Back to Journals » Cancer Management and Research » Volume 11
Serum hepatitis B viral (HBV) DNA is a predictive biomarker for survival in non-small cell lung cancer patients with chronic HBV infection
Authors Fu Y, Yang X, Liang H, Wu X
Received 18 December 2018
Accepted for publication 26 February 2019
Published 31 May 2019 Volume 2019:11 Pages 5091—5100
DOI https://doi.org/10.2147/CMAR.S198714
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Harikrishna Nakshatri
Yumei Fu,1 Xiaoyi Yang,1 Huifen Liang,1 Xingping Wu2
1Laboratory Department of Liwan Hospital of The Third Affiliated Hospital of Guangzhou Medicine University, Guangzhou, 510175, People’s Republic of China; 2State Key Laboratory of Oncology in South China, Collaborative Innovation Center for Cancer Medicine, Sun Yat-sen University Cancer Center, Guangzhou 510060, People’s Republic of China
Purpose: To study the association between pretreatment serum hepatitis B viral (HBV) DNA copy numbers and clinical outcome of non-small cell lung cancer (NSCLC) patients with chronic HBV infection.
Patients and methods: We retrospectively evaluated the medical records of NSCLC HBV (+) patients between January 2008 and December 2010. The HBV DNA copy numbers and other prognostic factors including albumin (ALB), C-reactive protein (CRP), platelet, neutrophil–lymphocyte ratio (NLR), platelet–lymphocyte ratio (PLR), and Glasgow Prognostic Score (GPS) were obtained before any antitumor treatment. Kaplan–Meier curves and the log-rank test were used to calculate prognostic significance. A multivariable Cox proportional hazard regression was modeled to analyze the independent prognostic factors for NSCLC HBV (+) patients. All independent prognostic factors in the Cox multivariable analysis were used to build a nomogram. The predictive accuracy of HBV DNA, TNM stage and nomogram was evaluated with the concordance index (C-index) and time-dependent receiver operating characteristics (ROC) curves, and simultaneously compared with traditional TNM staging system respectively.
Results: A total of 188 patients were recruited in this study; the median age was 56 years, and the median overall survival (OS) was 34 months. Cox multivariate analysis results showed independent factors for OS including TNM stage (P=0.028), treatment (P=0.002), HBV DNA (P<0.001), and GPS (P=0.026). The nomogram model for survival was built based on four prognostic factors. The C-index for HBV DNA was 0.67, 0.69 for TNM stage, and 0.76 for the nomogram model. There was no statistical difference between HBV DNA and TNM stage (P=0.48). However, the C-index values of nomogram model were statistically higher both than HBV DNA (0.76 vs 0.67, P<0.001), and TNM stage (0.76 vs 0.69, P<0.001).
Conclusion: Pretreatment serum HBV DNA copy numbers can act as a prognostic marker of survival for NSCLC patients with chronic HBV infection.
Keywords: non-small cell lung cancer, hepatitis B, hepatitis B viral DNA, HBV DNA, nomogram, prognosis
Introduction
Lung cancer is the commonest and most fatal of all cancers worldwide. The 5-year overall survival (OS) rate is estimated at 18%.1 Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, and accounts for approximately 85% of all lung cancer cases.2
A chronic hepatitis B (CHB) infection typically induces prolonged inflammation.3 Chronic inflammation can lead to carcinogenesis; there is evidence that inflammation can affect the proliferation and survival of malignant cells, angiogenesis, tumor initiation and metastasis, and tumor response to drugs.4,5 Moreover, hepatitis B viral (HBV) infection is associated with poorer outcomes for hepatocellular,6 pancreatic carcinoma,7 nasopharyngeal carcinoma,8 and NSCLC cases.9 This finding may imply that investigation of a new prognostic model is required in HBV-positive NSCLC patients.
Many studies have reported that the serum level of HBV DNA correlates with the progression of hepatocellular carcinoma (HCC).10 No studies, however, have explored the influence of serum HBV DNA on the progression of NSCLC. Therefore, it is necessary to determine the ability of HBV DNA to predict the outcome of HBV-positive NSCLC patients, to both improve and optimize therapies.
