Back to Journals » Infection and Drug Resistance » Volume 13
Serum Hepatitis B Virus RNA Levels Predict HBeAg Seroconversion and Virological Response in Chronic Hepatitis B Patients with High Viral Load Treated with Nucleos(t)ide Analog
Authors Ji X , Xia M, Zhou B, Liu S, Liao G, Cai S , Zhang X, Peng J
Received 8 March 2020
Accepted for publication 28 May 2020
Published 22 June 2020 Volume 2020:13 Pages 1881—1888
DOI https://doi.org/10.2147/IDR.S252994
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Joachim Wink
Xin Ji,* Muye Xia,* Bin Zhou, Shi Liu, GuiChan Liao, Shaohang Cai, Xiaoyong Zhang, Jie Peng
State Key Laboratory of Organ Failure Research, Guangdong Provincial Key Laboratory of Viral Hepatitis Research, Department of Infectious Diseases, Nanfang Hospital, Southern Medical University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jie Peng
Department of Infectious Diseases and Hepatology Unit, Nanfang Hospital, Southern Medical University, No. 1838, North Guangzhou Avenue, Guangzhou 510515, People’s Republic of China
Tel +86 20 6278 7428
Fax +86 20 8771 9653
Email [email protected]
Background and Aim: Hepatitis B virus (HBV) RNA has attracted increasing attention as a novel serum marker for intrahepatic HBV replication. However, the predictive value of the serum level of HBV RNA for hepatitis B e-antigen (HBeAg) seroconversion and viral response among patients with a high viral load (HVL) is unclear. We evaluated the role of the serum level of HBV RNA as a predictor of treatment response in chronic HBV (CHB) patients with an HVL.
Patients and Methods: The study cohort was 66 HBeAg-positive CHB patients with an HVL (serum HBV DNA > 1.9× 106 IU/mL) at baseline from our previous prospective cohort study treated with lamivudine (LAM) and adefovir dipivoxil(ADV) (N=31) or entecavir alone (N=35) for ≤ 96 weeks. The serum HBV RNA level was quantified by TaqMan® probe-based reverse transcription real-time quantitative polymerase chain reaction at four time points.
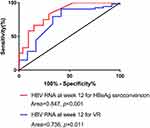
Results: The baseline serum HBV RNA level (in log10 copies/mL) in patients treated with LAM+ADV and ETV monotherapy was 8.97± 1.22 and 9.15± 0.92, respectively. After nucleos(t)ide analog (NA) therapy, the serum HBV RNA level decreased steadily in all patients (week 0 vs week 12, p< 0.001; week 12 vs week 24, p=0.010; week 24 vs week 48, p< 0.001). Fifty-three (80.3%) patients achieved a virologic response (VR), and 12 (18.2%) achieved HBeAg seroconversion after 96 weeks. Multivariate analyses revealed that the serum HBV RNA level at week 12 could predict HBeAg seroconversion (OR 3.560, 95% CI: 1.39– 9.110, p=0.008) and VR (1.908, 1.115– 3.265, 0.018) at 96 weeks. Analyses of receiver operating characteristic curves indicated that the serum HBV RNA level 12 weeks after NA treatment had predictive value for HBeAg seroconversion (AUC=0.847, p< 0.001) and VR (AUC=0.736, p=0.011).
Conclusion: The serum level of HBV RNA at 12 weeks could predict HBeAg seroconversion and a VR during NA treatment in CHB patients with an HVL.
Keywords: HBV RNA, HBeAg seroconversion, virological response, high viral load, nucleos(t)ide analogs
Background
Hepatitis B virus (HBV) infection is a major public health concern, especially in the Asia–Pacific region. A test for HBV DNA load is the only recognized laboratory indicator for monitoring the number of circulating viral particles in peripheral blood, which reflects the activity generated by the HBV in the liver.1,2 However, after long and extensive use of nucleos(t)ide analogs (NAs), the serum level of HBV DNA in most patients with chronic hepatitis B (CHB) infection is below the lower limit of detection, and the disappearance of HBV DNA in serum represents only effective inhibition of the reverse transcription of HBV.
