Back to Journals » International Journal of General Medicine » Volume 16
SEPTIN3 Promotes Progression of Triple-Negative Breast Cancer via Activating Wnt Pathway
Authors Wang GZ , Yang LH, Gao C
Received 22 May 2023
Accepted for publication 5 September 2023
Published 12 September 2023 Volume 2023:16 Pages 4155—4164
DOI https://doi.org/10.2147/IJGM.S419827
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Guo-Zhou Wang,1,* Li-Hua Yang,1,* Chao Gao2
1Department of Breast Tumor Surgery, Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University, Edong Healthcare Group, Huangshi, Hubei Province, 435000, People’s Republic of China; 2Department of General Practitioner, Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University, Edong Healthcare Group, Huangshi, Hubei Province, 435000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chao Gao, Department of General Practitioner, Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University, Edong Healthcare Group, 141 Tianjin Road, Huangshigang District, Huangshi, Hubei Province, 435000, People’s Republic of China, Email [email protected]
Background: There is a lack of targeted therapies for triple-negative breast cancer (TNBC), necessitating the search for novel targets. Patients with TNBC exhibit elevated expression of neuron-specific septin-3 (SEPTIN3), leading to poor prognosis. This study aimed to investigate the modulation of SEPTIN3 expression in TNBC cells.
Methods: The relative expression levels of SEPTIN3 in TNBC tissues and cell lines were determined using Western blotting and qRT-PCR. We generated lentivirally transduced TNBC cell lines so such that SEPTIN3 was overexpressed or knocked down. Next, the effect of SEPTIN3 on the biological behavior of TNBC cells was detected using a series of functional assays, including CCK8, colony formation, scratch, and transwell assays. We monitored the tumorigenicity of SEPTIN3 overexpressed cells and performed Ki-67 immunostaining in mice. The mechanism mediated by SEPTIN3 was studied using functional enrichment analysis and Western blotting.
Results: Protein and mRNA expression levels of SEPTIN3 were observed to be increased in TNBC tissues and cell lines. SEPTIN3 knockdown reduced cell growth, invasion, and migration, whereas SEPTIN3 overexpression exerted the opposite effects. SEPTIN3 was observed to favor cell growth and tumorigenicity in vivo. In addition, SEPTIN3 promoted TNBC cell aggressiveness and proliferation via activation of the Wnt signaling pathway.
Conclusion: SEPTIN3 emerged as an oncogene that accelerates tumor progression by regulating the Wnt signaling pathway.
Keywords: triple-negative breast cancer, SEPTIN3, proliferation, invasion, Wnt signaling pathway
Introduction
Breast cancer (BC) is the most common malignant tumor in women and a leading cause of cancer-related death.1 BC is classified according to its distinct biological and epigenetic features.2 Among the types of BC, triple-negative BC (TNBC) is characterized by the low expression or no amplification of the following three biomarkers at the gene level: estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). TNBC accounts for approximately 15% of BC cases.3,4 TNBC is a more aggressive phenotype of BC that usually progresses rapidly, develops drug resistance, metastasizes, relapses, and is difficult to treat in clinical practice.5 TNBC-based transcriptome sequencing studies have failed to identify novel recurrent mutations beyond TP53, PIK3CA, and PTEN,6 suggesting that the malignant biological phenotype of TNBC may be driven by non-genetic alterations. New omics studies have suggested that multiple differential genes may play important roles in the progression of breast cancer.7 Therefore, there is an urgent need to identify new molecular markers to guide treatment decisions.
