Back to Journals » OncoTargets and Therapy » Volume 13
Sequential Whole Exome Sequencing Reveals Somatic Mutations Associated with Platinum Response in NSCLC
Authors Guo AX, Xiao F, Shao WH, Zhan Y, Zhang L, Xiong J, Gao Y, Yin JY
Received 24 March 2020
Accepted for publication 28 May 2020
Published 3 July 2020 Volume 2020:13 Pages 6485—6496
DOI https://doi.org/10.2147/OTT.S254747
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Ao-Xiang Guo,1– 4 Fan Xiao,1– 4 Wei-Hua Shao,1– 4 Yan Zhan,1– 4 Le Zhang,5 Jing Xiong,6 Yang Gao,1,2,7,8 Ji-Ye Yin1– 4,9
1Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410078, People’s Republic of China; 2Institute of Clinical Pharmacology, Central South University, Hunan Key Laboratory of Pharmacogenetics, Changsha 410078, People’s Republic of China; 3Engineering Research Center of Applied Technology of Pharmacogenomics, Ministry of Education, Changsha 410078, People’s Republic of China; 4National Clinical Research Center for Geriatric Disorders, Changsha 410008, Hunan, People’s Republic of China; 5Department of Neurology, Xiangya Hospital, Central South University, Changsha, Hunan, People’s Republic of China; 6Department of Gynaecology and Obstetrics, The Second Xiangya Hospital, Central South University, Changsha 410011, People’s Republic of China; 7Department of Thoracic Surgery, Xiangya Hospital, Central South University, Changsha 410078, People’s Republic of China; 8Hunan Engineering Research Center for Pulmonary Nodules Precise Diagnosis & Treatment, Changsha 410078, People’s Republic of China; 9Hunan Key Laboratory of Precise Diagnosis and Treatment of Gastrointestinal Tumor, Changsha 410078, People’s Republic of China
Correspondence: Yang Gao
Department of Thoracic Surgery, Xiangya Hospital, Central South University, Changsha 410078, People’s Republic of China
Tel +86 731 89753003
Fax +86 731 82354476
Email [email protected]
Ji-Ye Yin
Department of Clinical Pharmacology, Xiangya Hospital, Central South University, Changsha 410078, People’s Republic of China
Tel +86 731 84805380
Fax +86 731 82354476
Email [email protected]
Purpose: Resistance is one of the main limitations of successful platinum treatment in non-small-cell lung cancer (NSCLC) patients. In this study, we aimed to identify somatic mutations associated with platinum response.
Patients and Methods: A total of 57 patients who received platinum-based chemotherapy only and 13 patients who received neoadjuvant chemotherapy (NAC) were enrolled. Somatic mutations were obtained from targeted and whole exome sequencing (WES).
Results: Somatic mutations in a total of 225 genes were observed. Nonsynonymous variants in EGFR, TTN, TP53 and KRAS, and copy number variations (SCNVs) in chromosome 8q24.3 and 22q11.21 were identified to be associated with platinum response. Based on these mutations, the mutational signature associated with the failure of DNA double-strand break and calcium signaling pathways were identified to be associated with platinum response. Besides, we observed a decrease in tumor mutational burden after chemotherapy. We also evaluated the mutation spectrum consistency between cell-free DNA (cfDNA) and tissue DNA. Somatic mutations detected in cfDNA were consistent with that in tDNA, which indicated that plasma might be used for somatic mutation detection.
Conclusion: These results support that somatic mutations can affect platinum drug response and provide potential clinical biomarkers for NSCLC treatment.
Keywords: platinum, cfDNA, NSCLC, TTN, TP53
Introduction
Lung cancer is one of the most leading cancers around the world.1 Non-small-cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancer patients.2 Currently, platinum-based chemotherapy is one of the primary treatments for NSCLC, especially for patients in advanced stage.3,4 Recent studies further showed that platinum-based chemotherapy combined with immunotherapy including pembrolizumab and atezolizumab could enhance therapeutic effects.5,6 However, patients show different responses to platinum, which becomes one of the major limitations in the clinical treatment. Therefore, it is necessary to identify biomarkers with the capability of predicting platinum response.