NAs are inhibitors of viral polymerase and cannot achieve complete eradication of covalently closed circular DNA (cccDNA). Furthermore, percutaneous liver biopsy for cccDNA detection is difficult because of its invasive nature, the uneven distribution of cccDNA in the liver, and lack of a standardized assay for cccDNA detection.3 Consequently, it is important that novel serum biomarkers related to the natural history, prognosis, and treatment responses of CHB are identified.
Pre-genomic RNA (pgRNA) is an intermediate product in HBV replication and is transcribed using cccDNA as a template in the viral nucleocapsid.4 In 2016, Wang et al reported that HBV RNA in the serum of HBV-infected patients was the pgRNA of the encapsidated virus.5 There is evidence that the HBV RNA level is related to the virologic response and cccDNA level in the liver, and could, in theory, be used to reflect the hepatic level of cccDNA.6–8
Increasing attention has been paid to the relationship between the serum level of HBV RNA and HBV replication. Several studies have shown that the HBV RNA level can reflect the efficacy of NAs, and that the HBV RNA level 12 weeks after treatment can be used as an independent predictor of the viral response (VR) or hepatitis B e-antigen (HBeAg) seroconversion at the outcome of treatment.9,10,23 Other studies have suggested that the reduction in the serum level of HBV RNA at the early stage of NA treatment is closely correlated with subsequent HBeAg seroconversion.12
HBeAg-positive CHB patients with a high viral load (HVL) comprise a unique group with relative immune tolerance. Recent evidence has suggested that CHB patients with an HVL are less likely to achieve a VR or HBeAg seroconversion and more likely to be associated with drug-related resistance and treatment failure compared with CHB patients with a normal level.13,14 At present, the value of using the HBV RNA level at different time points as a predictor of the VR or HBeAg seroconversion in HBeAg-positive CHB patients with an HVL at baseline is limited.
The primary purpose of the present study was to evaluate the role of the serum HBV RNA level as a predictor for the treatment response in HBeAg-positive CHB patients with an HVL at baseline. We also aimed to explore the dynamic changes in the HBV RNA level that occur during NA treatment.
Patients and Methods
Ethical Approval of the Study Protocol
This was a prospective, multicenter, controlled trial. The study protocol was approved by the Ethics Review Board of Nanfang Hospital, Southern Medical University (ZHF2011206) in Guangzhou, China. This study was undertaken in accordance with the Declaration of Helsinki 1975 and its later amendments. Patients provided written informed consent to participate in the study, and data were analyzed anonymously. The design and methods of this study were as described previously.15,16
Patients
The study cohort consisted of 66 HBeAg-positive CHB patients with an HVL (serum HBV DNA >1.9×106 IU/mL) at baseline, treated with lamivudine (LAM) and adefovir dipivoxil (ADV) (N = 31) or entecavir alone (ETV) (N = 35). All patients received ≥96 weeks of treatment.
All patients were HBsAg-positive for ≥6 months, and HBeAg-positive with an HVL at baseline. All patients were naive to NA treatment, and did not present spontaneous HBeAg seroconversion after 3–6 months of an increased level of alanine aminotransferase (ALT) (≥2 weeks apart, with the latest ALT ≥2 × the upper limit of normal).
The exclusion criteria were patients: (i) who had received treatment with interferon (IFN) or NA previously; (ii) who were pregnant; (iii) had a history of hepatic metabolic diseases; (iv) with diagnostic markers of co-infection with the hepatitis-C virus, hepatitis-D virus, or human immunodeficiency virus; (v) with autoimmune hepatitis; (vi) with liver cirrhosis; (vii) with liver failure; (viii) with hepatocellular carcinoma.
