Back to Journals » Biologics: Targets and Therapy » Volume 17
Serum LINC00152 and UCA1 in HCV-Induced Hepatocellular Carcinoma: Clinical Significance and Prognostic Value
Authors Shehab-Eldeen S , Essa A , Arafat ES, Sleem AS, Alhosary AA, Darwish E , Essa A, Al-Omair OA , Al-Khoufi EA, Al Abdulqader AK, Nada A
Received 4 August 2023
Accepted for publication 7 October 2023
Published 13 October 2023 Volume 2023:17 Pages 137—149
DOI https://doi.org/10.2147/BTT.S433872
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Doris Benbrook
Somaia Shehab-Eldeen,1,2 Abdallah Essa,1,2 Eman Salah Arafat,3 Asmaa Shaaban Sleem,4 Amal Abdelrasoul Alhosary,5 Ehab Darwish,6,7 Ali Essa,8 Omar Ahmed Al-Omair,1 Emad Ali Al-Khoufi,1 Abdulrhman Khaled Al Abdulqader,1 Ali Nada9
1Internal Medicine Department, College of Medicine, King Faisal University, Al-Ahsa, Kingdom of Saudi Arabia; 2Tropical Medicine Department, Faculty of Medicine, Menoufia University, Shebin Elkom, Egypt; 3Medical Biochemistry and Molecular Biology Department, Faculty of Medicine, Menoufia University, Shebin Elkom, Egypt; 4Medical Microbiology and Immunology Department, Faculty of Medicine, Menoufia University, Shebin Elkom, Egypt; 5Clinical Pathology Department, Faculty of Medicine, Menoufia University, Shebin Elkom, Egypt; 6Tropical Medicine Department, Faculty of Medicine, Zagazig University, Zagazig, Egypt; 7Gastroenterology and Infectious Diseases Unit, College of Medicine, King Faisal University, Al-Ahsa, Kingdom of Saudi Arabia; 8Medical Student, Faculty of Medicine, Menoufia University, Shebin Elkom, Egypt; 9Hepatology and Gastroenterology Department, National Liver Institute, Menoufia University, Shebin Elkom, Egypt
Correspondence: Somaia Shehab-Eldeen, Internal Medicine Department, College of Medicine, King Faisal University, Al-Ahsa, 31982, Kingdom of Saudi Arabia, Tel +966507913675 ; +201117251523, Email [email protected]; [email protected]
Background: Despite significant advancements in the molecular characterization of hepatocellular carcinoma (HCC), no oncogene addiction has been discovered. Long noncoding RNAs (lncRNAs) have a lot of promise as cancer biomarkers. LINC00152 and UCA1 have shown potential as diagnostic, prognostic, and therapeutic targets for human cancers.
Aim: To investigate the diagnostic and prognostic potential of serum LINC00152 and UCA1 in hepatocellular carcinoma (HCC).
Methods: The expression levels of LINC00152 and UCA1 in blood samples from 120 patients (60 with HCC, 60 with liver cirrhosis) and 40 healthy subjects were assessed using real-time qRT-PCR.
Results: Serum LINC00152 and UCA1 expression were considerably higher in HCC patients compared to patients with liver cirrhosis and the healthy controls (p< 0.001 and p< 0.001 respectively). And their expressions in the liver cirrhosis group were significantly higher than in healthy controls. Both lncRNAs performed well in the ROC analysis, distinguishing HCC patients from patients with liver cirrhosis. Higher levels of LINC00152 expression were linked to lesions in both lobes of the liver (p=0.02), while higher levels of UCA1 expression were linked to vascular invasion and the late stage (p=0.01, p=0.03 respectively). The multivariate analysis showed that a high level of LINC00152 in the blood was an independent indicator of a bad outcome for HCC patients (HR=2.23, 95% CI= 1.30– 5.29, p=0.03).
Conclusion: Serum LINC00152 and UCA1 expression were upregulated in patients with HCC, suggesting their use as non-invasive biomarkers for HCC. Furthermore, LINC00152 has the potential to serve as a prognostic indicator.
