Back to Journals » Infection and Drug Resistance » Volume 14
Serotype Distribution of Streptococcus pneumoniae Isolates Causing Invasive and Non-Invasive Infections Using Whole-Genome Sequencing in Ethiopia
Authors Sharew B , Moges F , Yismaw G, Mihret A , Abebe W, Fentaw S, Tessema B
Received 25 November 2020
Accepted for publication 18 February 2021
Published 2 March 2021 Volume 2021:14 Pages 787—794
DOI https://doi.org/10.2147/IDR.S293578
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 5
Editor who approved publication: Professor Suresh Antony
Bekele Sharew,1,2 Feleke Moges,1 Gizachew Yismaw,1 Adane Mihret,3 Wondiwossen Abebe,1 Surafal Fentaw,4 Belay Tessema1
1Department of Medical Microbiology, School of Biomedical and Laboratory Sciences, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia; 2Department of Medical Laboratory Sciences, College of Medicine and Health Sciences, Wollo University, Dessie, Ethiopia; 3Armauer Hansen Research Institute, Addis Ababa, Ethiopia; 4Ethiopian Public Health Institute, Addis Ababa, Ethiopia
Correspondence: Bekele Sharew
Department of Medical Microbiology, School of Biomedical and Laboratory Sciences, College of Medicine and Health Sciences, University of Gondar, P.O. Box 45, Gondar, Ethiopia
Tel +251 911 165431
Email [email protected]
Background: In Ethiopia, pneumococcal conjugate vaccine 10 (PCV10) was introduced in 2011 in the national vaccination program. This study was aimed to assess serotype distribution of invasive and non-invasive Streptococcus pneumoniae isolates using whole-genome sequencing.
Methods: A hospital-based prospective study was conducted from 2018 to 2019 at Addis Ababa and Amhara region referral hospitals, from all patients. Clinical Samples were collected and initially cultured onto 5% sheep blood agar at 37°C in a 5% CO2 atmosphere. Sequencing was done using the Illumina NextSeq 500 and SeroBA was used to predict serotypes from whole-genome sequencing raw data.
Results: Of the 57 S. pneumoniae isolates, there were 32 circulating serotypes. The most common serotypes were 15A/B/C (n=5, 8.8%), 6A (n=4, 7.0%), 10A/F (n=4, 7.0%), 23A (n=4, 7.0%) and 7C (n=3, 5.3%). The serotype coverage of PCV10 and PCV13 were 12.3% and 26.3% respectively. The most common invasive serotypes were 15A/B/C (n=5, 8.8%) and 6A (n=4, 7.0%), and non-invasive serotypes were 23A (n=4, 7.0%) and 10A/F (n=3, 5.3%). The most prevalent serotype obtained from PCV10 eligible children was 3 (n=2, 3.5%). The prevalent serotype obtained from PCV10 non-eligible patients were type 23A (n=4, 7%) and type 6A (n=3, 5.2%). The most common serotypes among children ≤ 18 years old were 10A/F, 7C, 35A/B, 16F, 19A, 3 and 38. However, the proportions of some non-vaccine serotypes (11A/B and 15A/B/C) were higher in adult patients.
Conclusion: In this study a shift in the distribution of non-vaccinated S. pneumoniae serotypes increases in the population, and PCV10 serotype coverage was reduced as compared to PCV13. Therefore, it is important to continue monitoring serotype changes among all patients in addition to assessing the impact and effectiveness brought by vaccines and provides a foundation for prevention strategies and vaccine policies.