Serology
Qualitative determination of the serum level of the surface antigen of the hepatitis B virus (HBsAg), HBeAg, and anti-HBe was done using the ARCHITECT i2000SR platform (Abbott Laboratories, Chicago, IL, USA). The serum HBV DNA level was measured using Cobas Ampliprep® and Cobas TaqMan®, v2.0 (Roche Diagnostics, Basel, Switzerland) with a limit of detection ranging from 20 IU/mL to 1.7×108 IU/mL. The serum ALT level was measured at local laboratories according to standard procedures.
Quantitative determination of HBV RNA was carried out according to the method of Fan and colleagues.17 HBV RNA was isolated from 200 μL of serum using the QIAamp MinElute Virus Spin kit (Qiagen, Hilden, Germany) according to manufacturer instructions, and treated subsequently with DNase I (Thermo Fisher Scientific, Waltham, MA, USA). DNase I-digested HBV RNA was quantified by TaqMan probe-based one-step reverse transcription real-time quantitative polymerase chain reaction in a LightCycler® 480 Instrument II system (Roche Diagnostics). Detailed procedures were as described previously.17
A “virologic response” was defined as HBV DNA < 20 IU/mL. A “biochemical response” was defined as normalization of the ALT level. “HBeAg seroconversion” was defined as a change from HBeAg-negative status to HBeAb-positive status.
Statistical Analyses
Data analyses were done using SPSS v21.0 (IBM, Armonk, NY, USA). Data are expressed as the mean ± standard deviation or median (interquartile range). The χ2 test was used for categorical variables. The Student’s t-test or Mann–Whitney test was employed for continuous variables when appropriate. The linear relationship between various viral markers was assessed by Pearson’s correlation coefficient and the significance of the correlation coefficient (p-value) was tested. Categorical variables were evaluated using Spearman correlation coefficient. A receiver operating characteristic (ROC) curve was used to calculate the predictive value.
Results
Patient Characteristics
Seventy-four HBeAg-positive patients were enrolled and divided into two groups according to the treatment plan. The serum samples of eight patients were lost, so 66 patients were included in our study and completed 96-week treatment. The baseline characteristics of the patients are shown in Table 1. Fifty-three patients achieved a virologic response and 12 achieved HBeAg seroconversion at 96 weeks. Of the 66 patients at baseline, 31 (mean age, 31.68±10.88 years) received LAM plus ADV combination therapy, whereas the remaining 35 cases (mean age, 30.57±8.38 years) received ETV monotherapy. The baseline serum HBV RNA level (in log10 copies/mL) in patients treated with LAM+ADV combination therapy and ETV monotherapy was 8.97±1.22 and 9.15±0.92, respectively. There was no significant difference between the two groups in the distribution of demographics or clinical characteristics at baseline.
 |
Table 1 Demographic and Baseline Characteristics of Patients |
Predictive Value of the HBV RNA Level for HBeAg Seroconversion in CHB Patients Treated with NAs
The serum HBV RNA level (in log10 copies/mL) was significantly lower at baseline (8.63 vs 9.17, p = 0.043), week 12 (5.64 vs 7.72, p < 0.001), week 24 (5.52 vs 7.09, p = 0.015), and week 48 (2.03 vs 5.60, p = 0.002) in patients with HBeAg seroconversion than in those without HBeAg seroconversion at 96 weeks (Figure 1A). We used univariate and multivariate analyses to determine the relationship between the serum HBV RNA level and HBeAg seroconversion (odds ratio (OR) 3.560, 95% confidence interval (CI): 1.39–9.110, p = 0.008) (Table 2). Based on univariate and multivariate analyses, the HBV RNA level at 12 weeks after treatment was an independent predictor for HBeAg seroconversion in HBeAg-positive patients after 96 weeks. Moreover, analyses of ROC curves indicated that the HBV RNA level 12 weeks after treatment had a certain predictive value for HBeAg seroconversion (area under the ROC curve (AUC) = 0.847, p < 0.001), with a cutoff value of 6.18 log10 copies/mL (sensitivity = 0.81, specificity = 0.80) (Figure 2). The percentage HBeAg seroconversion was 10% and 31.8% in patients with a serum HBV RNA level (in log10 copies/mL) ≤6.18 and >6.18 at week 12, respectively (p = 0.037) (Figure 3).