Keywords: LINC00152, UCA1, hepatocellular carcinoma, predictor, mortality
Introduction
Hepatocellular carcinoma (HCC) is among the most prevalent types of cancer. It accounts for approximately 80–90% of liver cancer cases. Furthermore, it is characterized by a high incidence and mortality rate.1 HCC epitomizes a giant public health problem in Egypt, occupying the 1st and 2nd cancers to occur in men and women, respectively, based on data from the National Cancer Registry Program of Egypt.2,3
HCC is usually caused by liver cirrhosis, which is caused by infections with the hepatitis B or C viruses, exposure to aflatoxins, persistent alcohol use, or nonalcoholic fatty liver disease (NAFLD), all of which enhance the risk of HCC.4
The common HCC screening methods include imaging techniques or serum biomarkers such as alpha-fetoprotein (AFP).5 Serum AFP is the most commonly used biomarker for HCC screening, diagnosis, assessment of treatment effectiveness, and prognosis.4 However, in many cases of HCC reporting negative AFP, further AFP may be elevated in liver cirrhosis or viral hepatitis.5 The sensitivity and specificity of AFP are limited, especially with early-stage HCC. As a result, it is critical to develop novel and noninvasive biomarkers with high sensitivity and specificity for HCC diagnosis and prognosis.6,7
Long non-coding RNAs, also known as lncRNAs, are a subclass of non-coding RNAs (ncRNAs) that are distinguished by their length, which is more than 200 nucleotides.8 They are mostly located in the cytosol, where they target mRNAs and down‐regulate protein translation.9
Many studies have shown the direct and indirect regulatory effects of lncRNAs on cancer biology.10 It has been shown that these lengthy molecules are commonly dysregulated in a range of malignancies and that certain lncRNAs are related to cancer recurrence, metastasis, and poor prognosis in several cancers.11 As a result, it is believed that the particular lncRNA biomarkers associated with the prognosis and diagnosis of HCC are of tremendous clinical value.12
LINC00152, also known as long intergenic non-coding RNA 152, is a lncRNA that is 828 base pairs long and is found on chromosome 2p11.2. It has been shown to influence genes in a variety of different ways, such as epigenetic changes and interactions between lncRNA-miRNA23, and lncRNA-protein.13 Intergenic lncRNA LINC00152, or CYTOR (cytoskeleton regulator RNA), was first discovered to operate as a non-coding oncogene by regulating cell cycle progression by interaction with a network of proteins involved with the M phase of the cell cycle.14
LINC00152 shows promise as a diagnostic, prognostic, and therapeutic target for human malignancies.14 It is overexpressed in a wide variety of malignancies, including glioma, retinoblastoma, lung cancer, kidney cancer, colon cancer, and gastric carcinoma.15 Most LINC00152 transcripts in hepatocellular carcinoma were found in the nucleus of the cells. It stimulates the mechanistic target of the rapamycin (mTOR) pathway, which is critical in the control of cancer cell proliferation, division, and carcinogenesis.13
The long noncoding RNA Urothelial cancer associated 1 (UCA1), also known as CUDR (cancer upregulated drug resistant), is situated at chromosome 19p13.12. Its upregulation has been documented in many cancers.16 Evidently, significant roles in colorectal, prostate, stomach, and bladder cancers have been attributed to it.17
On top of that, UCA1 may be used as a negative biomarker for predicting malignant phenotypes and prognosis. UCA1 may increase tumor cell tamoxifen resistance in breast cancer.18 This implies that UCA1 might also have a special function in the management of certain cancers. UCA1 expression has been linked to many clinical characteristics and malignant behaviors in HCC tissues and cell lines.18
The purpose of this research was, therefore, to evaluate the diagnostic and prognostic functions that LINC00152 and UCA1 play in HCC.
Subjects and Methods
Subjects
The present case-control study includes a total of 160 individuals. They were divided into the following categories: HCV-induced HCC (60 patients), HCV-induced liver cirrhosis (60 patients), and age- and gender-matched healthy controls (40 individuals). All the patients were selected from the inpatient and/or outpatient clinics of the National Liver Institute, Menoufia University, during the period from May 2019 to December 2019. A thorough history was taken from each participant in this study, followed by a clinical examination.
Clinical evaluation, laboratory analysis for viral and tumor markers, abdominal ultrasonography, and contrast-enhanced computed tomography were all used for the diagnosis of liver cirrhosis and HCC. Hepatocellular carcinoma was graded according to the Barcelona Clinic Liver Cancer (BCLC) staging criteria.19 The overall survival time was calculated from inclusion in our study to death or last recorded contact.
Patients with liver illnesses not caused by viral hepatitis C, patients who had already received therapy for HCV or HCC, and patients with other malignant tumors were not allowed to participate in the study.
Methods
A vacutainer needle was used to take 8 mL of blood from a vein. 4 mL of blood was put into a vacutainer tube with a red cap, and 2 mL of blood was put into a vacutainer tube with EDTA and 1.8 mL of blood into a sodium citrate vacutainer tube for prothrombin time measurement. Serum obtained from the plain tube after being centrifuged for 10 min at 4000 RPM was stored at −80 °C for further analysis of the serum AFP level, liver function tests, liver enzymes, and hepatitis viral markers. The EDTA samples were used for RNA extraction, HCV RT-PCR, LINC00152, and UCA1 gene expression assessment.