The Wnt/β-catenin signaling pathway is an important extracellular signaling pathway that is abnormally dysregulated in multiple types of cancers.8,9 The Wnt signaling pathway is a key regulator that controls tumor cell self-renewal and differentiation, tumor metastasis, and tumor treatment resistance.10 Canonical Wnt signaling activation is initiated by Wnt ligands that bind to Frizzled and LRP5/6, thereby disrupting the complex including axins, APC, and GSK3, leading to the stabilization of β-catenin. GSK-3β is a widely expressed serine/threonine protein kinase.11 Its phosphorylation at Tyr 216 is required to phosphorylate β-catenin, induce degradation of β-catenin, and inhibit Wnt signaling.12 Subsequently, β-catenin accumulates in the cytoplasm, translocates to the nucleus, and activates the Wnt transcriptional program. Abnormal activation of the Wnt/β-catenin pathway is a characteristic of TNBC that synergizes with tumorigenesis, metastasis, cancer stemness, and other processes.13,14 TNBC patients with abnormal Wnt/β-catenin signaling pathways are more likely to develop lung and brain metastases.
Neuron-specific septin-3 (SEPTIN3), a member of the septin family, is primarily expressed in the brain and testis.15 SEPTIN3 is a soluble, membrane-bound presynaptic protein. Previous studies have shown that a lack of SEPTIN3 has no effect on neuronal development, viability, or protrusion function.16,17 SEPTIN3is connected to autophagy and can bind to microtubule-associated protein 1 light chain 3 B (LC3B), the most studied homolog of the autophagy protein, Atg8.18 However, few studies have focused on the role of SEPTIN3 in cancer development. Our previous study revealed that SEPTIN3 is overexpressed in TNBC and is associated with poor prognosis in patients. In this study, we aimed to investigate the functional role and regulatory mechanisms of SEPTIN3 in TNBC.
Materials and Methods
Bioinformatics Analysis
We used the Gene Expression Profiling Interactive Analysis (GEPIA, http://gepia.cancer-pku.cn/) online database to analyze differences in SEPTIN3 expression between TNBC and normal tissues. Gene set enrichment analysis (GSEA) was performed using the KEGG pathways. Gene set enrichment analysis (GSEA) is commonly used to estimate changes in pathways and biological processes. The gene set was downloaded from the MsigDB database19 for GSEA analysis, and FDR < 0.25 and P < 0.05 was considered significantly enriched.
Clinical Tissues and Cell Lines
TNBC tissues and matched para-tumors were surgically collected from patients at Huangshi Central Hospital Medical and preserved in liquid nitrogen. Informed consent was obtained from all patients. This procedure was approved by the Ethics Committee of Huangshi Central Hospital (2022–14). The normal human breast epithelial cell line (MCF-10A) and TNBC cell lines (BT-549, SK-BR-3, HCC1937, and MDA-MB-453) were acquired from The Shanghai Cell Bank. Cell lines were cultured according to the instructions with the use of Dulbecco’s modified Eagle medium and 10% fetal bovine serum.
qRT-PCR
All tissues and cells were washed with PBS. Total RNA was isolated from cells and tissues using the TRIzol reagent (Invitrogen). RNA was reverse-transcribed to cDNA using reverse transcriptase according to the manufacturer’s instructions. GAPDH mRNA was used as an endogenous control. The primers used are listed in Table S1.
Western Blot
Lysis buffer containing proteinase inhibitors for cells and tissues was used to extract total protein from the cells. All proteins were separated on 10% polyacrylamide gels. Next, the proteins were transferred to the PVDF membranes and incubated with blocking buffer. The membranes were incubated with primary antibodies. Subsequently, the membranes were incubated with secondary antibodies. ECL chemiluminescence was used for visualization.
Lentiviral Transfection
Lentivirus-mediated short hairpin RNA (shRNA) was used to knockdown SEPTIN3. MDA-MB-453 and BT549 cells were transfected with lentiviruses obtained from GenePharm (Shanghai, China) to suppress SEPTIN3, or increase SEPTIN3 expression. Stable TNBC cells were generated via puromycin (2 ug/mL).