Keywords: Streptococcus pneumoniae, serotypes, whole-genome sequencing
Introduction
Streptococcus pneumoniae is a major pathogen that causes community-acquired pneumonia (CAP), acute exacerbations of chronic bronchitis, meningitis, sinusitis, otitis media, and sepsis; which are divided into invasive and non-invasive infections.1 Isolates of S. pneumoniae are traditionally characterized in terms of the antigenicity of their capsular polysaccharides (CPS) and to date, there are nearly 100 known serotypes described for S. pneumoniae based on the differences in the antigenic properties of the capsule.2
Serotyping methods of S. pneumoniae can be grouped into two different categories: phenotype-based methods and genotype-based methods.3 The phenotype-based methods (based on antisera reactions) remain the Gold Standard method used in most laboratories.4 However, this method is expensive, lengthy, and not fully reliable. Genotype-based methods were developed to provide cost-effective and reliable assays for the serotyping of S. pneumoniae.3,5 Among these methods, Whole-genome sequencing (WGS) became a suitable method for serotyping with the improvement in accuracy and a decrease in cost which has allowed the identification of serotype by comparing cps loci sequences.6–9
Serotype surveillance projects around the world showed an increase in S. pneumoniae disease due to non-vaccine serotypes that are caused by serotype replacement.10–14 Therefore, it is very important to survey the circulating serotypes, to observe the epidemiological trends of S. pneumoniae before and after vaccination. Globally, serotypes 19A (28%), 19F (10%), and 14 (9%) were the most common in children under 5 years. In adults over 16 years, 19A (13%), 3, 6A and 7F (all 7%) were most common.15
A higher number of serotypes are responsible for an equal proportion of invasive and non-invasive pneumococcal diseases in Africa and Asia than in North America, suggesting a greater diversity of serotypes causing disease in Africa and Asia continents.16 As a result, the World health organization (WHO) recommended the inclusion of pneumococcal conjugate vaccines in national immunization programs throughout the world.17
In Ethiopia, Pneumococcal conjugate vaccine 10 (PCV10) was introduced in 2011 in the national vaccination program as a 3 dose primary series at 6, 10, and 14 weeks of age and no booster dose (3p + 0).18 The coverage of PCV10 in Ethiopia is 63% by WHO and UNICEF and 95% by Ethiopian Minister of Health estimates of immunization coverage, 2019 report.19 Although the serotypes that circulate in different communities were not known only two studies have previously reported on the prevalence of pneumococcal and Haemophilus serotypes.20,21 These studies, however, had limitations in which the sample sources were CSF using the quellung method and the target population was children only and also one study performed two decades ago. So, more recent studies with an advanced method of characterization from different sample sources are mandatory to generalize the serotype distribution of S. pneumoniae for the entire country.
Information on serotype distribution is important to change the current PCV10 to more vaccine number coverage of PCVs. Therefore, the main goal of this study was to assess the serotype distribution of S. pneumoniae isolates which are responsible for invasive and non-invasive infections in children and adults from clinical isolates of suspected pneumococcal infections in Ethiopia using WGS assay.
Methods
Patients and Clinical Isolates
A hospital-based prospective study was conducted from January 2018 to 2019 at Addis Ababa (Yekatite12 Hospital, Alert Hospital, and International Clinical Private Laboratory) and Amhara Region (University of Gondar Comprehensive Specialized Hospital, Felege Hiwot Comprehensive Specialized Hospital and Dessie Regional Laboratory) Referral Hospitals, Ethiopia. During the study period, a total of 70 phenotypically confirmed S. pneumoniae were isolated from pediatric and adult patients suspected of pneumococcal infections. Clinical samples (cerebrospinal fluid, blood, sputum, eye discharge, ear discharge, pleural and peritoneal fluids) were collected from appropriate collection sites and were initially cultured onto 5% sheep blood agar plates overnight at 37°C in a 5% CO2 atmosphere. S. pneumoniae was identified and confirmed by typical colony morphology, alpha-hemolysis, Gram staining, optochin susceptibility, and bile solubility test. The strains were stocked in Skimmed Milk-Trypticase Soy-Glucose-Glycerol (STGG) medium and transported to Norwegian Institute of Public Health Microbiology Laboratory, Oslo, Norway using dry ice for serotyping using WGS.
PCV status was determined on the base of patients’ age at the date when the pneumococcal strain was isolated. Each patient was categorized according to his/her immunization status at pneumococcal disease presentation as either “PCV10 vaccinated” or “PCV10 non-vaccinated”. All children born on September 1st, 2011, or thereafter were eligible for vaccination regardless of whether they had received the vaccine or not. We defined them as a group of vaccinated children who had received 3 doses of PCV10. Another group of children born less than 14 weeks of age and adults not eligible for PCV10 vaccination (those born before September 2011) from the data recorded on the request of cultures to laboratories was defined as non-vaccinated individuals. We compared serotype distribution rates for both groups of patients (PCV10 eligible children and PCV10 non-eligible children and adults).