The American Joint Committee on Cancer (AJCC) TNM classification is the current standard for determining treatment schemes for many cancer patients. However, because of the heterogeneous nature of cancer, differences in clinical outcomes may arise in cancer patients with the same TNM stage receiving similar treatment strategies.11 This indicates that further research is needed to explore more functional biomarkers which are able to improve prognosis prediction of cancer. Numerous risk factors, such as age,12 sex,13 smoking status,14 albumin (ALB), C-reactive protein (CRP),15 neutrophil–lymphocyte ratio (NLR),16 platelet–lymphocyte ratio (PLR),16 and Glasgow Prognostic Score (GPS)17 have been reported to be prognostic factors for lung cancer. Nomograms are widely used to predict cancer prognosis for an individual patient.18 Moreover, a nomogram could have several advantages over the TNM stage.19–21
The purpose of the present study is to evaluate the ability of HBV DNA in combination with inflammatory markers and clinicopathological prognostic factors to predict outcomes for NSCLC patients with HBV infection. Notably, we develop a nomogram-based risk prediction model and compare the predictive power of HBV DNA and the nomogram model with conventional TNM staging.
Material and methods
Patients and study design
The present study was approved by the Clinical Research Ethics Committee of Liwan Hospital of the Third Affiliated Hospital of Guangzhou Medicine University and Sun Yat-sen University Cancer Center (Guangzhou, China). Informed consent was provided by each included patient. We performed a retrospective review of NSCLC patients with chronic HBV infection from Liwan Hospital of the Third Affiliated Hospital of Guangzhou Medicine University and Sun Yat-sen University Cancer Center between January 2008 and December 2010. The inclusion criteria as follows. All patients were newly diagnosed and histologically confirmed as NSCLC; all patients had HBsAg (+); and had no fibrosis, steatosis or cirrhosis of the liver. Patients were excluded if they were diagnosed with malignancies other than NSCLC, had acute hepatitis, or had hepatitis A or hepatitis C infection or had incomplete clinical pathological data.
The following information was obtained from the patients’ medical records: patient’s name, gender, age, family history of cancer, smoking status, TNM stage (according to the 7th edition of AJCC TNM staging manual), histological type, tumor diameter, and treatment. Patients’ serum HBV DNA, ALB, CRP, platelet, NLR, PLR, and GPS were obtained before anti-tumor treatment.
The inflammation-based prognostic factors were defined and calculated as follows: GPS: patients with both CRP >10 mg/L and ALB <35 g/L were allocated a score of 2; patients with CRP >10 mg/L or ALB <35 g/L were allocated a score of 1; and patients with neither of these abnormalities were categorized as having a score of 0.22
All continuous variables were transformed into categorical variables based on the optimal cutoff values. These were each 10-fold increase in HBV DNA level or the median values for age (56 years), tumor size (4.0 cm), ALB (42.0 g/L), CRP (4.0 mg/L), platelet (229×109/L), NLR (2.23), and PLR (121.5).
Overall survival (OS) was defined time from the date of the diagnosis until the date of death, or the deadline. Patient follow-up was complete up to January 2016.
Statistical analyses
Statistical analyses were performed with SPSS Statistics 19.0 (IBM Corporation, Armonk, NY, USA) and R version 3.5.1 (Institute of Statistics and Mathematics, Vienna, Austria). Survival curves were depicted using the Kaplan–Meier curve with log-rank test, to calculate the prognostic significance. All prognostic factors with P<0.05 in the univariable analysis were included as covariates in the multivariable Cox proportional hazard regression to determine the independent prognostic factors for NSCLC patients with chronic HBV infection. The nomogram was built on the results of the Cox regression. The predictive accuracy and discrimination of HBV DNA, TNM stage and nomogram were compared using Harrell’s concordance index (C-index)23 and time-dependent receiver operative characteristics (ROC) curve.24 A larger C-index and area under curve (AUC) of ROC indicated more accurate prognostic stratification. A two-sided P<0.05 was of statistical significance.
Results
Baseline clinical and characteristics
In accordance with our selection criteria, a total of 188 HBV-positive NSCLC patients were eligible for this study. Of these, 144 (76.6%) were male and 44 (23.4%) were female. The median age and OS were 56 years (27–82 years) and 34 months (range =1–82 months), respectively. Based on the concentrations of HBV DNA (copies/mL), the 188 patients were divided into five groups containing <103 (60, 31.9%), 103–9,999 (49, 26.1%), 104–99,999 (30, 16.0%), 105–999,999 (22, 11.7%), and ≥106 (27, 14.3%) respectively. Baseline characteristics of these patients are listed in Table 1.
