that in the other groups. The expressions of proteins in Cluster 2 were markedly downregulated in HC than in the other groups. A pattern consistent with the total protein in cluster 3 could not be identified. The expressions of proteins in Cluster 4 were markedly downregulated in RC than those in the other groups.
Differentially Expressed Proteins in CDE
Among the identified 89 proteins, 22 were differentially expressed between CDE and the other groups (Table 2), with more than a 1.2-fold change and p < 0.05. The expressions of five proteins were upregulated and while those of another five proteins were downregulated in CDE than in RCM (Figure 2a). The expressions of seven proteins were upregulated, and those of six proteins were downregulated in CDE than those in RC (Figure 2b). The expression of one protein was upregulated and while that of seven proteins were downregulated in CDE than those in HC (Figure 2c).
 |
Table 2 Differentially Expressed Proteins with Statistical Significance (P<0.05) |
Meanwhile, although the expressions of 22 proteins were up- or downregulated in CDE than those in the other three control groups (Figure 2a–c), the expressions of two proteins, apolipoprotein A-IV (P06727) and fibronectin (P02751), were significantly downregulated in CDE than those in all the three control groups (p < 0.05) (Figure 2d). Apolipoprotein A-IV and fibronectin were therefore selected as candidate biomarkers for distinguishing CDE from those who do not have present clinical depressive episodes or healthy controls. Twenty-two proteins were clustered according to their expression patterns using heatmap analysis, and apolipoprotein A-IV and fibronectin were indicated by black arrows (Figure 2e). The average protein relative intensity of each group was used for clustering.
MRM Analysis for Verification of Candidate Biomarkers
For validation, MRM analysis was conducted for absolute quantification. One peptide each of apolipoprotein A-IV and fibronectin was selected and analyzed (Table 3). We found that apolipoprotein A-IV expression was significantly downregulated in CDE than that in all the other control groups (p < 0.05) (Figure 3a). Fibronectin expression was also significantly downregulated in CDE than in HC who had no history of clinical depressive mood state and had a BDI score of <13 (p < 0.05) (Figure 3b). Compared to the subjects who had a history of clinical depression, fibronectin levels were not significantly downregulated in CDE.
 |
Table 3 MRM Parameters for Candidate Protein Verification |
Apolipoprotein A-IV expression was significantly lower in CDE than that in all the three control groups. The study’s approach of utilizing remission patients (RCM and RC) as controls was crucial in excluding biomarkers influenced by past inflammatory reactions. By selecting remission patients who had experienced depression but were currently asymptomatic and subdividing them into groups with and without drug treatment effects, the study effectively identified biomarkers that are specific to the current depressive state and free from drug-related interference.
Meanwhile, the fibronectin levels of CDE were only different compared to those of HC, with lower levels in those with a CDE. The decrease in fibronectin expression was significantly more pronounced in HC than in that remission patients (RCM and RC), which suggest that remission patients (RCM and RC) have not yet recovered to the same status as HC. Compared with the remission patients (RCM and RC), the HC showed depression levels closer to zero. Therefore, fibronectin levels may be useful in determining whether the extent of depressive mood had a sufficiently low depressive mood score (closest to 0) and whether the patients are at risk of relapse.
Discussion
It has been reported that high levels of inflammatory and immune responses are accompanied by a clinically relevant depressive mood state.2,11–13 Antidepressant drugs that reduce the inflammatory response have been used to suppress depression. The use of antidepressants that target the inflammatory response demonstrates a direct association of inflammation with depression. Inflammation levels also reduce the effectiveness of antidepressants.12 As such, it can be inferred that depression and the inflammatory response are directly related and that if the level of inflammation is high, it is difficult to recover from depression by treatment with antidepressants. Some studies have shown that inflammation levels do not return to normal, and retain at a high inflammatory response, equal to that in a disease state not only in patients with current episodes of depression, but also in remission patients in whom the depression has disappeared.6,7 For example, levels of inflammatory markers, such as those of C-reactive protein or cytokines, are high even in a remission state. Functional analysis in this study confirmed that most of the identified proteins that showed similar expression levels between samples with and without current clinical depressive moods were related to the pro-inflammatory response. The inflammatory response is not only activated in the current depressive mood but also indicative of the past depressive mood.
Therefore, inflammation-related proteins that reflect the disappearance of current depressive episodes can be used as a beneficial indicator to evaluate the current depressive episode. The expression of apolipoprotein A-IV, which is involved in the inflammatory response, was decreased in CDE in this study. Apolipoprotein A-IV plays an anti-inflammatory role.14 Depression coincides with an upsurge in the pro-inflammatory response, and this inflammation might persist even when the condition is in remission.6,7 However, as the ongoing depressive episode abates, it can be assumed that the anti-inflammatory response normalizes, accompanied by an augmentation of anti-inflammatory proteins, such as apolipoprotein A-IV. Our finding of the difference in apolipoprotein A-IV expression in all the three control groups (RCM, RC, and HC) compared to CDE indicates that the apolipoprotein A-IV-mediated anti-inflammatory response is reactivated after the depressive mood disappears.