DNA Extraction of Streptococcus pneumoniae
Isolates were recovered on a blood agar plate with 5% sheep blood and incubated overnight at 37°C in a 5% CO2 atmosphere. Bacteria were collected with two full inoculating loops and suspended in the ATL master mix. Samples were then frozen at −20°C until extraction. DNA extraction was performed with the QIAamp@DNA Mini Kit, QIAGEN, and QIAcubeTM BioRobot machine work station according to the manufacturer’s instructions. DNA concentrations were measured using Qubit 4 fluorometer and QubitTM 1X dsDNA HS Assay Kit (Thermo Fisher Scientific); 40 ng of genomic DNA was used for sample preparation.
Pneumococcal Serotyping
Pneumococcal serotypes/groups were determined for the 70 isolates using WGS. Based on the WGS result, 57 (81.4%) isolates were further confirmed as S. pneumoniae. Libraries for WGS were prepared with the KAPA HyperPlus DNA library preparation kit and sequenced using an Illumina MiSeq reagent kit v3 (600 cycles, paired ends) following the manufacturer’s instructions. Illumina Nextera DNA libraries were constructed and sequenced using the Illumina NextSeq 500. SeroBA was used to predict serotypes, by identifying the cps locus, directly from raw whole genome sequencing read data with 98% concordance using a kmer-based method, can process 10, 000 samples in just over 1 day using a standard server, and can call serotypes at coverage as low as 15–21X. SeroBA is implemented in Python3 and is freely available under an open-source GPLv3 license from https://github.com/sanger-pathogens/seroba.6 If the serotype was not detected by the method mentioned above, the strain was classified as non-typable due to the lack of a capsular operon. Afterward, the coverage rates of PCV10 and PCV13 were estimated by calculating the percentage of isolates expressed the serotypes included in the vaccines.
Data Analysis
Data were entered and analyzed using the Statistical Package for the Social Science (version 20; SPSS Inc, Chicago, IL, USA). The rates of serotype distribution among vaccinated and unvaccinated patients, source of samples, and age group were compared using tables and figures. Discrete variables were expressed as percentages and proportions.
Results
Demographic and Clinical Data of Patients
We analyzed 70 phenotypically confirmed S. pneumoniae isolates recovered from specimens of children and adults with invasive and non-invasive infections. However, using whole-genome sequencing method, 57 (81.4%) isolates were S. pneumoniae, 6 (8.6%) were S. mitis, 1 (1.4%) was S. oralis, 1 (1.4%) was S. peroris and 1 (1.4%) was Enterococcus avium. Among 57 S. pneumoniae isolates, 37 (65%) were obtained from children aged between 38 days to 18 years; the remaining 20 strains (35%) were recovered from adult patients. Out of the 37 children, 20 had received PCV10 vaccination (age-based vaccine status), covering 35.1% out of all patients with invasive and non-invasive infection (Table 1). Invasive disease refers to isolation of S. pneumoniae from a normally sterile body sites like blood, cerebrospinal fluid (CSF) or pleural fluid and non-invasive pneumococcal disease included non-bacteremic pneumonia cases and those with isolates from non-sterile sites such as sputum, middle ear fluid and eye discharge. The clinical sources of isolates were cerebrospinal fluid (CSF) 20 (35.1%), blood 12 (21.1%), eye discharge 10 (17.5%), sputum 10 (17.5%), pleural fluid 2 (3.5%), ear discharge 2 (3.5%) and peritoneal fluid 1 (1.8%).
 |
Table 1 Distribution of S. pneumoniae Serotypes According to Source of Strain Isolation, Vaccination Status and Age Group at Addis Ababa and Amhara Region Referral Hospitals, Ethiopia, 2018–2019 |
Serotype Distribution and Conjugate Vaccine Coverage of S. pneumoniae Isolates
Serotype distribution of both invasive and non-invasive isolates according to source of the strain isolation, vaccine status and age group is presented in Table 1. There were thirty-two (32) circulating serotypes amongst the isolates: 18 were found in invasive isolates only, 10 from non-invasive isolates only and 4 from both invasive and non-invasive isolate (Figures 1 and 2). Generally, the most common serotypes were 15A/B/C (n=5, 8.8%), 6A (n=4, 7.0%), 10A/F (n=4, 7.0%), 23A (n=4, 7.0%) and 7C (n=3, 5.3%). All of these serotypes, except type 6A, are non-vaccine serotypes. In the whole isolate collection, the serotype coverage of PCV10 and PCV13 were 12.3% (n= 7) and 26.3% (n = 15) respectively (Table 1).