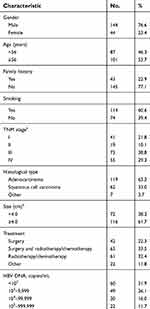 | Table 1 Baseline clinical and characteristics of patients |
Univariable and multivariable analysis
Univariable survival analysis showed that TNM stage, tumor size, treatment, HBV DNA, ALB, CRP, NLR, and GPS were associated with the OS of HBV-positive NSCLC patients (Table 2). In the multivariable Cox proportional hazard regression, results continued to demonstrate that independent factors for OS including TNM stage (HR: 1.36; 95% CI: 1.03–1.78, P=0.028), treatment (HR: 1.57; 95% CI: 1.17–2.09, P=0.002), HBV DNA (HR: 1.43; 95% CI: 1.23–1.67, P<0.001), and GPS (HR: 1.69; 95%CI: 1.07–2.69, P=0.026) (Table 2). A forest plot showing the hazard ratios and 95% confidence intervals for OS according to the multivariate Cox proportional hazard results is presented in Figure 1.
 | Table 2 Univariable and multivariable analysis for OS |
Survival analysis
Kaplan–Meier curves and the log-rank test were used to calculate prognostic significance. The results showed that serum HBV DNA levels significantly affected OS (P<0.001, Figure 2D). The 1-year OS rates for the patients with serum HBV DNA <103, 103–9,999, 104–99,999, 105–999,999, and ≥106 were 90.0%, 83.7%, 83.3%, 68.2%, and 55.6%, respectively (P=0.005, Figure 2A). The 3-year OS rates were 73.3%, 44.9%, 56.7%, 18.2%, and 14.8%, respectively (P<0.001, Figure 2B). The 5-year OS rates were 60.0%, 28.6%, 23.3%, 4.5%, and 7.4%, respectively (P<0.001, Figure 2C).
Prognostic nomogram for OS
We constructed a prognostic nomogram model incorporating all significant independent factors for OS based on multivariable Cox proportional hazards results, and further visualized it as shown in Figure 3. Each prognostic factor was assigned a point. By summing up the risk points from all the variables and locating it to the total points scale, we could determine the 1-, 3- and 5-year OS probability of a specific patient by drawing a vertical line from “Total Points” towards the axis labeled “1-, 3- and 5-year survival”. The prognostic nomogram model showed that serum HBV DNA was considered to be the most promising potential biomarker to complement TNM stage.
Comparison of predictive accuracy between independent factors and nomogram model
The C-indices for TNM stage, treatment, HBV DNA, GPS, and the nomogram model were 0.69 (95% CI: 0.64–0.73), 0.68 (95% CI: 0.63–0.74), 0.67 (95% CI: 0.61–0.72), 0.61 (95% CI: 0.56–0.66), and 0.76 (95% CI: 0.72–0.81), respectively. The nomogram model showed much higher results than the TNM stage (0.76 vs 0.69, P<0.001) and HBV DNA (0.76 vs 0.67, P<0.001). These results indicated the nomogram model had greater predictive accuracy. However, there was no statistical difference between HBV DNA and TNM stage (0.67 vs 0.69, P=0.48), treatment (0.67 vs 0.68, P=0.50), and GPS (0.67 vs 0.61, P=0.12). The results are listed in Table 3. No significant difference was observed when comparing the area under curves (AUCs) of time-dependent ROC curve between HBV DNA and TNM stage, treatment, and GPS (Figure 4).
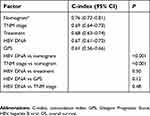 | Table 3 The C-index of independent factors and nomogram for prediction of OS* |
Discussion
In this study, we retrospectively evaluated the prognostic role of serum HBV DNA in NSCLC patients with chronic HBV infection. Our results indicated that HBV DNA is an independent prognostic marker for the HBV-positive NSCLC patients based on both univariable and multivariable analyses. Patients with high levels of HBV DNA have poor prognosis. Accordingly, this finding highlights the importance of baseline serum HBV DNA for predicting outcomes, and thus suggests that cancer therapy should be combined with antiviral therapy in NSCLC patients with hepatitis B. In addition, we also create a prognostic nomogram model for OS in HBV-positive NSCLC patients, which including HBV DNA, TNM stage, treatment, and GPS. This nomogram has more predictive accuracy than TNM stage.