Hepatitis C Virus Antibody (ANTI-HCV) was measured by electrochemiluminescence immunoassay (ECLA), employed in Cobas immunoassay analyzer, and Hepatitis B Surface Antigen (HBsAg) was determined by using an enzyme-linked immunosorbent assay (ELISA).
A kinetic UV-optimized method (IFCC ELTEC Kit, England) was used to test ALT and AST in the blood.20 Serum total bilirubin estimation was performed using the DIAMOND Diagnostics Kit, Germany.21 Quantitative enhanced specificity of bromocresol green colorimetric by Diamond Diagnostics kit, Germany was used to test the albumin in the blood.22
Serum AFP estimation was performed by ELISA using the IMMULITE 1000 system with a kit supplied by Siemens Medical Solutions Diagnostics, USA.23 Prothrombin time was measured by the STA-Stago Compact CT autoanalyzer.24 HCV RT-PCR was performed, and nucleic acid extraction was carried out using the QIAGEN viral RNA mini extraction kit.
LINC00152 and UCA1 quantitative real-time PCR were done in the following steps:25 first RNA extraction [QIAamp RNA Blood Mini Kit (Cat. No./ID: 52304) from Qiagen, USA], then RNA was reverse transcribed into cDNA using QuantiTect Reverse Transcription Kit (Qiagen, USA), according to their manual, followed by the real-time PCR step; amplification of cDNA using the QuantiTect SYBR Green PCR Kit (Qiagen, USA).
Each PCR reaction had a final volume of 20 μL, containing 10 μL SYBR Green 2× QuantiTect PCR Master Mix, 3 μL cDNA, 1 μL of each of the forward primer and reverse primer, and 5 μL RNase‑free H2O. PCR conditions were as follows: a 3-minute activation step at 95°C, then 55 cycles of 30 seconds at 94°C, then 30 seconds at 55°C followed by 30 seconds at 72°C. The following primers (Midland, Texas) were used:
- LINC00152 Forward, 5′-GACTGGATGGTCGCTGCTTT-3′;
- LINC00152 reverse, 5′ CCCAGGAACTGTGCTGTGAA-3′;
- UCA1 forward, 5′ TGCACCCTAGACCCGAAACT-3′;
- UCA1 reverse, 5′ CAAGTGTGACCAGGGACTGC3′;
- Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH; endogenous Housekeeper gene) forward and reverse primers, 5′-CGGAGTCAACGGATTGGTCGTAT-3′ and 5′AGCCTTCTCCATGGTGGTGAAGAC-3′ respectively.
Lastly, the 7500 ABI PRISM (Applied Biosystems, USA, v.2.0.1) was used for fluorescence detection and data processing. Relative expression of the LINC00152 and UCA1 was calculated using the ΔΔCT method.26 To confirm the specificity of the amplification and the absence of primer dimers, melting curve analysis was employed (Supplementary Figure 1).
Ethical Considerations
Each participant in the research voluntarily agreed to participate after receiving an adequate explanation of the study’s purpose and risks. The research was conducted in accordance with the principles outlined in the Helsinki Declaration of 1975, and it was approved by the ethical review board of the Faculty of Medicine, Menoufia University (MICRO-35).
Statistical Analysis
The data was analyzed using the IBM SPSS software program version 20.0 (Armonk, NY: IBM Corp). Normal distribution was determined for each variable by using the Kolmogorov–Smirnov test. The Chi-square test was used for the examination of relationships between categorical variables. The t-test was used to compare two groups when the variables follow a normal distribution, while the Mann–Whitney test was used when they did not follow a normal distribution. Receiver operating characteristic (ROC) curves were created by plotting sensitivity versus 1- specificity. The ideal cut-off values for the ROC curves were established using the Youden index (YI = sensitivity + specificity 1), and the statistical differences between ROC curves were examined using the DeLong technique. Kaplan-Meier curves were drawn to show the influence on survival, and the difference between subgroups was assessed by Kaplan-Meier curve analysis and a Log rank test. Cox proportional hazard models were used to examine risk variables for death. The obtained results were considered significant at p ≤ 0.5.