A large number of studies demonstrated that genetic alterations were associated with drug response.7,8 Thus, they could be served as effective biomarkers for platinum response prediction. Previously, our and other studies have identified some germline mutations significantly associated with platinum response in NSCLC patients.9–12 Based on these findings, we established a prediction model integrating both germline mutations and clinical factors by using data mining techniques. This model achieved an overall accuracy of 0.76 with the receiver operating characteristic (ROC) area under curve (AUC) of 0.80.9 However, the performance of this model still needs to be improved. We propose that germline mutations cannot completely predict platinum drug response. Other biomarkers are still waiting to be discovered.
Mutations accumulated in somatic cells can lead to cancer.13 Some somatic mutations have been successfully developed as cancer treatment targets. Thus, various types of somatic mutations including single nucleotide variants (SNVs), insertion-deletion (InDel) and somatic copy number variations (SCNVs) can be used as biomarkers to predict drug response. Previously, somatic mutations were mainly used in targeted drug therapy, such as epidermal growth factor receptor-tyrosine kinase inhibitor (EGFR-TKI).14 In recent years, some studies found that somatic mutations were also associated with the response of chemotherapeutic drugs such as anthracycline.15 Therefore, we hypothesized that they might also affect platinum response in NSCLC patients and could be developed as new pharmacogenomics (PGx) biomarkers for platinum.
In this study, we aimed to evaluate the association of somatic mutations with platinum response in Chinese NSCLC patients. These results may provide more information to support platinum drug response prediction.
Patients and Methods
Ethical Statement
The study was approved by the Ethics Committee of Xiangya Hospital, Central South University (permission NO.201904103). All participants signed informed consent prior to inclusion in the study. This study was conducted in accordance with the Declaration of Helsinki.
Patient Samples
We prospectively enrolled 72 patients diagnosed with primary NSCLC at Xiangya Hospital, Central South University (Changsha, Hunan, China). All patients received at least 2 cycles of platinum-based chemotherapy and were not given radiotherapy and/or biological therapy before and during chemotherapy. As indicated in Figure S1, we got pre-chemotherapy samples from 57 patients who received chemotherapy only, including formalin fixed paraffin embedded (FFPE) samples from 43 patients, plasma cell-free DNA (cfDNA) from 8 patients, and matched FFPE samples and plasma cfDNA from 6 patients. In addition, we got samples from 13 patients who received platinum-based neoadjuvant chemotherapy (NAC), including matched pre- and post-chemotherapy samples from 4 patients and post-chemotherapy samples from 9 patients. According to the Response Evaluation Criteria in Solid Tumors (RECIST) guideline, we regarded complete response (CR) and partial response (PR) as responders, while stable disease (SD) and progressive disease (PD) were non-responders.
DNA Extraction and Sequencing
Genomic DNA was extracted from venous blood using Wizard Genomic DNA Purification Kit (Promega, WI, USA). Fresh frozen tissue DNA, FFPE DNA and cfDNA were extracted using DNeasy Tissue Kit (Qiagen, Hilden, Germany), GeneRead DNA FFPE Kit (Qiagen) and QIAamp Circulating Nucleic Acid Kit (Qiagen), respectively. Extracted DNA was quantified by NanoDrop ND-1000 spectrophotometer (Thermo Scientific, Delaware, USA) and Qubit 2.0 Fluorometer (Life Technologies, Massachusetts, USA). The integrity of DNA was examined by agarose gel electrophoresis. Targeted sequencing depth of DNA from FFPE samples and cfDNA was more than 1000× and 5000×, respectively. Whole exome sequencing (WES) depth of DNA from blood, fresh frozen tissue and FFPE samples was 100×, 200× and 500×, respectively. Tumor cell percentage was assessed based on the hematoxylin-eosin (HE) staining. The proportion of tumor cells was more than 10% in FFPE samples used for targeted sequencing and was more than 30% in FFPE samples for WES. Targeted sequencing and WES was performed on Illumina Nextseq500 (Illumina, CA, USA) and Novaseq platform (Illumina), respectively.