HBV infection, as an independent prognostic factor, has been reported in patients with hepatocellular, pancreatic carcinoma, nasopharyngeal carcinoma, and NSCLC. Chen et al established a prognostic nomogram model for OS in HBV-positive NSCLC patients based on liver function markers25 and inflammation-based factors,26 but did not include the HBV DNA levels. Interestingly, our study incorporated HBV DNA copy numbers, clinicopathologic factors, and inflammation-based prognostic factors into the TNM staging system to establish a nomogram model, which has a better predictive accuracy and discriminative ability than HBV DNA alone and TNM stage. Our results could help clinicians with decision-making and clinical consultations.
The molecular mechanisms underlying the influence of HBV on the outcome of NSCLC patients are not understood. There are several possible explanations. Firstly, the interaction of HBV and the host immune system could lead to nonresolving inflammation,27 which could activate transcription factors (NF-κB, STAT3), and then promote the production of cytokines and chemokines.28,29 These aid in the proliferation and survival of malignant cells, and promote angiogenesis and metastasis.30 Secondly, HBV had been detected in extrahepatic tissue and diverse cell types, which means HBV could also exist in the lung.31 Prolonged exposure to HBV may promote cancer proliferation, invasion, and metastasis.32,33 Finally, the hepatitis B virus X protein (HBx) is a small 17-kDa soluble protein in the nucleus and cytoplasm of host cells,34 that is closely related to the carcinogenesis of HCC.35 Some studies have shown significant alteration in expression of lncRNA, microRNA and mRNA induced by HBx in liver cell lines.36,37 Many of the above molecules displayed diverse biological functions, such as gene expression, DNA replication, DNA repair, cell cycle, cell division, cell–cell signaling, cell adhesion, signal transduction, and cell surface receptor signaling pathway.37 These molecules may enter the blood circulation to the lung, where they could promote tumor growth and progression.
There are several limitations to our study. First of all, it was a retrospective study, and thus a selection bias cannot be fully excluded. Secondly, all the data came from a single institute with a relatively small sample size. Therefore, external multi-institute validation is needed in the future. Third, information on post-antiviral drug intake was insufficient. Finally, other potential prognostic factors such as cytokeratin 19 fragment (CYFRA21-1), carcinoembryonic antigen (CEA), and epidermal growth factor receptor (EGFR) were not assessed.38,39 In the future, more studies are needed to confirm the mechanisms underlying the observed influence of HBV on the survival outcomes of NSCLC patients.
In conclusion, our study demonstrates that pretreatment serum HBV DNA is an independent prognostic marker for NSCLC patients with chronic HBV infection. Higher copy numbers were linked with poor prognosis. The predictive accuracy of HBV DNA was equivalent to TNM stage. A prognostic nomogram based on HBV DNA, TNM stage, treatment, and GPS could enhance the ability to predict OS for HBV-positive NSCLC patients.
Acknowledgments
We thank the staff at the Laboratory Department of Liwan Hospital of The Third Affiliated Hospital of Guangzhou Medicine University, and the Director of Clinical Laboratories, Sun Yat-sen University Cancer Center for providing support on research conditions in this study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi:10.3322/caac.21387
2. Sève P, Reiman T, Dumontet C. The role of betaIII tubulin in predicting chemoresistance in non-small cell lung cancer. Lung Cancer. 2010;67(2):136–143. doi:10.1016/j.lungcan.2009.09.007
3. Xie Y. Hepatitis B virus-associated hepatocellular carcinoma. Adv Exp Med Biol. 2017;1018:11–21. doi:10.1007/978-981-10-5765-6_2