 |
Table 2 Univariate and Multivariate Analyses of Associations Between the HBV RNA Level and HBeAg Seroconversion at Week 96 |
 |
Figure 3 Probability of HBeAg seroconversion based on the serum HBV RNA level at week 12. |
Predictive Value of the HBV RNA Level for the Virologic Response in CHB Patients Treated with NAs
The serum HBV RNA level (in log10 copies/mL) was significantly lower at week 12 (7.02 vs 8.53, p < 0.011), week 24 (6.61 vs.7.69, p = 0.021), and week 48 (4.35 vs 7.53, p = 0.021) in patients with a VR than in those without a VR (Figure 1B). However, the serum RNA level (in log10 copies/mL) was similar and non-significant between the two groups at baseline (8.63 vs 9.12).
The HBV RNA level 12 weeks after treatment had a certain predictive value for a VR at 96 weeks after treatment (OR 1.908, 95% CI: 1.115–3.265, p = 0.018) (Table 3). The HBV RNA level 12 weeks after treatment was an independent predictor for a VR (AUC = 0.736, p = 0.011), with a cutoff value of 9.05 log10 copies/mL (sensitivity = 0.583, specificity = 0.91) (Figure 2).
 |
Table 3 Univariate and Multivariate Analyses of Associations Between the HBV RNA Level and Virologic Response at Week 96 |
Dynamic Change in the HBV RNA Level During NA Therapy
After NA therapy, the serum HBV RNA level decreased steadily in all patients (Figure 4A). The level of HBV RNA (in log10 copies/mL) at different time points was significantly lower than that at preceding time points (week 0 vs week 12: 9.19 vs.7.31, p < 0.001; week 12 vs week 24: 7.31 vs 6.90, p = 0.010; week 24 vs week 48: 6.90 vs 4.86, p < 0.001). Patients with HBeAg seroconversion at 96 weeks showed a significant reduction in the HBV RNA level (in log10 copies/mL) at week 12 (3.28 vs 1.52, p = 0.005) and at week 48 (3.79 vs 1.38, p = 0.008) compared with patients without HBeAg seroconversion (Figure 4B). We found a similar result when predicting the VR. Compared with patients without a VR, those with a VR at week 96 showed a significant reduction in the HBV RNA level (in log10 copies/mL) at week 12 (2.19 vs 0.64, p < 0.001) and at week 48 (2.23 vs 0.06, p < 0.001) (Figure 4C).
Association Between the Serum HBV RNA Level and Clinical Parameters
The HBV RNA level was not correlated significantly with age, sex, or different options of drug treatment (p > 0.05 for all, data not shown). The serum HBV RNA level was significantly and positively correlated with HBV DNA and ALT levels (r = 0.653, p < 0.001 and r = 0.537, p < 0.001, respectively) (Supplementary Figure 1) in all patients during the whole treatment, whereas no significant relationship was found between HBV DNA and ALT levels at each time point evaluated (data not shown).
Discussion
This was the first longitudinal study to analyze dynamic changes in the HBV RNA level after NA treatment. As such, it revealed whether the HBV RNA level has value in predicting HBeAg seroconversion and the virologic response in HBeAg-positive CHB patients with an HVL. We found that the HBV RNA level decreased significantly in CHB patients with an HVL after NA treatment. Moreover, the HBV RNA level could also predict HBeAg seroconversion and the VR at week 12 in these patients.
As a new marker of hepatitis B, HBV RNA is transcribed from cccDNA and correlates equally well with intrahepatic cccDNA in CHB patients.4,18,19 These observations suggest that the HBV RNA level can be considered a surrogate marker of the cccDNA level in infected cells.