Bioinformatics Analysis
Sequence Alignment and Variant Calling
Trimmomatic was used for quality control of raw data in FASTQ file format.16 The adapter contamination, low quality and unrecognizable nucleotides were discarded. Remained clean reads were aligned by Burrows Wheeler Aligner (BWA).17 SAM tools18 and Picard (http://broadinstitute.github.io/picard/) were used to sort BAM files and remove duplicates. GATK was employed for local realignment and base quality recalibration.19 Then, the generated final BAM files were subjected to variant calling. Only high-quality reads were inputted. SNVs and InDels were called by Mutect20 and Strelka.21 Finally, filtered variants were annotated by ANNOVAR.22 In the current study, only somatic mutations in exons and splice sites were further analyzed. Control-FREEC was utilized to detect SCNVs.23 Tumor mutation burden (TMB) was defined as the total number of nonsynonymous SNV and InDel variants.
Mutational Signature, Functional Classification and Pathway Analysis
R package of “deconstructSigs” was used to access mutational signature contribution. All discovered signatures were compared to the 30 signatures in COSMIC database.24 The enriched functional groupings Gene Ontology (GO) classification and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways were analyzed by using Database for Annotation Visualization and Integrated Discovery (DAVID).25
Statistical Analysis
SPSS software (IBM, NY, USA) was used for statistical analysis. Independent samples t-test was used to compare the variant allelic fractions (VAF) of somatic mutations between responders and non-responders. For focal GO and KEGG pathway analysis, P value was corrected for multiple hypothesis testing using the Benjamini–Hochberg method. A two-sided P value less than 0.05 was considered to be statistically significant in all analyses.
Results
Patient Characteristics
The clinical characteristics of this cohort are summarized in Table 1. The median age of the patients was 57 years at the time of treatment initiation. Most of them (91.43%) were stage III or IV at diagnosis. 88.57% of the patients were men and 77.14% of them had a history of smoking. We performed targeted sequencing on the pre-chemotherapy samples of 57 patients who received chemotherapy only and WES on the samples of 13 patients who received NAC.
 |
Table 1 The Clinical Characteristics of Patients |
cfDNA-tDNA Mutation Spectrum Consistence Analysis
Tissues were required for detection of somatic mutations. However, they were not easy to sequentially obtain during the treatment. Therefore, we evaluated the consistency between the detected mutations in tumor and plasma. The VAF of mutations detected in plasma cfDNA was significantly lower than that in tissue DNA (tDNA) (Figure 1A) and showed a poor correlation (Figure 1B). However, the mutation counts detected in tDNA and cfDNA were highly correlated (Figure 1C). Then we examined the consistency of each individual variant detected in tDNA and cfDNA of the same patient. Most of the tDNA mutations with high VAF could also be detected in cfDNA, although some mutations with low VAF were not found. On the contrary, it was interesting to note that most of the cfDNA mutations with high VAF could not be detected in tDNA (Figure 1D). Then we characterized the spectrum of somatic variants in matched samples of each patient. Two-thirds of patients shared exactly the same mutations in matched tDNA and cfDNA (Figure 1E). Based on these results, we found that even though the VAF of somatic mutations detected in tDNA was higher than that in cfDNA, cfDNA could cover the main information of tDNA. More importantly, cfDNA could provide additional mutation information which could not get from tDNA. Previous other studies also supported this conclusion.26 Thus, we concluded that somatic mutations detected in cfDNA were consistent with that in tDNA. They were included in the subsequent somatic mutation analysis.
Somatic SNVs/InDels
SNVs/InDels Identified in Patients Who Received Chemotherapy Only
Next, we investigated the association between somatic mutations and clinical outcomes using targeted sequencing data from 57 patients who received chemotherapy only. A total of 26 cancer-related genes were included in this analysis. 8 somatic mutations in 6 genes were detected only in responders, while 27 somatic mutations in 12 genes were detected only in non-responders (Figure 2). However, mutations located in these genes occurred in too few patients. The top 3 genes with highest mutation frequency were TP53, EGFR and KRAS (Figure 2). In addition, their variant distributions were disequilibrium between two groups. Thus, they were further analyzed.