Cell Proliferation Assay
CCK8 assay and colony formation assays were performed to detect cell growth. Equal numbers of TNBC cells were seeded in 96-well plates and cultured for 4 days. Each well was treated with CCK-8 (Dojindo, Japan) solution at 37 °C for 1 h, which was detected by the absorbance at 450 nm. One thousand cells were seeded in petri dishes to test their colony-forming ability. After 21 days of incubation, the colonies were visualized using crystal violet and counted.
Cell Migration and Invasion Assays
24-Transwell inserts (Corning, Corning, NY, USA) pre-coated with Matrigel (BD Biosciences) were used to detect cell migration. Cells were seeded in the inserts supplemented with no FBS and the medium containing 10% FBS was added to the lower chamber. The incubation temperature was set to 37 °C. Subsequently, non-invading cells were eliminated and the number of invading cells was observed under a microscope. For wound healing assay, a 200 ul pipette was utilized to form artificial wound on cells seeded in six-well plates. Images were acquired to visualize the wound healing process at 0 and 24 h.
Xenograft Assay
Female nude mice (Vitalstar, China) aged 6–8 weeks were randomly divided into two groups. 2×106 MDA-MB-231 cells with SEPTIN3 knockdown were injected subcutaneously into the right flank of the mice. Tumor growth was monitored weekly and the tumors were harvested 4 weeks after tumor cell injection. The xenograft assay was approved by the Ethics Committee of Huangshi Central Hospital (2023–3) and performed in accordance with institutional guidelines.
Immunohistochemical (IHC) Staining
Samples from the xenografts were prepared or subjected to IHC and hematoxylin–eosin (HE) staining. Ki-67 is expressed during mitosis and is considered as an indicator of cell proliferation. Therefore, we determined the expression of Ki-67. All the samples were sectioned after paraffin embedding, deparaffinized, hydrated, and incubated with a Ki-67 primary antibody (1:200; Abcam, USA), followed by incubation with an HRP-conjugated secondary antibody. Finally, 3.3 ‘-diaminobenzidine and hematoxylin were used for visualization. Sections were stained with hematoxylin and eosin.
Statistical Analysis
All Data are represented as the mean ± standard deviation, followed by statistical analysis based on the GraphPad Prism 9 software. Student’s t-test was used to determine significance between two groups, whereas one-way analysis of variance was used for comparisons between multiple groups. Statistical significance was set at p < 0.05.
Results
High Expression Level of SEPTIN3 in TNBC
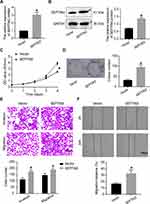
We integrated the database and histological data to determine the differential expression of SEPTIN3. Compared to that in normal tissues, the mRNA expression of SEPTIN3 in TNBC was increased (Figure 1A). Subsequently, we validated this finding in 10 pairs of tissues (Figure 1B). We also observed a significant increase SEPTIN3 in TNBC cell lines, both at protein and mRNA levels (Figure 1C and D). The BT-549 and MDA-MB-453 cells were used for subsequent analyses.
SEPTIN3 Knockdown Inhibited the Malignant Behaviors of TNBC Cells
As presented in Figure 2A and B, the lentivirus successfully knocked down the expression of SEPTIN3 in MDA-MB-453 cells at both gene and protein levels. Compared with control (sh-NC) cells, the growth rate of TNBC cells was greatly suppressed by SEPTIN3 knockdown (Figure 2C). The colony formation assay showed that SEPTIN3 knockdown resulted in a decrease in colony number (Figure 2D). These findings suggest that SEPTIN3 regulates TNBC cell proliferation. To assess cell invasion, a Transwell assay was performed. As shown in Figure 2E, the number of invasive sh-NC cells was significantly higher than that of SEPTIN3 knockdown cells. In the wound healing assay, the knockdown of SEPTIN3 expression slowed cell migration (Figure 2F).