This raises the question of the relationship between the inflammatory response and the neurological changes that cause a depressive mood state. It has been reported that inflammatory and immune responses cause neurological changes and that neurological changes are followed by psychological symptoms.12,15 Activation of the inflammatory response in patients with depressive mood states, including MDD and BD, does not result from the depressive mood itself; rather, it triggers the depressive mood and subsequent neurological changes and results in psychological symptoms.12,15 Apolipoprotein A-IV cannot cross the blood–brain barrier.14 Therefore, it could be inferred that the depressive mood state occurs due to neurological changes in the brain initiated by the deactivation of an apolipoprotein A-IV-mediated anti-inflammatory response in the blood. Decreased apolipoprotein A-IV expression, as a blood biomarker, thus has the potential to trigger neurological changes that cause psychotic symptoms by inducing an inflammatory response. The decrement of this anti-inflammatory protein is an interesting finding, given that the pro-inflammatory substances are generally similar in remission patients. Apolipoprotein A-IV is expected to be used as an objective biomarker to discriminate clinical depressive mood.
In addition, environmental risk factors such as stress are involved in the hypothalamic–pituitary–adrenal (HPA) axis that is hyperactivated in depression.2,16 Chronic stress can dysregulate the HPA axis and contribute to developing mood and cognitive disorders. Research has shown that individuals with mood and cognitive disorders have alterations in the HPA axis function, including the stimulated release of the stress hormone cortisol.1–3 Dysregulation of the HPA axis has been associated with a range of psychiatric disorders, including MDD, BD, anxiety, and ADHD. Interestingly, the previous report that apolipoprotein A-IV represses HPA suggests that lowered apolipoprotein A-IV may destroy the homeostasis of the brain system following depressive mood disorder.4 It has been reported that a decrease in apolipoprotein A-IV level in the blood is associated with several diseases, such as coronary artery disease and diabetes.13,17,18 An association with the apolipoprotein family of proteins in depression has also been reported previously.19–21 However, to our best knowledge, there have been no reports of a decrease in serum apolipoprotein A-IV levels in patients with depressive mood, including MDD and BD, to date.
This study has some limitations. First, this study did not investigate the mechanism by which a deficiency in apolipoprotein A-IV could lead to psychotic symptoms, as it aimed to identify biomarkers for active clinical depressive mood. These mechanisms should be investigated further in the future. Second, although the extent of depressive mood in the healthy control group is evaluated based on the BDI score, which is the most widely used questionnaire for this purpose, and although the healthy controls were only included if their BDI score was <13, they were not evaluated by a psychiatrist. Thus, they may not have been truly normal healthy controls. However, the present study holds several strengths. First, the study’s findings with regards to apolipoprotein A-IV are unique and noteworthy. The decrease in apolipoprotein A-IV expression in CDE, as compared to RCM, RC, and HC, provides valuable insights into the potential role of this anti-inflammatory protein in depression in the inflammatory response and the neurological changes that cause a depressive mood state. The study’s investigation of the relationship between the inflammatory response and neurological changes contributing to a depressive mood state adds a new insight to our understanding of the underlying mechanisms of depression. Second, the study’s thorough consideration of various factors, such as chronic stress and the involvement of the HPA axis, in relation to depression helps in establishing a comprehensive framework for studying the complex interplay between biological and environmental factors in depressive mood disorders.
Conclusion
Apolipoprotein A-IV levels in the blood are decreased in patients with a CDE. This implies that apolipoprotein A-IV-mediated inflammation is involved in a clinical depressive mood, possibly by inducing neurological changes in the brain. Meanwhile, fibronectin levels are decreased and thus may be useful to determine whether the extent of depressive mood had a sufficiently low depressive mood score (close to 0) and whether the patient is at risk of a relapse. The findings of this study aims to contribute to the development of precise and reliable methods for diagnosing depression and to provide valuable insights into the biological mechanisms underlying current depressive episodes across various mood-related disorders. Ultimately, these findings may lead to improved treatments and personalized approaches for managing depression.
Abbreviations
BDI, Beck Depression Inventory; MDD, major depressive disorder; BD, bipolar disorder; ADHD, Attention deficit hyperactivity disorder; CDE, Clinical depressive episode; HC, Healthy controls; RCM, Remission controls with medication treatment; RC, Remission controls with no medication treatment; LC-MS/MS, liquid chromatography–tandem mass spectrometry; MRM, multiple reaction monitoring; sPLS-DA, Sparse partial least squares discriminant analysis; HPA, hypothalamic–pituitary–adrenal.
Data Sharing Statement
The data presented in this study are available on request from the corresponding author.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Institutional Review Board of EULJI UNIVERSITY (protocol code EUIRB2022-051, August 1, 2022)
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Acknowledgments
This work was supported by a National Research Foundation of Korea (NRF) grant funded by the Korean Government (MSIT) (No. 2020R1C1C1009196).