4. Gomes M, Teixeira AL, Coelho A, et al. The role of inflammation in lung cancer. Adv Exp Med Biol. 2014;816(816):1–23.
5. Morrison WB. Inflammation and cancer: a comparative view. J Vet Intern Med. 2012;26(1):18–31. doi:10.1111/j.1939-1676.2011.00836.x
6. Chang TS, Lee KF, Tung SY, et al. Abstract 1792: association of enhanced IGF-1R signaling and HBV/inflammation with poor HCC prognosis. Cancer Res. 2011;70(8 Supplement):1792. doi:10.1158/1538-7445.AM10-1792
7. Wei XL, Qiu MZ, Chen WW, et al. The status of HBV infection influences metastatic pattern and survival in Chinese patients with pancreatic cancer. J Transl Med. 2013;11(1):249–257. doi:10.1186/1479-5876-11-249
8. Liu X, Li X, Jiang N, et al. Prognostic value of chronic hepatitis B virus infection in patients with nasopharyngeal carcinoma: analysis of 1301 patients from an endemic area in China. Cancer. 2014;120(1):68–76. doi:10.1002/cncr.28377
9. Peng JW, Liu DY, Lin GN, et al. Hepatitis B virus infection is associated with poor prognosis in patients with advanced non small cell lung cancer. Asian Pac J Cancer Prev. 2015;16(13):5285–5288.
10. Ohkubo K, Kato Y, Ichikawa T, et al. Viral load is a significant prognostic factor for hepatitis B virus-associated hepatocellular carcinoma. Cancer. 2010;94(10):2663–2668. doi:10.1002/cncr.10557
11. Mao YP, Xie FY, Liu LZ, et al. Re-evaluation of 6th edition of AJCC staging system for nasopharyngeal carcinoma and proposed improvement based on magnetic resonance imaging. Int J Radiat Oncol Biol Phys. 2009;73(5):1326–1334. doi:10.1016/j.ijrobp.2008.07.062
12. Foucher P, Coudert B, Arveux P, et al. Age and prognosis of non-small cell lung cancer. Usefulness of a relative survival model. Eur J Cancer. 1993;29A(13):1809–1813.
13. Salmerón D, Chirlaque MD, Izarzugaza MI, et al. Lung cancer prognosis in Spain: the role of histology, age and sex. Respir Med. 2012;106(9):1301–1308. doi:10.1016/j.rmed.2012.06.006
14. Parsons A, Daley A, Begh R, et al. Influence of smoking cessation after diagnosis of early stage lung cancer on prognosis: systematic review of observational studies with meta-analysis. BMJ. 2010;340(7740):251–257. doi:10.1136/bmj.b5569
15. Cavit C, Mustafa Fuat A, Ata O, et al. Prognostic impact of intratumoral C-reactive protein expression in patients with clear cell renal cell carcinoma. Urol Int. 2014;92(3):270–275. doi:10.1159/000353401
16. Diem S, Schmid S, Krapf M, et al. Neutrophil-to-Lymphocyte ratio (NLR) and Platelet-to-Lymphocyte ratio (PLR) as prognostic markers in patients with non-small cell lung cancer (NSCLC) treated with nivolumab. Lung Cancer. 2017;111:176–181. doi:10.1016/j.lungcan.2017.07.024
17. Kawashima M, Murakawa T, Shinozaki T, et al. Significance of the Glasgow Prognostic Score as a prognostic indicator for lung cancer surgery. Interact Cardiovasc Thorac Surg. 2015;21(5):637–643. doi:10.1093/icvts/ivv223
18. Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26(8):1364–1370. doi:10.1200/JCO.2007.12.9791
19. Galsky MD, Erin Moshier MS, Susan Krege MD, et al. Nomogram for predicting survival in patients with unresectable and/or metastatic urothelial cancer who are treated with cisplatin-based chemotherapy. Cancer. 2013;119(16):3012–3019. doi:10.1002/cncr.28146
20. Kim SH, Shin KH, Kim HY, et al. Postoperative nomogram to predict the probability of metastasis in Enneking stage IIB extremity osteosarcoma. BMC Cancer. 2014;14(1):1–10. doi:10.1186/1471-2407-14-1
21. Lee CK, Goldstein D, Gibbs E, et al. Development and validation of prognostic nomograms for metastatic gastrointestinal stromal tumour treated with imatinib. Eur J Cancer. 2015;51(7):852–860. doi:10.1016/j.ejca.2015.02.015
22. Forrest L, McMillan D, McArdle C, et al. Evaluation of cumulative prognostic scores based on the systemic inflammatory response in patients with inoperable non-small-cell lung cancer. Br J Cancer. 2003;89(6):1028–1030. doi:10.1038/sj.bjc.6601242
23. Pencina MJ, Da RB. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat Med. 2010;23(13):2109–2123. doi:10.1002/sim.1802
24. Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000;56(2):337–344.