 |
Figure 1 Distribution of S. pneumoniae serotypes isolated from invasive pneumococcal diseases at Addis Ababa and Amhara Region Referral Hospitals, Ethiopia, 2018–2019. |
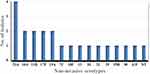 |
Figure 2 Distribution of Streptococcus pneumoniae serotypes isolated from non-invasive pneumococcal diseases at Addis Ababa and Amhara Region Referral Hospitals, Ethiopia, 2018–2019. |
Depending of source of pneumococcal isolates, the most common serotypes among invasive cases were 15A/B/C (n=5, 8.8%), 6A (n=4, 7.0%) 7C, 8, 14, 16F, 1 and 3 (n=2, 3.5% each). The PCV10 and PCV13 vaccine coverage rates were 7 (20.0%) and 13 (37.1%) for patients with invasive cases, respectively. Out of the 35 invasive isolates, fourteen were obtained from PCV10 eligible children (40%). Among PCV10 eligible children, seven were from vaccine serotypes (14, 19F, 23F, 1, 7F, 3, 6A) of both PCV10 and PCV13. However, the most common serotypes among non-invasive cases were 23A (n=4, 7.0%), 10A/F (n=3, 5.3%), 11A/B, 35A/B, 17F, and 19A (n=2, 3.5% each), and the vaccine coverage rate of PCV13 was 9.1% but not vaccine coverage for PCV10. Out of the 22 non-invasive isolates, six were obtained from PCV10 eligible children (27.3%). Among them, one was vaccine serotypes 19A. The proportions of vaccinated children among 35 invasive S. pneumoniae isolates were higher (40.0%) than the proportion of vaccinated children with non-invasive strains (27.3%).
Serotypes distribution of the isolates among the study population was also analyzed according to the vaccination status of the study participants. In isolates obtained from PCV10 eligible children (n = 20), the most prevalent serotype was 3 (n=2, 3.5%). In this group of vaccinated eligible children, 2 (10%) and 6 (30%) of the serotypes were vaccine serotypes included in PCV10 and PCV13, respectively. Among pneumococci obtained from PCV10 non-eligible patients with invasive and non-invasive isolates (n = 37), the most prevalent serotypes were 23A (n=4, 7.0%), 6A (n=3, 5.2%), 10A, 7C, 15B, 11B, 35B, 8, 16F, 1 and 38 (n=2, 3.5% each). In this group of non-eligible patients, PCV10 and PCV13 were 13.5% and 24.3% serotype coverages, respectively. The proportions of some non-vaccine serotypes (15A/B/C, 23A, 38, 16F, 8, 11A/B, 7C, 10A/F) were higher in PCV10 non-eligible patients than in the PCV10 eligible children. Similarly, some vaccine serotype isolates (6A, 1) were more often found in the non-eligible patients than in the eligible ones. However, only one vaccine serotype (3) showed higher rates in PCV10 eligible children.
Another serotype distribution in the study population was analyzed according to age groups. The most common serotypes among children ≤18 years old were 10A/F, 7C, 35A/B, 16F, 19A, 3, and 38. Serotype 19A and 3 were included in PCV13 but not included in PCV10, the vaccine currently included in the national vaccination program in Ethiopia. In this group of children, PCV10 and PCV13 were 10.8% and 27.0% serotype coverages, respectively. However, the proportions of some non-vaccine serotypes (11A/B and 15A/B/C) were higher in adult patients. The vaccine coverage rate of PCV10 and PCV13 in adults was 15% and 25%, respectively.