Somatic mutations in TP53 were identified in responders and non-responders with a frequency of 60% and 72.22%, respectively. The VAF of TP53 in non-responders was significantly higher than that in responders (Figure S2A). Then we analyzed the composition of mutation types of TP53 in responders, non-responders and both groups (Figure S2B). While somatic EGFR mutations were less frequently occurred in non-responders (21.05%) than that in responders (47.37%). Their locations in the gene were shown in Figure S2C. Among them, p.L858R was identified as gain of function mutation.27 For KRAS, somatic mutations were more frequently identified in non-responders (21.05%) than in responders (5.26%). It was noteworthy that all KRAS mutations in responders and non-responders were missense mutation and located at the same site of protein functional domains (Figure S2D). As a conclusion, somatic mutations in TP53 and KRAS may be associated with platinum resistance. While somatic mutations in EGFR may be associated with platinum sensitivity.
SNVs/InDels Identified in Patients Who Received NAC
To explore more mutations, we employed WES data from 13 patients receiving NAC. We hypothesized that somatic mutations related to platinum response would have a significant change after being exposed to platinum. After treatment, tumor cells carrying sensitive mutations would be killed. Thus these alterations would disappear. On the contrary, tumor cells carrying resistant somatic mutations could survive during chemotherapy. In addition, some acquired mutations may occur during chemotherapy.
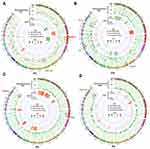
Therefore, the mutations uniquely existed in pre- or post-chemotherapy tumors may be associated with platinum sensitivity or resistance, respectively. Firstly, we explored SNVs in matched pre- and post-chemotherapy samples of 4 patients. Totally, 880 and 101 SNVs were, respectively, identified only in pre- or post-chemotherapy samples (Figure 3 and Tables S1 and S2). For each individual patient, the SNV with highest VAF existed only in pre- or post-chemotherapy sample is highlighted in Figure 3A–D. However, no SNV occurred in two or more patients.
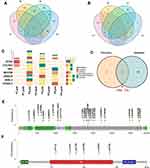
Thus, we analyzed SNV/InDel located genes. The genes existed only in pre-chemotherapy samples of two or more patients may be associated with platinum sensitivity. As a result, 124 genes were identified (Figure 4A). The genes shared in both pre- and post-chemotherapy samples and genes existed only in post-chemotherapy samples of at least 2 patients may contribute to platinum resistance. Six genes were identified (Figure 4B). Two genes were shared in both sensitivity and resistance groups and thus be excluded from sensitivity group. Among the other 122 platinum-sensitive genes, the top 7 genes with highest mutation occurrence frequencies were RYR3, COL5A3, GP2, MUC5B, OBSCN, ASXL2 and PCNXL2 (Figure 4C). The other genes were shown in Figure S3.
Even though lacking pre-chemotherapy control, platinum-resistant mutations remained in post-chemotherapy samples after platinum treatment. Therefore, another 9 post-chemotherapy samples were used for validation of platinum-resistant genes. It was considered as platinum-resistant gene if mutations in this gene occurred in at least 2 patients. As a result, a total of 101 genes were identified (Figure S4). Among them, two genes, TTN and TP53, overlapped with the results identified in matched pre- and post-chemotherapy samples (Figure 4D). TTN and TP53 had a top occurrence frequency of 55.56% and 66.67% in the validation samples, respectively (Figure S4). The distribution of TTN mutation sites was relatively scattered and almost all of them were missense mutations except for two truncating mutations (Figure 4E). For TP53, all mutation sites were concentrated in P53 domain and all of them were missense mutations (Figure 4F). In addition, TP53 was identified as a platinum-resistant gene in patients who received chemotherapy only (Figure S2A). In conclusion, somatic mutations in TTN and TP53 were associated with platinum resistance.
SCNVs
In addition to SNVs/InDels, SCNVs were also important for cancer development and may affect the sensitivity of anti-cancer drug.28 Therefore, we explored the association of SCNVs with platinum response by using sequential WES data. Firstly, we analyzed the change of SCNVs in 4 matched samples and found that there was an overall decrease in SCNVs after chemotherapy in all patients (Figure 3). SCNVs in chromosome 8q24.3 occurred only in all 4 pre-chemotherapy samples, indicating that it may be associated with platinum sensitivity. SCNVs in 3q26.1, 3q29, 22q11.21 and 21q21.3 occurred only in at least two post-chemotherapy samples. Copy number gain in 22q11.21 was validated in another 9 post-chemotherapy samples, which may indicate that it was associated with platinum resistance (Figure 5). As a conclusion, SCNVs in 8q24.3 and 22q11.21 were identified to be associated with platinum sensitivity and resistance, respectively.