Since the identification of pgRNA in HBV-infected patients,5 the serum HBV RNA level has attracted increasing attention as a potential biomarker in CHB management.4–6,20 Recent reports have suggested that HBV RNA in serum is associated with responses to NA and pegylated-IFN therapy.5,9,12,15,21,22 However, relatively few studies have evaluated the performance of serum HBV RNA among HBeAg-positive CHB patients with an HVL. This was the first longitudinal study to investigate the value of HBV RNA in HBeAg-positive CHB patients with an HVL. We found that baseline serum DNA and RNA loads were high in these patients. The HBV RNA level was higher at baseline in our HBeAg-positive patients with an HVL than that reported for HBeAg-positive patients in other studies.5,14,23 This finding may be related to our selection of patients with an HVL. During NA treatment, the serum HBV RNA level of all patients decreased gradually from 0 to 48 weeks of therapy and was significantly lower than that at previous time points. Also, the serum level of HBV RNA was significantly lower in patients with HBeAg seroconversion than in that without HBeAg seroconversion from baseline to week 48 of treatment. Furthermore, the degree of decline in the HBV RNA level in HBeAg-positive patients with an HVL was comparable with that reported for HBeAg-positive patients.23 This observation was most likely because NAs inhibit reverse transcription of HBV polymerase, thereby suppressing the replication of the HBV.
Multivariate analysis demonstrated that the serum level of HBV RNA at week 12 was an independent prognostic factor for HBeAg seroconversion in CHB patients with an HVL after NA treatment. An ROC curve of the serum HBV RNA level at 12 weeks was generated to predict HBeAg seroconversion at 96 weeks (AUC: 0.847). We proposed a cutoff level of HBV RNA of 6.18 log10 copies/mL at week 12 for the identification of HBeAg seroconversion, which led to sensitivity prediction of 81% and specificity prediction of 80%. Then, we analyzed the relationship between the HBV RNA level and the VR, and showed that the HBV RNA level at week 12 was also associated with the VR. The HBV RNA level decreased more rapidly in patients with a VR than in those without a VR, reaching significance at weeks 12, 24, and 48. In patients without a VR, there was a non-significant decrease. Multivariate analysis also indicated that the HBV RNA level at week 12 was associated with a VR. Luo and colleagues demonstrated that the serum level of HBV RNA 24 weeks after NA treatment can predict the response of NA treatment in HBeAg-positive patients (n = 25).23 However, we found that the serum HBV RNA level at week 12 was an independent indicator for predicting the response of NA treatment in HBeAg-positive patients with an HVL (n = 66). These results suggested that early suppression of HBV RNA in responders may reflect the silencing of cccDNA activity, which might also represent the establishment of durable immune control over HBV infection.
Several studies have demonstrated that the serum HBV RNA level is correlated significantly with HBV markers, such as HBV DNA and HBsAg,11,19,24,25 which supports the notion that the serum level of HBV RNA could be a marker to reflect the intrahepatic activity of HBV replication. We found a correlation between the total serum level of HBV RNA and total serum level of HBV DNA, with a correlation coefficient of 0.653. However, we did not observe a significant difference in the serum level of HBV DNA, HBV RNA, or ALT at any of the evaluated time points. Bömmel and colleagues demonstrated that the serum level of HBV RNA is correlated strongly with the serum level of HBV DNA before treatment, but the correlation becomes weaker after starting NA treatment.12 We also found a correlation between the total serum level of HBV RNA levels and total serum level of ALT. Gu et al discovered that the serum level of HBV RNA was associated with T-helper type 1 (Th1)/Th2-related immune responses, which play important parts in liver inflammation, potentially contributing to the association between the serum level of HBV RNA and serum level of ALT.25
We provided a comprehensive description of HBV RNA kinetics in CHB patients with an HVL receiving NAs, and this was designed to be a prospective treatment study. However, our study had two main limitations. First, we suffered from a lack of liver specimens. Second, the quantification of HBV RNA in serum can be influenced by several viral factors, including HBV genotypes,26 and we did not include analyses of HBV genotypes. All patients were Asian and were most likely to have been infected with genotype B or C. Therefore, further studies are needed, particularly among ethnically diverse populations and different HBV genotypes, to confirm our observations.