Discussion
S. pneumoniae serotyping has become more important since the release of the different vaccines for the monitoring of emerging non-vaccine serotypes. However, the information on pneumococcal serotype might not directly benefit the clinical diagnosis or the immediate treatment of the patient. The knowledge of pneumococcal serotypes circulating in carriage and disease is required to correctly estimate the impact of pneumococcal vaccines locally and globally and is needed by vaccine policymakers. In this study, we present serotype-specific variant profiles that can be used to distinguish 32/57 serologically distinct serogroups/serotypes and SeroBA, a bioinformatics tool that uses the capsular typing variant database to predict capsular type from WGS raw data.
Our present study demonstrated that the most common serotypes were 15A/B/C, 6A, 10A/F, 23A, and 7C, and the serotype coverages of PCV10 and PCV13 were 12.3% and 26.3%, respectively. All of these serotypes, except type 6A, were non-vaccine serotypes. Which was similar to other recent studies in Malawi,7 Russia,8 Germany,13 Japan,22 and China23 but the ranking orders varied. Vaccine coverage was lower in our study compared to other countries. In other words, there is a change in the relative rates of the most common serotypes observed due to a reduction of PCV10/13 vaccine types and a concurrent increase in non-vaccine serotypes. Because pneumococcal vaccine was included in the National vaccination schedule in September 2011 and all of the serotypes were not included in the PCV10 vaccine.
In this study, serotype 15A/B/C was isolated most frequently among all invasive isolates, followed by types 6A, 7C, 8, 14, 16F, 1, and 3. Similar findings have reported in recent studies.7,8,13,14,23,24 However, marked differences between countries, with serotype 15A/B/C dominating in Germany and China, serotype 6A dominating in China and Russia, serotype 7C dominating in China, and England and wales, serotype 3 dominating in Russia, and serotype 1 dominating in Malawi and other 11 African countries. The proportion of strains with serotypes included in PCV10 and PCV13 vaccines was 20% and 37.1%, respectively. Because five isolates were included in PCV10 and seven isolates were included in PCV13.
The most common serotypes among non-invasive isolates were 23A, 10A/F, 11A/B, 35A/B, 17F, and 19A, which is consistent with other countries in the world.7,22 The vaccine coverage rate of PCV13 was 9.1% but zero coverage rates for PCV10. This is because only six isolates were received from vaccinated children and the serotypes were not included in PCV10, only one serotype (19A) was included in PCV13.
Also, serotype 3 was more prevalent among vaccinated children with vaccine coverage of 10% for PCV10, and 30% for PCV13. Serotype 23A, 6A, 10A, 7C, 15B, 11B, 35B, 8, 16F, 1 and 38 were the most prevalent serotypes among non-vaccinated patients with a vaccine coverage of 13.5% for PCV10 and 24.3% for PCV13. As expected we observed that the introduction of vaccine was followed by a decrease in the rate of vaccine serotype accompanied by an increase in the diversity of non-vaccine serotype for both vaccinated and non-vaccinated individuals,25 a result that is in line with findings in other countries.7,8,23,24
In the present study, we reported the increase of 10A/F, 7C, 35A/B, 16F, 19A, 3, and 38 serotypes prevalence among pediatrics (≤ 18 years of age) and 11A/B and 15A/B/C were more prevalent among adults (> 18 years old). The vaccine coverage of pediatrics was 10.8% for PCV10 and 27% for PCV13, and 15% for PCV10 and 25% for PCV13 among adults. Our findings similar to the findings of other studies done in other countries with a rank difference like serotype 3 and 38 dominating in Russia,8 7C dominating in England and Wales,14 10A dominating in Japan,22 and 15A/B/C dominating in China.23 After the introduction of PCV10 into our childhood immunization program, the majority of the circulating pneumococci among vaccine eligible children were non-vaccine type which is included in PCV13 such as serotype 6A, 3, and 19A.
Compared to global data and local studies, our results demonstrated that in the total pneumococcal isolates studied, a serotype replacement was observed following the introduction of PCV10. Overall, serotype 6A appeared as the most common vaccinated serotype among all patients, and serotype 23A, 15B, 10A, and 7C also appeared as the most common non-vaccinated serotypes among all patients. Vaccine coverage was lower in our study as compared to other countries mentioned above. Although the vaccine is not widely used, we still observed the phenomenon of serotype changes, which suggests that the changes in serotype may be directly related to vaccination (serotype replacement)10,12,14 or not directly related to vaccination (pneumococci was able to change their capsular serotype by exchanging the capsular locus genes).26 The limitation of this study is the small sample size, and also lake of pre-vaccination data, descriptive study and not a vaccine effectiveness study in Ethiopia. We will perform continuous surveillance in Ethiopia to draw a reasonable analysis of the distribution of serotypes causing invasive and non-invasive infections.