Mutational Signatures
To better understand the role of somatic mutations in platinum response, we performed mutational signature analysis based on all SNVs of matched pre- and post-chemotherapy samples. Similar to the SNV analysis strategy, we hypothesized that a specific mutational signature sensitive to platinum might disappear after chemotherapy and vice versa. Mutational signatures 3 and 16 were uniquely existed in the post-chemotherapy sample of patient 1 (Figures S5A and S6A). Mutational signatures 3, 13, 21 and 30 were uniquely existed in the post-chemotherapy samples of patient 3 (Figures S5B and S6B). Because almost no SNV was identified in post-chemotherapy samples of patients 2 and 4, the mutational signature of them could not be identified. There was no overlapped mutational signature uniquely existed only in pre-chemotherapy samples of patient 1 or 3 (Figure S5C). Mutational signature 3 was uniquely existed in the post-treatment samples of both patients (Figure S5D). Then we used the other 9 post-chemotherapy samples to further verify the role of mutational signature 3 in platinum resistance and found it was occurred with a high frequency of 66.67% (Figure S5E). Mutational signature 3 was reported to be associated with the failure of DNA double-strand break (DSB) repair by homologous recombination (HR). However, it accounted for a small proportion in all mutational signatures of nearly all post-chemotherapy samples (Figure S5A, B and E).
Gene Ontology and Pathway Analysis
Based on our results mentioned above, 1 and 122 platinum-sensitive genes were identified from patients receiving chemotherapy only and patients receiving NAC, respectively. Besides, 2 and 104 platinum-resistant genes were identified from patients receiving chemotherapy only and patients receiving NAC, respectively. To further understand how these somatic alternations affected platinum response, we performed GO and KEGG analysis based on these identified genes. In GO analysis, platinum-sensitive and platinum-resistant genes were most enriched in smooth endoplasmic reticulum and homophilic cell adhesion via plasma membrane adhesion molecules, respectively (Figure S7A). In KEGG pathway analysis, platinum-sensitive genes were most enriched in calcium signaling pathway (Figure S7B). Platinum-resistant genes were not enriched in any pathway. Thus, calcium signaling pathway may be associated with platinum sensitivity.
TMB Change
TMB was regarded as one important predictor for immunotherapy response. One recent research suggested that TMB combined with PD-L1 expression could predict platinum response.29 Therefore, we hypothesized that TMB may relate to platinum response. We analyzed the TMB change based on the WES data from matched samples. After chemotherapy, the TMB had a significant decrease in patients 2, 3 and 4, who had a better response to platinum-based chemotherapy (Figure S8A, B and D). On the contrary, the TMB had nearly no change between pre- and post-treatment samples of patient 1 who was the non-responder (Figure S8C). These results suggested that the decrease of TMB may relate to platinum sensitivity.
Validation of Identified Genes
Somatic mutations in TTN, TP53, KRAS and EGFR were identified to be associated with platinum response. To further verify this result, we analyzed the IC50 values in different cancer cells treated by cisplatin from the Genomics of Drug Sensitivity in Cancer (GDSC) database (https://www.cancerrxgene.org/). The IC50 values in cells with TP53 mutation or KRAS mutation were higher than that in wild type cells (Figure S9A and B), which indicated that TP53 and KRAS mutations were related to cisplatin response. However, there was no statistical difference between the IC50 values in EGFR-mutated and EGFR-wild type cells (Figure S9C). The association of TTN mutation and cisplatin response was not analyzed due to the lacking of data. These results confirmed our conclusion that somatic mutations in TP53 and KRAS were associated with platinum resistance.
Discussion
In this study, we explored the association of somatic mutations with platinum response in NSCLC patients. Somatic mutations in TTN, TP53, KRAS and EGFR and SCNVs in chromosome 8q24.3 and 22q11.21 were found to be associated with platinum response. Calcium signaling pathway was identified to be associated with platinum sensitivity. Besides, we found that mutational signature 3 was associated with platinum resistance and the decrease of TMB tended to be correlated with better platinum response. These results indicated that somatic mutations could affect platinum response.