Results
Demographic, Clinical, and Laboratory Data of the Study Groups
The study groups were age and sex-matched, and their demographic, clinical, and laboratory features are shown in Table 1. All HCC cases were on top of liver cirrhosis, and most of the patients in the HCC and liver cirrhosis groups have no ascites and are in class A according to the Child-Pugh classification. There was a significant difference between the studied groups regarding liver function tests and serum alpha-fetoprotein levels.
 |
Table 1 Demographic, Clinical, and Laboratory Data of the Studied Groups |
LINC00152 and UCA1 Expression Level
Our results showed a significant increase in LINC00152 and UCA1 expression levels in the sera of the HCC group compared to the liver cirrhosis group and the healthy controls (P<0.001, and P<0.001 respectively). Moreover, their expression level in the liver cirrhosis group were significantly higher than in healthy controls (P<0.001) as shown in Figure 1.
 |
Figure 1 LINC00152 and UCA1 expression level in the studied groups. ***P≤ 0.001. |
Diagnostic Performance of LINC00152 and UCA1 in HCC
ROC curves were generated to determine and compare the diagnostic accuracy of alpha-fetoprotein with that of the LncRNAs. ROC curve analysis revealed that LINC00152 at the cut-off point of 1.585 has an accuracy of 72.5% with a sensitivity of 81.7% and a specificity of 63.3% with an AUC of 0.84 (p-value 0.001, 95% confidence interval: 0.77–0.92) in discriminating patients with HCC from patients with liver cirrhosis, While UCA1 at the cut-off point of 1.68 has an accuracy of 84.2% with a sensitivity of 85% and specificity of 83.3% with an AUC of 0.91 (p-value <0.001, 95% confidence interval: 0.86–0.96). Combination of both LncRNAs with alpha-fetoprotein results in a robust increase in the accuracy of diagnosis of HCC as shown in Table 2 and Figure 2.
 |
Table 2 Validity of Alpha-Fetoprotein, LINC00152, and UCA1 in Distinguishing HCC Cases from Cirrhotic Cases |
 |
Figure 2 ROC curve analysis of AFP, LINC00152, and UCA1 to differentiate HCC cases from cirrhotic cases. |
LINC00152 and UCA1 Expression Level in Relation to the Clinical and Pathological Characteristics of HCC
Results revealed that higher levels of LINC00152 expression were related to tumors involving both liver lobes, while higher levels of UCA1 expression were related to vascular invasion and BCLC stages (Table 3).
 |
Table 3 The Relation Between the Studied LncRNAs and Tumor Characteristics Among HCC Cases |
Overall Survival of the Studied Cases in Relation to Different Parameters
According to the Kaplan-Meier survival analysis, individuals who have no jaundice, BCLC stage B, no lymph node metastasis and low LINC00152 expression had considerably greater overall survival than their peers (Table 4 and Figure 3).
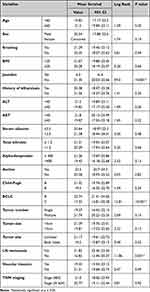 |
Table 4 Overall Survival of HCC Cases in Relation to Different Demographic Data, Laboratory Investigations, and Tumor Characteristics |
 |
Figure 3 Kaplan–Meier survival curve in relation to (A) LINC00152, (B) UCA1 expression levels in patients with HCC. |
The multivariate COX regression analysis demonstrated that serum LINC00152 expression is an independent prognostic factor in HCC patients with a hazard ratio of 2.23, 95% CI: 1.3–5.29, and p=0.03 (Table 5).
 |
Table 5 Cox Regression Analysis for Independent Factors Affecting Overall Survival Among HCC Cases |
Discussion
HCC is the most prevalent malignant tumor with the lowest five-year survival rate in the world because it is difficult to detect, diagnose, and treat early.27,28 Chronic hepatitis C virus (HCV) infection is linked to about one-third of all HCC cases and one-fifth of all HCC deaths.29 Most HCC diagnosis currently involves using radiology and measuring serum AFP 6–12 months apart, but AFP has low sensitivity for detecting very small lesions. In addition, AFP’s specificity is also not satisfactory.30–34 Consequently, we aimed to discover novel, extremely effective, sensitive, and specific biomarkers for detecting and monitoring HCC in its early stages.
Recent research shows that long noncoding RNAs (lncRNAs) play a key role in the development and growth of HCC. Some lncRNAs linked to HCC have been shown to have abnormal expression and play a role in carcinogenesis (such as proliferation, resistance to apoptosis, increased vessel formation, and invasion) by binding to DNA, RNA, or proteins, or by encoding small peptides.35 It is simple to identify circulating LncRNAs since they are stable in blood and other bodily fluids.36 More and more researchers are looking at circulating LncRNAs as a potential molecular marker for cancer, despite the fact that they have less specificity and sensitivity than more conventional tumor markers.37 A better knowledge of lncRNA dysregulation would therefore bring new insights into HCC etiology as well as potential techniques for early detection and therapy of HCC, as AFP has inadequate sensitivity and specificity.38
The present study showed a significant upregulation of LINC00152 expression levels in the sera of patients with liver cirrhosis and HCC compared to the healthy controls, and HCC patients had the highest relative expression level when compared to individuals with liver cirrhosis. Moreover, higher levels of LINC00152 expression were linked to tumors involving both liver lobes. It could distinguish HCC patients from those with liver cirrhosis with an AUC of 0.84. Interestingly, it was shown that LINC00152 expression level was an independent factor of survival in HCC patients. To the best of our knowledge, this is the first study that looked at the significance of serum LINC0052 in the prognosis of HCV-induced HCC, and it is the first to report that the expression level of LINC00152 in the serum is a reliable indicator of survival in HCV-induced HCC.