Conclusion
We revealed the dynamic changes that occur in the serum level of HBV RNA in CHB patients with an HVL during NA therapy. We also showed that, following NA treatment, the serum HBV RNA level at 12 weeks can predict HBeAg seroconversion and a VR in HBeAg-positive CHB patients with an HVL.
Acknowledgments
We thank Dr Yegui Jiang for providing participants from Southwest Hospital, Third Military Medical University; Dr Yonghong Zhang for providing participants from Second Xiangya Hospital, Central South University; Dr Fangfang Lv for providing the participants from Sir Run Run Shaw Hospital, Zhejiang University. Xin Ji and Muye Xia are co-first authors for this study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. European Association for the Study of the Liver. EASL 2017 clinical practice guidelines on the management of hepatitis B virus infection. J Hepatol. 2017;67(2):370–398. doi:10.1016/j.jhep.2017.03.021
2. Terrault NA, Lok ASF, McMahon BJ, et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Clin Liver Dis (Hoboken). 2018;12(1):33–34. doi:10.1002/cld.728
3. Kock J, Theilmann L, Galle P, Schlicht HJ. Hepatitis B virus nucleic acids associated with human peripheral blood mononuclear cells do not originate from replicating virus. Hepatology. 1996;23(3):405–413. doi:10.1002/hep.510230303
4. Liu S, Zhou B, Valdes JD, Sun J, Guo H. Serum hepatitis B virus RNA: a new potential biomarker for chronic hepatitis B virus infection. Hepatology. 2019;69(4):1816–1827. doi:10.1002/hep.30325
5. Wang J, Shen T, Huang X, et al. Serum hepatitis B virus RNA is encapsidated pregenome RNA that may be associated with persistence of viral infection and rebound. J Hepatol. 2016;65(4):700–710. doi:10.1016/j.jhep.2016.05.029
6. Wang J, Yu Y, Li G, et al. Relationship between serum HBV RNA levels and intrahepatic viral as well as histologic activity markers in entecavir-treated patients. J Hepatol. 2017;pii:
7. Giersch K, Allweiss L, Volz T, Dandri M, Lutgehetmann M. Serum HBV pgRNA as a clinical marker for cccDNA activity. J Hepatol. 2017;66(2):460–462. doi:10.1016/j.jhep.2016.09.028
8. Carey I, Gersch J, Wang B, et al. Pre-genomic HBV RNA and HBcrAg predict outcomes in HBeAg negative chronic hepatitis B patients suppressed on nucleos(t)ide analogue therapy. Hepatology. 2019. doi:10.1002/hep.31026
9. Huang YW, Takahashi S, Tsuge M, et al. On-treatment low serum HBV RNA level predicts initial virological response in chronic hepatitis B patients receiving nucleoside analogue therapy. Antivir Ther. 2015;20(4):369–375. doi:10.3851/IMP2777
10. Ma G, Lou B, Lv F, Zhao D, Zhang Z, Chen Y. HBcrAg and pgRNA and the therapeutic effect in HBeAg-positive patients receiving anti-viral therapy, baseline serum HBV-RNA is a powerful predictor of response. J Viral Hepat. 2020. doi:10.1111/jvh.13299
11. Farag MS, van Campenhout MJH, Pfefferkorn M, et al. Hepatitis B virus RNA as early predictor for response to PEGylated interferon alfa in HBeAg negative chronic hepatitis B. Clin Infect Dis. 2020;pii:ciaa013. doi:10.1093/cid/ciaa013
12. Bömmel FV, Bartens A, Mysickova A, et al. Serum hepatitis B virus RNA levels as an early predictor of hepatitis B envelope antigen seroconversion during treatment with polymerase inhibitors. Hepatology. 2015;61(1):66–76. doi:10.1002/hep.27381