25. Chen S, Lai Y, He Z, et al. Establishment and validation of a predictive nomogram model for non-small cell lung cancer patients with chronic hepatitis B viral infection. J Transl Med. 2018;16(1):116–125. doi:10.1186/s12967-018-1496-5
26. Chen S, Li X, Lv H, et al. Prognostic dynamic nomogram integrated with inflammation-based factors for non-small cell lung cancer patients with chronic hepatitis B viral infection. Int J Biol Sci. 2018;14(13):1813–1821. doi:10.7150/ijbs.27260
27. Zi-Xiong LI, Zhang Q, Lin J, et al. Possible mechanisms of malignant transformation of nonresolving inflammation caused by chronic infection with hepatitis B virus. Progr Biochem Biophys. 2014;41(1):9–16.
28. Yu LX, Ling Y, Wang HY. Role of nonresolving inflammation in hepatocellular carcinoma development and progression. Npj Precis Oncol. 2018;2(1):6–14. doi:10.1038/s41698-018-0048-z
29. Nathan C, Ding A. Nonresolving inflammation. Cell. 2010;140(6):871–882. doi:10.1016/j.cell.2010.02.029
30. Mantovani A, Allavena P, Sica A, et al. Cancer-related inflammation. Nature. 2008;454(7203):436–444. doi:10.1038/nature07205
31. Mason A, Wick M, White H, et al. Hepatitis B virus replication in diverse cell types during chronic hepatitis B virus infection. Hepatology. 2010;18(4):781–789. doi:10.1002/hep.1840180406
32. Mitri MSD, Cassini R, Bernardi M. Hepatitis B virus-related hepatocarcinogenesis: molecular oncogenic potential of clear or occult infections. Eur J Cancer. 2010;46(12):2178–2186. doi:10.1016/j.ejca.2010.03.034
33. Engels EA, Cho ERJee SH. Hepatitis B virus infection and risk of non-Hodgkin lymphoma in South Korea: a cohort study. Lancet Oncol. 2010;11(9):827–834. doi:10.1016/S1470-2045(10)70167-4
34. Chao W, Wen Y, He-Xin Y, et al. Hepatitis B virus X (HBx) induces tumorigenicity of hepatic progenitor cells in 3,5-diethoxycarbonyl-1,4-dihydrocollidine-treated HBx transgenic mice. Hepatology. 2012;55(1):108–120. doi:10.1002/hep.24675
35. Yonghong L, Zhangwei T, Ting L, et al. Hepatitis B virus X protein stabilizes amplified in breast cancer 1 protein and cooperates with it to promote human hepatocellular carcinoma cell invasiveness. Hepatology. 2012;56(3):1015–1024. doi:10.1002/hep.25751
36. Hou Z, Xu X, Fu X, et al. HBx-related long non-coding RNA MALAT1 promotes cell metastasis via up-regulating LTBP3 in hepatocellular carcinoma. Am J Cancer Res. 2017;7(4):845–856.
37. Chen RC, Wang J, Kuang XY, et al. Integrated analysis of micro RNA and m RNA expression profiles in HBx-expressing hepatic cells. World J Gastroenterol. 2017;23(10):1787–1795. doi:10.3748/wjg.v23.i10.1787
38. Romero-Ventosa EY, Blanco-Prieto S, Gonzalez-Pineiro AL, et al. Pretreatment levels of the serum biomarkers CEA, CYFRA 21-1, SCC and the soluble EGFR and its ligands EGF, TGF-alpha, HB-EGF in the prediction of outcome in erlotinib treated non-small-cell lung cancer patients. Springerplus. 2015;4:171–183. doi:10.1186/s40064-015-0891-0
39. Baek AR, Seo HJ, Lee JH, et al. Prognostic value of baseline carcinoembryonic antigen and cytokeratin 19 fragment levels in advanced non-small cell lung cancer. Cancer Biomark. 2018;22(1):1–8. doi:10.3233/CBM-170885
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