Previous studies also explored the effect of somatic mutations on platinum response. Many of them focused on some special genes such as DNA repair genes.30 One study explored the association of somatic mutations with platinum response in muscle-invasive bladder cancer (MIBC) by using WES, which mainly focused on mutational signature and tumor evolution.31 However, few studies explored somatic mutations associated with platinum response in lung cancer. We explored somatic mutations in matched pre- and post-chemotherapy samples of NSCLC patients who received NAC at whole exome level. These results provided more specific somatic mutations correlated with platinum response. Of them, somatic mutations in TTN were identified to be associated with platinum resistance. TTN consists of 364 exons and produces the largest known protein.32 Previous studies reported that somatic mutations in TTN frequently occurred in a number of cancer types.33 We firstly reported that it was also associated with platinum resistance. In addition, somatic mutations in TP53 were identified to be associated with platinum resistance in patients receiving chemotherapy only and patients receiving NAC, which was consistent with the results of previous other studies.34 To further confirm these results, we performed bioinformatics analysis and found that cells with TP53 mutation or KRAS mutation had higher IC50 values of cisplatin in cancer cells.
Based on all identified genes with somatic mutations, calcium signaling pathway was found to be associated with platinum sensitivity. This result further supported the important role of calcium signaling pathway in platinum response.35 Besides, we found that mutational signature 3 was associated with platinum resistance. However, this signature is correlated with failure of HR. HR deficiency was reported to promote responding to platinum in breast cancer.36 This inconsistency indicates that the role of mutational signature 3 in platinum response may still need to be further determined. One possible explanation may be that nucleotide excision repair (NER) rather than HR is the most primary way to repair DNA damages induced by platinum.37
In addition, we found that SCNVs had a decrease after chemotherapy, which was not consistent with previous studies.31 This may result from different tumor types and purity. For TMB, we found that patients with a decrease of TMB after chemotherapy tended to have a better response to platinum. One recent research suggested that TMB tended to have a decrease after platinum chemotherapy.38 We propose that it may become a predictor for platinum response, which still needs to be further verified by larger size of patients. Therefore, in addition to single mutation, other factors such as SCNVs and TMB also should be taken into account when evaluating the effect of somatic mutations on platinum response.
Our study supports that somatic mutations contribute to platinum response. Somatic mutations only occur in some somatic cells, while germline mutations exist in all somatic cells. One previous study made a direct comparison between germline and somatic mutation rates in humans. It found that somatic mutation rate was much higher than germline mutation rate. In addition, it was reported that somatic mutations may interact with germline mutations.39 Therefore, taking somatic mutations into consideration may improve the performance of platinum drug response model we previously established.9 However, biopsy tissues are not easy to obtain from patients. Thus, we attempted to detect somatic mutations in cfDNA instead of tDNA. We found that mutations detected in cfDNA were consistent with that in tDNA. While the VAF of cfDNA was much lower than tDNA, which may result from low levels and short half-life of cfDNA. Previous studies focused less on the VAF difference of mutations detected in cfDNA and tDNA. While one study showed that the average VAF of cfDNA from NSCLC patients was at a low level of 12.8%, which supported our results.40 Besides, cfDNA could provide more information than tDNA, which was consistent with previous investigations.41 This may result from the tumor heterogeneity. Therefore, we could detect somatic mutations in cfDNA instead of cfDNA to monitor somatic mutations and predict platinum response.
There are several limitations in our study. One limitation of our study is that the sample size is small, especially matched pre- and post-chemotherapy samples of patients who received NAC. In addition, the biological function and mechanism of these identified genes in platinum response are still need to be further studied. In conclusion, our results confirmed that somatic mutations contributed to platinum response and provide more information for guiding the clinical use of platinum.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (81773823, 81573463, 81571151, 81641039), National Science and Technology Major Project of China (2017ZX09304014, 2019ZX09201-002-006), National Science & Technology Fundamental Resources Investigation Program of China (2018FY100900), Key Research and Development Program of Hunan Province (2016JC2039, 2019SK2251).