These findings were corroborated by the work of Li et al, who found, for the first time, that plasma LINC00152 was highly up regulated in Chinese patients with HCC. They next examined LINC00152 expression levels in homologous tissues of the same individuals and found a significant association between circulating and tissue levels, indicating that hepatic tissue overexpression is the cause of increased plasma levels. Moreover, the expression of LINC00152 was strongly correlated with tumor size, TNM stage, grade of differentiation, and tumor capsular invasion.39 These findings are also in line with the results of another study conducted in China.6 Abdelrahman et al also found that serum LINC00152 was significantly higher in Egyptian patients with HCC than in cirrhotic people without HCC.40 In addition, its area under the curve (AUC) in these studies for distinguishing HCC patients from healthy individuals was more than 0.8, which is quite good. So, they anticipated that LINC00152 may serve as new biomarkers for HCC.
Wang et al found that LINC00152 levels were much higher in HCC tissues compared to non-tumorous tissues. They also found that high levels of LINC00152 were linked to worse outcomes for patients with HBV-induced HCC.41 Deng et al discovered a correlation between LINC00152 and HBx expression in HCC tissues and between high LINC00152 expression and a bad prognosis for patients with HBV-induced HCC. They proposed that HBx increased the expression of LINC00152 and that LINC00152 inhibition could be a potential therapeutic target for HCC.42
LINC00152 increases the expression of semaphorin-4C through altering the function of miR-125b, allowing HCC cells to proliferate and multiply.43 Meanwhile, Ji et al demonstrated that LINC00152 increases HCC cell proliferation in vitro and tumor formation in vivo by activating the mammalian target of rapamycin (mTOR) signaling pathway through a cis-regulatory combination of EpCAM promoters.44 According to Hu et al, altering LINC00152 may affect the miR-125b-5p/KIAA1522 axis, which controls hepatocellular carcinoma cell growth, cell cycle progression, and apoptosis.45 Wang et al showed that blocking LINC00152 prevented the growth of HCC via altering miR-215 to activate CDK13.46 Thus, the increased knowledge of LINC00152 implies that targeting it may be a novel treatment approach for hepatocellular cancer.
Regarding UCA1 expression, it showed the same findings as LINC00152. Additionally, higher levels of UCA1 expression were related to vascular invasion and BCLC stages. It had the ability to distinguish HCC patients from those with liver cirrhosis with an AUC of 0.91. However, there was no substantial link between its levels and the overall survival of HCC patients.
Urothelial cancer associated 1 (UCA1) was first discovered in patients with bladder cancer.47 Then, it has been extensively studied as a non-invasive biomarker for several forms of cancer.48–51 In HCC tissues, it was up-regulated and linked with numerous clinical characteristics and malignant tendencies.52,53 The link between UCA1 expression and tumor size, metastasis, and overall survival was supported by a meta-analysis of seven studies.18 Some studies have shown that UCA1 is highly expressed in the sera of HCC patients. And its AUC for discriminating HCC patients from healthy people in these studies was over 0.8, which is considered good.6,54–56
Unlike our results, few studies have found UCA1 to be an independent predictor of survival in HCC patients.33,55 This difference could be because the causes of HCC are different. In our cases, HCC was caused only by HCV infection.
This study has a few limitations due to the small patient population, the unicentral design, and HCV as the sole contributing factor for HCC. Hence, more extensive research is required to understand the molecular mechanisms underlying the action of these LncRNAs and to pinpoint prospective HCC treatment targets.
Conclusions
LINC00152 and UCA1 expression levels were markedly higher in the serum of patients with HCC compared to those with liver cirrhosis and healthy controls, and their expressions were substantially greater in liver cirrhosis patients than healthy controls. Higher levels of LINC00152 expression were linked to tumors involving both liver lobes, while higher levels of UCA1 expression were related to vascular invasion and BCLC stages. Both LncRNAs have good sensitivity and specificity for HCC, making them effective diagnostic biomarkers for the disease. Furthermore, LINC00152 has the potential to be a prognostic marker for HCC. To corroborate our results, more research is required, ideally in the form of large-scale clinical studies, to identify potential targets for the therapy of HCC and to understand the molecular processes underlying the impact of these LncRNAs.