Funding
This research was funded by the NATIONAL RESEARCH FOUNDATION OF KOREA (NRF) funded by the Korean Government (MSIT), grant number 2020R1C1C1009196. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. Malhi GS, Mann JJ. Depression. Lancet. 2018;392(10161):2299–2312. doi:10.1016/S0140-6736(18)31948-2
2. Iob E, Kirschbaum C, Steptoe A. Persistent depressive symptoms, HPA-axis hyperactivity, and inflammation: the role of cognitive-affective and somatic symptoms. Mol Psychiatry. 2020;25(5):1130–1140. doi:10.1038/s41380-019-0501-6
3. Severe J, Greden JF, Reddy P. Consequences of recurrence of major depressive disorder: is stopping effective antidepressant medications ever safe? Focus. 2020;18(2):120–128. doi:10.1176/appi.focus.20200008
4. Choi H, Mun S, Joo EJ, Lee KY, Kang HG, Lee J. Serum proteomic analysis of major depressive disorder patients and their remission status: novel biomarker set of zinc-alpha-2-glycoprotein and keratin type II cytoskeletal 1. Int J Biol Macromol. 2021;183:2001–2008. doi:10.1016/j.ijbiomac.2021.05.172
5. van Haeringen M, Milaneschi Y, Lamers F, Penninx BWJH, Jansen R. Dissection of depression heterogeneity using proteomic clusters. Psychol Med. 2023;53(7):2904–2912. doi:10.1017/S0033291721004888
6. Carlier A, Berkhof JG, Rozing M, et al. Inflammation and remission in older patients with depression treated with electroconvulsive therapy; findings from the MODECT study✰. J Affect Disord. 2019;256:509–516. doi:10.1016/j.jad.2019.06.040
7. Dittrich K, Boedeker K, Kluczniok D, et al. Elevated inflammatory markers in women with remitted major depressive disorder and the role of early life maltreatment. Brain Behav Immun. 2021;97:219–225. doi:10.1016/j.bbi.2021.07.024
8. Dubovsky SL, Ghosh BM, Serotte JC, Cranwell V. Psychotic depression: diagnosis, differential diagnosis, and treatment. Psychother Psychosom. 2021;90(3):160–177. doi:10.1159/000511348
9. McCarron RM, Shapiro B, Rawles J, Luo J. Depression. Ann Intern Med. 2021;174(5):ITC65–ITC80. doi:10.7326/AITC202105180
10. Mun S, Lee J, Park M, Shin J, Lim MK, Kang HG. Serum biomarker panel for the diagnosis of rheumatoid arthritis. Arthritis Res Ther. 2021;23(1):31. doi:10.1186/s13075-020-02405-7
11. Köhler CA, Freitas TH, Maes M, et al. Peripheral cytokine and chemokine alterations in depression: a meta-analysis of 82 studies. Acta Psychiatr Scand. 2017;135(5):373–387. doi:10.1111/acps.12698
12. Lee CH, Giuliani F. The role of inflammation in depression and fatigue. Front Immunol. 2019;10:1696. doi:10.3389/fimmu.2019.01696
13. Serafini G, Costanza A, Aguglia A, et al. The role of inflammation in the pathophysiology of depression and suicidal behavior: implications for treatment. Med Clin North Am. 2023;107(1):1–29. doi:10.1016/j.mcna.2022.09.001
14. Wang F, Kohan AB, Lo CM, Liu M, Howles P, Tso P. Apolipoprotein A-IV: a protein intimately involved in metabolism. J Lipid Res. 2015;56(8):1403–1418. doi:10.1194/jlr.R052753
15. Miyata S, Ishino Y, Shimizu S, Tohyama M. Involvement of inflammatory responses in the brain to the onset of major depressive disorder due to stress exposure. Front Aging Neurosci. 2022;14:934346. doi:10.3389/fnagi.2022.934346
16. Sheng JA, Bales NJ, Myers SA, et al. The hypothalamic-pituitary-adrenal axis: development, programming actions of hormones, and maternal-fetal interactions. Front Behav Neurosci. 2020;14:601939. doi:10.3389/fnbeh.2020.601939
17. Li J, Song M, Qian D, et al. Decreased plasma apolipoprotein A-IV levels in patients with acute coronary syndrome. Clin Invest Med. 2013;36(4):E207–E215. doi:10.25011/cim.v36i4.19954
18. Wang Z, Wang L, Zhang Z, Feng L, Song X, Wu J. Apolipoprotein A-IV involves in glucose and lipid metabolism of rat. Nutr Metab (Lond). 2019;16:41. doi:10.1186/s12986-019-0367-2
19. Hui L, Han M, Du XD, et al. Serum ApoB levels in depressive patients: associated with cognitive deficits. Sci Rep. 2017;7:39992. doi:10.1038/srep39992
20. Sadeghi M, Roohafza H, Afshar H, et al. Relationship between depression and apolipoproteins A and B: a case-control study. Clinics. 2011;66(1):113–117. doi:10.1590/S1807-59322011000100020
21. Wang WW, Liu XL, Ruan Y, Wang L, Bao TH. Depression was associated with apolipoprotein E ε4 allele polymorphism: a meta-analysis. Iran J Basic Med Sci. 2019;22(2):112–117. doi:10.22038/ijbms.2018.30825.7436
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.