Disclosure
The authors declare no conflict of interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Xiong Y, Huang BY, Yin JY. Pharmacogenomics of platinum-based chemotherapy in non-small cell lung cancer: focusing on DNA repair systems. Med Oncol. 2017;34(4):48. doi:10.1007/s12032-017-0905-6
3. Wang LY, Cui JJ, Liu JY, et al. Gene-gene and gene-environment interaction data for platinum-based chemotherapy in non-small cell lung cancer. Sci Data. 2018;5:180284. doi:10.1038/sdata.2018.284
4. Yin JY, Shen J, Dong ZZ, et al. Effect of eIF3a on response of lung cancer patients to platinum-based chemotherapy by regulating DNA repair. Clin Cancer Res. 2011;17(13):4600–4609. doi:10.1158/1078-0432.CCR-10-2591
5. Langer CJ, Gadgeel SM, Borghaei H, et al. Carboplatin and pemetrexed with or without pembrolizumab for advanced, non-squamous non-small-cell lung cancer: a randomised, Phase 2 cohort of the open-label KEYNOTE-021 study. Lancet Oncol. 2016;17(11):1497–1508. doi:10.1016/S1470-2045(16)30498-3
6. Horn L, Mansfield AS, Szczesna A, et al. First-line atezolizumab plus chemotherapy in extensive-stage small-cell lung cancer. N Engl J Med. 2018;379(23):2220–2229. doi:10.1056/NEJMoa1809064
7. Ben-David U, Siranosian B, Ha G, et al. Genetic and transcriptional evolution alters cancer cell line drug response. Nature. 2018;560(7718):325–330. doi:10.1038/s41586-018-0409-3
8. Yin JY, Li X, Zhou HH, Liu ZQ. Pharmacogenomics of platinum-based chemotherapy sensitivity in NSCLC: toward precision medicine. Pharmacogenomics. 2016;17(12):1365–1378. doi:10.2217/pgs-2016-0074
9. Yin JY, Li X, Li XP, et al. Prediction models for platinum-based chemotherapy response and toxicity in advanced NSCLC patients. Cancer Lett. 2016;377(1):65–73. doi:10.1016/j.canlet.2016.04.029
10. Cui JJ, Wang LY, Zhu T, et al. Gene-gene and gene-environment interactions influence platinum-based chemotherapy response and toxicity in non-small cell lung cancer patients. Sci Rep. 2017;7(1):5082. doi:10.1038/s41598-017-05246-8
11. Sullivan I, Riera P, Andres M, et al. Prognostic effect of VEGF gene variants in metastatic non-small-cell lung cancer patients. Angiogenesis. 2019;22(3):433–440. doi:10.1007/s10456-019-09668-y
12. Chen J, Wang Z, Zou T, et al. Pharmacogenomics of platinum-based chemotherapy response in NSCLC: a genotyping study and a pooled analysis. Oncotarget. 2016;7(34):55741–55756. doi:10.18632/oncotarget.9688
13. Martincorena I, Campbell PJ. Somatic mutation in cancer and normal cells. Science (New York, NY). 2015;349(6255):1483–1489. doi:10.1126/science.aab4082
14. Bertotti A, Papp E, Jones S, et al. The genomic landscape of response to EGFR blockade in colorectal cancer. Nature. 2015;526(7572):263–267. doi:10.1038/nature14969
15. Guryanova OA, Shank K, Spitzer B, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med. 2016;22(12):1488–1495. doi:10.1038/nm.4210
16. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics (Oxford, England). 2014;30(15):2114–2120. doi:10.1093/bioinformatics/btu170
17. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics (Oxford, England). 2009;25(14):1754–1760. doi:10.1093/bioinformatics/btp324
18. Li H, Handsaker B, Wysoker A, et al. The sequence alignment/map format and SAMtools. Bioinformatics (Oxford, England). 2009;25(16):2078–2079. doi:10.1093/bioinformatics/btp352
19. McKenna A, Hanna M, Banks E, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi:10.1101/gr.107524.110
20. Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31(3):213–219. doi:10.1038/nbt.2514
21. Saunders CT, Wong WS, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor-normal sample pairs. Bioinformatics (Oxford, England). 2012;28(14):1811–1817. doi:10.1093/bioinformatics/bts271
22. Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. doi:10.1093/nar/gkq603