Conclusions
Vaccination has substantially reduced vaccine serotype pneumococcal invasive and non-invasive diseases among all patients’ ages after the introduction of PCV10 as a routine vaccine immunization program. However, a shift in the distribution of non-vaccinated S. pneumoniae serotypes increases in the population, and PCV10 serotype coverage was reduced as compared to PCV13. Therefore, it is important to continue monitoring serotype changes among all patients in addition to assessing the impact and effectiveness brought by vaccines and provides a foundation for prevention strategies and vaccine policies.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethics Approval and Consent to Participate
All procedures performed in this study were reviewed and approved by the ethical review board of the University of Gondar (No. O/V/P/RCS/05/377/2017) in accordance with the 1964 Helsinki declaration. Permission was obtained from each hospital laboratory for collecting the isolates. Written informed consent was obtained from all patients, and a parent or legal guardian of patients under the age of 18 years after explaining the purpose and objective of the study.
Acknowledgments
The authors would like to thank the study participants. We are also very thankful for the institutional support grant by the University of Gondar. We are indebted to thank Microbiology laboratory staff of the hospitals and the University of Gondar for collecting and rechecking the isolates. We would like to thank the staff of the Norwegian Institute of Public Health Microbiology Laboratory (especially Prof. Dominique A Caugant) for their assistance in doing the whole genome sequencing and serotyping.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This study was supported by the University of Gondar with Grant No.VP/RCS/05/2/43/2017 and PhD research grant.
Disclosure
The authors declare that they have no competing interests for the publication of this paper.
References
1. Pokrovskiy VI, Tvorogova MG, Shipulin GA, eds. Molecular Diagnostics of Infectious Diseases. Moscow: RIPOL klassic; 2018.
2. van Tonder AJ, Bray JE, Quirk SJ, et al. Putatively novel serotypes and the potential for reduced vaccine effectiveness: capsular locus diversity revealed among 5405 pneumococcal genomes. Microb Genom. 2016;2(10). doi:10.1099/mgen.0.000090
3. Jauneikaite E, Tocheva AS, Jefferies JMC, et al. Current methods for capsular typing of Streptococcus pneumoniae. J Microbiol Methods. 2015;113:41–49. doi:10.1016/j.mimet.2015.03.006
4. Sørensen UB. Typing of pneumococci by using 12 pooled antisera. J Clin Microbiol. 1993;31(8):2097–2100. doi:10.1128/JCM.31.8.2097-2100.1993
5. Bentley SD, Aanensen DM, Mavroidi A, et al. Genetic analysis of the capsular biosynthetic locus from all 90 Pneumococcal serotypes. PLoS Genet. 2006;2(3):e31. doi:10.1371/journal.pgen.0020031
6. Epping L, van Tonder AJ, Gladstone RA, Bentley SD, Page AJ, Keane JA; The Global Pneumococcal Sequencing Consortium. SeroBA: rapid high-throughput serotyping of Streptococcus pneumoniae from whole genome sequence data. Microb Genom. 2018;4. doi:10.1099/mgen.0.000186.