Acknowledgement
This study was funded by the Deanship of Scientific Research, King Faisal University, KSA (Project number GRANT4432).
Disclosure
The authors report no competing interest exists in this work.
References
1. Li Y, Wang X, Chen S, et al. Long non-coding RNA small nucleolar RNA host genes: functions and mechanisms in hepatocellular carcinoma. Mol Biol Rep. 2022;2022:1–10.
2. Ibrahim AS, Khaled HM, Mikhail NN, Baraka H, Kamel H. Cancer incidence in Egypt: results of the national population-based cancer registry program. J Cancer Epidemiol. 2014;2014:1–18. doi:10.1155/2014/437971
3. Gharib AF, Eed EM, Khalifa AS, et al. Value of serum miRNA-96-5p and miRNA-99a-5p as diagnostic biomarkers for hepatocellular carcinoma. Int J Gen Med. 2022;15(null):2427–2436. doi:10.2147/IJGM.S354842
4. Zhang H, Chen X, Yuan Y. Investigation of the miRNA and mRNA coexpression network and their prognostic value in hepatocellular carcinoma. Biomed Res Int. 2020;2020. doi:10.1155/2020/8726567
5. Wang T, Zhang K-H. New blood biomarkers for the diagnosis of AFP-negative hepatocellular carcinoma. Front Oncol. 2020;10:1316. doi:10.3389/fonc.2020.01316
6. Huang J, Zheng Y, Xiao X, et al. A circulating long noncoding RNA panel serves as a diagnostic marker for hepatocellular carcinoma. Dis Markers. 2020;2020:5417598. doi:10.1155/2020/5417598
7. Shehab-Eldeen S, Metwaly MF, Saber SM, El-Kousy SM, Badr EAE, Essa A. MicroRNA-29a and MicroRNA-124 as novel biomarkers for hepatocellular Carcinoma. Digest Liver Dis. 2022;55:283–290. doi:10.1016/j.dld.2022.04.015
8. Yang W-J, Sun Y-F, Jin A-L, et al. BCL11B suppresses tumor progression and stem cell traits in hepatocellular carcinoma by restoring p53 signaling activity. Cell Death Dis. 2020;11(10):1–13. doi:10.1038/s41419-020-03115-3
9. Khashkhashi Moghadam S, Bakhshinejad B, Khalafizadeh A, Mahmud Hussen B, Babashah S. Non‐coding RNA‐associated competitive endogenous RNA regulatory networks: novel diagnostic and therapeutic opportunities for hepatocellular carcinoma. J Cell Mol Med. 2022;26(2):287–305. doi:10.1111/jcmm.17126
10. Tang G, Luo L, Zhang J, et al. lncRNA LINC01057 promotes mesenchymal differentiation by activating NF-κB signaling in glioblastoma. Cancer Lett. 2021;498:152–164. doi:10.1016/j.canlet.2020.10.047
11. Zhou R, Sun H, Zheng S, et al. A stroma‐related lncRNA panel for predicting recurrence and adjuvant chemotherapy benefit in patients with early‐stage colon cancer. J Cell Mol Med. 2020;24(5):3229–3241.
12. Shi Y, Zhang -D-D, Liu J-B, et al. Comprehensive analysis to identify DLEU2L/TAOK1 axis as a prognostic biomarker in hepatocellular carcinoma. Mol Ther Nucleic Acids. 2021;23:702–718. doi:10.1016/j.omtn.2020.12.016
13. Yu Y, Yang J, Li Q, Xu B, Lian Y, Miao L. LINC 00152: a pivotal oncogenic long non‐coding RNA in human cancers. Cell Prolif. 2017;50(4):e12349. doi:10.1111/cpr.12349
14. Matis S, Rossi M, Brondolo L, et al. LINC00152 expression in normal and Chronic Lymphocytic Leukemia B cells. Hematol Oncol. 2022;40(1):41–48. doi:10.1002/hon.2938
15. Bian Z, Zhang J, Li M, et al. Long non-coding RNA LINC00152 promotes cell proliferation, metastasis, and confers 5-FU resistance in colorectal cancer by inhibiting miR-139-5p. Oncogenesis. 2017;6(11):1–11. doi:10.1038/s41389-017-0008-4
16. Ramli S, Sim MS, Guad RM, et al. Long noncoding RNA UCA1 in gastrointestinal cancers: molecular regulatory roles and patterns, mechanisms, and interactions. J Oncol. 2021;2021. doi:10.1155/2021/5519720
17. An M, Xing X, Chen T. Long non‑coding RNA UCA1 enhances cervical cancer cell proliferation and invasion by regulating microRNA‑299‑3p expression. Oncol Lett. 2021;22(5):1–9. doi:10.3892/ol.2021.13033
18. Qin LT, Tang RX, Lin P, et al. Biological function of UCA1 in hepatocellular carcinoma and its clinical significance: investigation with in vitro and meta-analysis. Pathol Res Pract. 2018;214(9):1260–1272. doi:10.1016/j.prp.2018.03.025
19. Reig M, Forner A, Rimola J, et al. BCLC strategy for prognosis prediction and treatment recommendation: the 2022 update. J Hepatol. 2022;76(3):681–693. doi:10.1016/j.jhep.2021.11.018
20. Bergmeyer H, Horder M, Rey J. Approved recommendation on IFCC methods for the measurement of catalytic enzymes. Part 3: IFCC method for alanine aminotransferase. J Clin Chem Clin Biochem. 1986;24:481–495.