23. Boeva V, Popova T, Bleakley K, et al. Control-FREEC: a tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics (Oxford, England). 2012;28(3):423–425. doi:10.1093/bioinformatics/btr670
24. Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–D811. doi:10.1093/nar/gku1075
25. Huang DW, Sherman BT, Tan Q, et al. The DAVID gene functional classification tool: a novel biological module-centric algorithm to functionally analyze large gene lists. Genome Biol. 2007;8(9):R183. doi:10.1186/gb-2007-8-9-r183
26. Adalsteinsson VA, Ha G, Freeman SS, et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat Commun. 2017;8(1):1324. doi:10.1038/s41467-017-00965-y
27. Lynch TJ, Bell DW, Sordella R, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi:10.1056/NEJMoa040938
28. Cheng L, Pandya PH, Liu E, et al. Integration of genomic copy number variations and chemotherapy-response biomarkers in pediatric sarcoma. BMC Med Genomics. 2019;12(Suppl 1):23. doi:10.1186/s12920-018-0456-5
29. Chen Y, Liu Q, Chen Z, et al. PD-L1 expression and tumor mutational burden status for prediction of response to chemotherapy and targeted therapy in non-small cell lung cancer. J Exp Clin Cancer Res. 2019;38(1):193. doi:10.1186/s13046-019-1192-1
30. Pennington KP, Walsh T, Harrell MI, et al. Germline and somatic mutations in homologous recombination genes predict platinum response and survival in ovarian, fallopian tube, and peritoneal carcinomas. Clin Cancer Res. 2014;20(3):764–775. doi:10.1158/1078-0432.CCR-13-2287
31. Liu D, Abbosh P, Keliher D, et al. Mutational patterns in chemotherapy resistant muscle-invasive bladder cancer. Nat Commun. 2017;8(1):2193. doi:10.1038/s41467-017-02320-7
32. Chauveau C, Rowell J, Ferreiro A. A rising titan: TTN review and mutation update. Hum Mutat. 2014;35(9):1046–1059. doi:10.1002/humu.22611
33. Kim N, Hong Y, Kwon D, Yoon S. Somatic mutaome profile in human cancer tissues. Genomics Inform. 2013;11(4):239–244. doi:10.5808/GI.2013.11.4.239
34. Ma X, Le Teuff G, Lacas B, et al. Prognostic and predictive effect of TP53 mutations in patients with non-small cell lung cancer from adjuvant cisplatin-based therapy randomized trials: a LACE-bio pooled analysis. J Thorac Oncol. 2016;11(6):850–861. doi:10.1016/j.jtho.2016.02.002
35. Florea AM, Varghese E, McCallum JE, et al. Calcium-regulatory proteins as modulators of chemotherapy in human neuroblastoma. Oncotarget. 2017;8(14):22876–22893. doi:10.18632/oncotarget.15283
36. Zhao EY, Shen Y, Pleasance E, et al. Homologous recombination deficiency and platinum-based therapy outcomes in advanced breast cancer. Clin Cancer Res. 2017;23(24):7521–7530. doi:10.1158/1078-0432.CCR-17-1941
37. Liu YB, Mei Y, Tian ZW, Long J, Luo CH, Zhou HH. Downregulation of RIF1 enhances sensitivity to platinum-based chemotherapy in epithelial ovarian cancer (EOC) by regulating nucleotide excision repair (NER) pathway. Cell Physiol Biochem. 2018;46(5):1971–1984. doi:10.1159/000489418
38. Sakai H, Takeda M, Sakai K, et al. Impact of cytotoxic chemotherapy on PD-L1 expression in patients with non-small cell lung cancer negative for EGFR mutation and ALK fusion. Lung Cancer. 2019;127:59–65. doi:10.1016/j.lungcan.2018.11.025
39. Wu J, Mamidi TKK, Zhang L, Hicks C. Integrating germline and somatic mutation information for the discovery of biomarkers in triple-negative breast cancer. Int J Environ Res Public Health. 2019;16(6):1055.
40. Rothwell DG, Ayub M, Cook N, et al. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25(5):738–743. doi:10.1038/s41591-019-0380-z
41. Wang DS, Liu ZX, Lu YX, et al. Liquid biopsies to track trastuzumab resistance in metastatic HER2-positive gastric cancer. Gut. 2019;68(7):1152–1161. doi:10.1136/gutjnl-2018-316522
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.