7. Everett DB, Cornick J, Denis B, et al. Genetic characterization of malawian Pneumococci prior to the roll-out of the PCV13 vaccine using a high-throughput whole genome sequencing approach. PLoS One. 2012;7(9):e44250. doi:10.1371/journal.pone.0044250
8. Mironov KO, Korchagin VI, Mihailova YV, et al. Characterization of Streptococcus Pneumoniae strains causing invasive infections using whole-genome sequencing. J Microbiol Epidemiol Immunobiol. 2020;97(2):113–118. doi:10.36233/0372-9311-2020-97-2-113-118
9. Kapatai G, Sheppard CL, Al-Shahib A, et al. Whole genome sequencing of Streptococcus pneumoniae: development, evaluation and verification of targets for serogroup and serotype prediction using an automated pipeline. PeerJ. 2016;4:e2477. doi:10.7717/peerj.2477
10. Gladstone RA, Jefferies JM, Tocheva AS, et al. Five winters of pneumococcal serotype replacement in UK carriage following PCV introduction. Vaccine. 2015;33(17):2015–2021. doi:10.1016/j.vaccine.2015.03.012
11. Metcalf BJ, Gertz RE
12. Weinberger DM, Malley R, Lipsitch M. Serotype replacement in disease following pneumococcal vaccination: a discussion of the evidence. Lancet. 2011;378(9807):1962–1973. doi:10.1016/S0140-6736(10)62225-8
13. Makarewicz O, Lucas M, Brandt C, et al. Whole genome sequencing of 39 invasive Streptococcus pneumoniae sequence Type 199 isolates revealed switches from serotype 19A to 15B. PLoS One. 2017;12(1):e0169370. doi:10.1371/journal.pone.0169370
14. Makwana A, Ladhani SN, Kapatai G, Campion E, Fry NK, Sheppard C. Rapid spread of pneumococcal nonvaccine serotype 7c previously associated with vaccine serotype 19F, England and Wales. Emerg Infect Dis. 2018;24(10):1919–1922. doi:10.3201/eid2410.180114
15. Hackel M, Lascols C, Bouchillon S, Hilton B, Morgenstern D, Purdy J. Serotype prevalence and antibiotic resistance in Streptococcus pneumoniae clinical isolates among global populations. Vaccine. 2013;314881–314887.
16. Johnson HL, Deloria-Knoll M, Levine OS, et al. Systematic evaluation of serotypes causing invasive pneumococcal disease among children under five: the pneumococcal global serotype project. PLoS Med. 2010;7(10):e1000348. doi:10.1371/journal.pmed.1000348
17. Pneumococcal conjugate vaccine for childhood immunization-WHO position paper. Wkly Epidemiol Rec. 2007;82(12):93–104.
18. Ministry of Health Federal Republic of Ethiopia. Introducing pneumococcal conjugate vaccine in Ethiopia: training manual for health workers. Addis Ababa, Ethiopia; 2011.
19. WHO and UNICEF estimates of immunization coverage: 2019 revision, Ethiopia
20. Muhe L, Klugman KP. Pneumococcal and Haemophilus influenzae meningitis in a children’s hospital in Ethiopia: serotypes and susceptibility patterns. Trop Med Int Health. 1999;4(6):421–427. doi:10.1046/j.1365-3156.1999.00417.x
21. Tegene B, Denekew K, Mesele G. Phenotypic Characterization and Serotypes Identification of CSF isolates in acute bacterial meningitis. Am J Infect Dis. 2017;5(3):100–105. doi:10.12691/ajidm-5-3-1
22. Chang B, Morita M, Lee K-I, Ohnishi M. Whole-genome sequence analysis of streptococcus pneumoniae strains that cause hospital-acquired pneumonia infections. J Clin Microbiol. 2018;56(5):e01822–17. doi:10.1128/JCM.01822-17
23. Zhao W, Pan F, Wang B, et al. Epidemiology characteristics of Streptococcus pneumoniae from children with pneumonia in Shanghai: a retrospective study. Front Cell Infect Microbiol. 2019;9:258. doi:10.3389/fcimb.2019.00258
24. Cornick JE, Chaguza C, Harris SR, et al.; for the PAGe Consortium. Region-specific diversification of the highly virulent serotype 1 Streptococcus pneumoniae. Microb Genom. 2015;1–13. doi:10.1099/mgen.0.000027
25. Morales M, Ludwig G, Ercibengoa M, et al. Changes in the serotype distribution of Streptococcus pneumoniae causing otitis media after PCV13 introduction in Spain. PLoS One. 2018;13(12):e0209048. doi:10.1371/journal.pone.0209048
26. Coffey TJ, Enright MC, Daniels M, et al. Recombinational exchanges at the capsular polysaccharide biosynthetic locus lead to frequent serotype changes among natural isolates of Streptococcus pneumoniae. Mol Microbiol. 1998;27(1):73–83. doi:10.1046/j.1365-2958.1998.00658.x
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