21. Doumas BT, Perry BW, Sasse EA, Straumfjord JV Jr. Standardization in bilirubin assays: evaluation of selected methods and stability of bilirubin solutions. Clin Chem. 1973;19(9):984–993. doi:10.1093/clinchem/19.9.984
22. Pinnell AE, Northam BE. New automated dye-binding method for serum albumin determination with bromcresol purple. Clin Chem. 1978;24(1):80–86. doi:10.1093/clinchem/24.1.80
23. Gitlin D. Normal biology of α‐fetoprotein. Ann N Y Acad Sci. 1975;259(1):7–16. doi:10.1111/j.1749-6632.1975.tb25397.x
24. Colman RW. Hemostasis and Thrombosis: Basic Principles and Clinical Practice. Lippincott Williams & Wilkins; 2006.
25. Dorak MT. Real-Time PCR. Taylor & Francis; 2007.
26. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
27. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi:10.3322/caac.20107
28. Zhao J, Greene CM, Gray SG, Lawless MW. Long noncoding RNAs in liver cancer: what we know in 2014. Expert Opin Ther Targets. 2014;18(10):1207–1218. doi:10.1517/14728222.2014.941285
29. Akinyemiju T, Abera S, Ahmed M, et al. The burden of primary liver cancer and underlying etiologies from 1990 to 2015 at the global, regional, and national level: results from the global burden of disease study 2015. JAMA Oncol. 2017;3(12):1683–1691. doi:10.1001/jamaoncol.2017.3055
30. Seo SI, Kim SS, Choi BY, et al. Clinical significance of elevated serum alpha-fetoprotein (AFP) level in acute viral hepatitis A (AHA). Hepato-Gastroenterology. 2013;60(127):1592–1596.
31. Wong CR, Garcia RT, Trinh HN, et al. Adherence to screening for hepatocellular carcinoma among patients with cirrhosis or chronic hepatitis B in a community setting. Dig Dis Sci. 2009;54(12):2712–2721. doi:10.1007/s10620-009-1015-x
32. Li H, Li Y, Liu D, Sun H, Liu J. miR-224 is critical for celastrol-induced inhibition of migration and invasion of hepatocellular carcinoma cells. Cell Physiol Biochem. 2013;32(2):448–458. doi:10.1159/000354450
33. Zhang Z, Li J, Wei Z, et al. Correlation between expression levels of lncRNA UCA1 and miR-18a with prognosis of hepatocellular cancer. Eur Rev Med Pharmacol Sci. 2020;24(7):3586–3591. doi:10.26355/eurrev_202004_20820
34. Zhou L, Liu J, Luo F. Serum tumor markers for detection of hepatocellular carcinoma. World J Gastroenterol. 2006;12(8):1175. doi:10.3748/wjg.v12.i8.1175
35. Huang Z, Zhou J-K, Peng Y, He W, Huang C. The role of long noncoding RNAs in hepatocellular carcinoma. Mol Cancer. 2020;19(1):77. doi:10.1186/s12943-020-01188-4
36. Li H, Yu B, Li J, et al. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget. 2014;5(8):2318. doi:10.18632/oncotarget.1913
37. Beylerli O, Gareev I, Sufianov A, Ilyasova T, Guang Y. Long noncoding RNAs as promising biomarkers in cancer. Noncoding RNA Res. 2022;7(2):66–70. doi:10.1016/j.ncrna.2022.02.004
38. DiStefano JK. Long noncoding RNAs in the initiation, progression, and metastasis of hepatocellular carcinoma. Non-Coding RNA Res. 2017;2(3–4):129–136. doi:10.1016/j.ncrna.2017.11.001
39. Li J, Wang X, Tang J, et al. HULC and Linc00152 Act as novel biomarkers in predicting diagnosis of hepatocellular carcinoma. Cell Physiol Biochem. 2015;37(2):687–696. doi:10.1159/000430387
40. Abdelrahman E, El-Aal A, Sobhy M, Shamsya A, Zanet Y, Bedewy EE. Serum long intergenic non-coding ribonucleic acid LINC00152 as a potential predictor of hepatocellular carcinoma in Egyptian patients. Afro-Egypt J Infect Enem Dis. 2020;10(3):264–270. doi:10.21608/aeji.2020.29616.1078