13. Liaw YF, Gane E, Leung N, et al. 2-year GLOBE trial results: telbivudine is superior to lamivudine in patients with chronic hepatitis B. Gastroenterology. 2009;136(2):486–495. doi:10.1053/j.gastro.2008.10.026
14. Tseng TC, Liu CJ, Hsu CY, et al. High level of hepatitis B core-related antigen associated with increased risk of hepatocellular carcinoma in patients with chronic HBV infection of intermediate viral load. Gastroenterology. 2019;157(6):1518–1529.e1513. doi:10.1053/j.gastro.2019.08.028
15. Cai S, Li Z, Yu T, Xia M, Peng J. Serum hepatitis B core antibody levels predict HBeAg seroconversion in chronic hepatitis B patients with high viral load treated with nucleos(t)ide analogs. Infect Drug Resist. 2018;11:469–477. doi:10.2147/IDR.S163038
16. Wu Y, Gao C, Cai S, et al. Circulating miR-122 is a predictor for virological response in CHB patients with high viral load treated with nucleos(t)ide analogs. Front Genet. 2019;10:243. doi:10.3389/fgene.2019.00243
17. Fan R, Zhou B, Xu M, et al. Association between negative results from tests for HBV DNA and RNA and durability of response after discontinuation of nucles(t)ide analogue therapy. Clin Gastroenterol Hepatol. 2019;18(3):719–727. doi:10.1016/j.cgh.2019.07.046
18. Lin N, Ye A, Lin J, et al. Diagnostic value of detection of pregenomic RNA in sera of hepatitis B virus-infected patients with different clinical outcomes. J Clin Microbiol. 2020;58(2):
19. Huang H, Wang J, Li W, et al. Serum HBV DNA plus RNA shows superiority in reflecting the activity of intrahepatic cccDNA in treatment-naive HBV-infected individuals. J Clin Virol. 2018;99–100:71–78. doi:10.1016/j.jcv.2017.12.016
20. Wu Y, Wen J, Xiao W, Zhang B. Pregenomic RNA: how to assist the management of chronic hepatitis B? Rev Med Virol. 2019;29(4):e2051. doi:10.1002/rmv.2051
21. van Campenhout MJH, van Bommel F, Pfefferkorn M, et al. Serum hepatitis B virus RNA predicts response to peginterferon treatment in HBeAg-positive chronic hepatitis B. J Viral Hepat. 2020. doi:10.1111/jvh.13272
22. van Bommel F, van Bommel A, Krauel A, et al. Serum HBV RNA as a predictor of peginterferon alfa-2a response in patients with HBeAg-positive chronic hepatitis B. J Infect Dis. 2018;218(7):1066–1074. doi:10.1093/infdis/jiy270
23. Luo H, Zhang XX, Cao LH, et al. Serum hepatitis B virus RNA is a predictor of HBeAg seroconversion and virological response with entecavir treatment in chronic hepatitis B patients. World J Gastroenterol. 2019;25(6):719–728. doi:10.3748/wjg.v25.i6.719
24. Wang J, Yu Y, Li G, et al. Natural history of serum HBV-RNA in chronic HBV infection. J Viral Hepat. 2018;25(9):1038–1047. doi:10.1111/jvh.12908
25. Gu Y, Chen L, Lian Y, et al. Serum HBV pregenomic RNA is correlated with Th1/Th2 immunity in treatment-naive chronic hepatitis B patients. J Med Virol. 2020;92(3):317–328. doi:10.1002/jmv.25612
26. van Campenhout MJH, van Bommel F, Pfefferkorn M, et al. Host and viral factors associated with serum hepatitis B virus RNA levels among patients in need for treatment. Hepatology. 2018;68(3):839–847. doi:10.1002/hep.29872
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.