41. Wang B, Yang S, Zhao W. Long non-coding RNA NRAD1 and LINC00152 are highly expressed and associated with prognosis in patients with hepatocellular carcinoma. Onco Targets Ther. 2020;13:10409–10416. doi:10.2147/ott.S251231
42. Deng X, Zhao X, Liang X, Chen R, Pan Y, Liang J. Linc00152 promotes cancer progression in hepatitis B virus-associated hepatocellular carcinoma. Biomed Pharmacother. 2017;90:100–108. doi:10.1016/j.biopha.2017.03.031
43. Tian Q, Yan X, Yang L, Liu Z, Yuan Z, Zhang Y. lncRNA CYTOR promotes cell proliferation and tumor growth via miR‑125b/SEMA4C axis in hepatocellular carcinoma. Oncol Lett. 2021;22(5):1–12. doi:10.3892/ol.2021.13057
44. Ji J, Tang J, Deng L, et al. LINC00152 promotes proliferation in hepatocellular carcinoma by targeting EpCAM via the mTOR signaling pathway. Oncotarget. 2015;6(40):42813. doi:10.18632/oncotarget.5970
45. Hu B, Yang X-B, Yang X, Sang X-T. LncRNA CYTOR affects the proliferation, cell cycle and apoptosis of hepatocellular carcinoma cells by regulating the miR-125b-5p/KIAA1522 axis. Aging. 2021;13(2):2626. doi:10.18632/aging.202306
46. Wang J, Zhang Y, Lu L, Lu Y, Tang Q, Pu J. Insight into the molecular mechanism of LINC00152/miR‐215/CDK13 axis in hepatocellular carcinoma progression. J Cell Biochem. 2019;120(11):18816–18825. doi:10.1002/jcb.29197
47. Wang F, Li X, Xie X, Zhao L, Chen W. UCA1, a non-protein-coding RNA up-regulated in bladder carcinoma and embryo, influencing cell growth and promoting invasion. FEBS Lett. 2008;582(13):1919–1927. doi:10.1016/j.febslet.2008.05.012
48. Duan W, Du L, Jiang X, et al. Identification of a serum circulating lncRNA panel for the diagnosis and recurrence prediction of bladder cancer. Oncotarget. 2016;7(48):78850. doi:10.18632/oncotarget.12880
49. Pan J, Xie X, Li H, Li Z, Ren C, Ming L. Detection of serum long non-coding RNA UCA1 and circular RNAs for the diagnosis of bladder cancer and prediction of recurrence. Int J Clin Exp Pathol. 2019;12(8):2951.
50. Wang J, Gao Y, Wang X, et al. Circulating lncRNAs as noninvasive biomarkers in bladder cancer: a diagnostic meta-analysis based on 15 published articles. Int J Biol Markers. 2020;35(2):40–48. doi:10.1177/1724600820926685
51. Wang W, Yin Z. Diagnostic value of long non-coding RNA H19, UCA1, and HOTAIR as promising biomarkers in human bladder cancer. Int J Clin Exp Pathol. 2017;10(12):11659.
52. Li J, Gao J, Kan A, Hao T, Huang L. SNHG and UCA1 as prognostic molecular biomarkers in hepatocellular carcinoma: recent research and meta-analysis. Minerva Med. 2017;108(6):568–574. doi:10.23736/S0026-4806.17.05094-7
53. Wang F, Ying H-Q, B-S H, et al. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6(10):7899. doi:10.18632/oncotarget.3219
54. El-Tawdi AH, Matboli M, El-Nakeep S, Azazy AE, Abdel-Rahman O. Association of long noncoding RNA and c-JUN expression in hepatocellular carcinoma. Expert Rev Gastroenterol Hepatol. 2016;10(7):869–877. doi:10.1080/17474124.2016.1193003
55. Zheng ZK, Pang C, Yang Y, Duan Q, Zhang J, Liu WC. Serum long noncoding RNA urothelial carcinoma-associated 1: a novel biomarker for diagnosis and prognosis of hepatocellular carcinoma. J Inter Med Res. 2018;46(1):348–356. doi:10.1177/0300060517726441
56. Kamel MM, Matboli M, Sallam M, Montasser IF, Saad AS, El-Tawdi AH. Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma. Translat Res. 2016;168:134–145. doi:10.1016/j.trsl.2015.10.002
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