Overexpression of SEPT Facilitated the Progression of TNBC Cells
We increased the expression of SEPTIN3 in BT-549 cells using a lentivirus (Figure 3A and B). Upregulation of SEPTIN3 significantly promoted cell proliferation and bacterial colonization (Figure 3C and D). Compared to the empty vector, SEPTIN3 overexpression enhanced the invasion and migration of TNBC cells (Figure 3E and F).
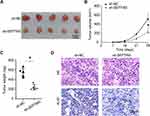
Knockdown of SEPTIN3 Inhibited Tumorigenicity of TNBC Cells
To evaluate the tumorigenic potential of SEPTIN3 in vivo, xenograft assays were performed. The results demonstrated that the tumor volume and weight of SEPTIN3 knockdown cells were significantly reduced (Figure 4A–C). One month of monitoring indicated that SEPTIN3 knockdown slowed the tumor growth rate in vivo (Figure 4B). Ki-67, a reliable marker of cell proliferation, was increased in control tumors (Figure 4D). These studies showed that SEPTIN3 affects cell proliferation and tumorigenic ability in vivo.
SEPTIN3 Activated Wnt Signaling Pathway
To further understand the regulatory mechanism of SEPTIN3 in TNBC, we performed enrichment analysis. As shown in Figure 5A and B, SEPTIN3 mainly affected cancer-related pathways such as the PI3K, cell cycle, and Wnt signaling pathways. Given that the aberrant activation of Wnt signaling is a hallmark of TNBC, we investigated the association between SEPTIN3 and Wnt signaling. Western blotting showed that SEPTIN3 significantly upregulated the expression of β-catenin and cyclinD1, and decreased the expression of Axin1 and GSK-3β (Figure 5C). In contrast, these effects were reversed by SEPTIN3 knockdown.
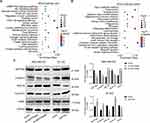 |
Figure 5 SEPTIN3 activated Wnt signaling pathway. (A and B) SEPTIN3 GSEA enrichment analysis. (C) Effects of SEPTIN3 on the Wnt signaling pathway by Western blot. *P<0.05. |
Discussion
TNBC has a poor clinical treatment outcome owing to its unique biological characteristics.20 The overexpression of septin family members has been investigated in multiple types of cancers and is associated with the malignant progression of various cancer types.21–23 SEPT9 induces cell migration by regulating the cytoskeleton.24 SEPT9 can promote epithelial-mesenchymal transition in glioma.25 Increased expression of SEPT2 facilitates tumor growth in vivo.26
However, the association between SEPTIN3 and cancer has not yet been reported. We investigated the role of SEPTIN3 in TNBC. In the present study, we found that SEPTIN3 protein and mRNA were highly expressed in TNBC tissues and cells. In addition, in vitro and in vivo functional assays demonstrated that elevated levels of SEPTIN3 enhanced cell proliferation, invasion, metastasis, and tumorigenesis in TNBC. These findings suggest that SEPTIN3 is a promising novel target for the treatment of TNBC.
Accumulating evidence suggests that the Wnt signaling pathway is involved in tumor progression, including gastric, pancreatic, lung, and breast cancer.8 Aberrant Wnt signaling is associated with tumor proliferation, metastasis, stemness, and drug resistance. To date, no study has reported that SEPTIN3 can activate this signaling pathway in TNBC. We explored the effects of SEPTIN3 on the Wnt signaling pathway. SEPTIN3 overexpression can upregulate the expression of-catenin signaling pathway-related genes β-catenin and CyclinD1, suggesting that SEPTIN3 can promote the progression of TNBC by activating-catenin signaling pathway. The post-transcriptional Wnt signaling pathway suppressed septin4 phosphate 1 to affect physiological development through GSK3.27 This may be a potential mechanism by which SEPTIN3 interacts with Wnt signaling. As a downstream regulator of Wnt signaling, CyclinD1 participates in cell cycle progression. Previous studies have demonstrated the involvement of septin in cell cycle regulatory.28 SEPTIN3 may activate signaling pathways by directly interacting with cell cycle regulators. However, this study has some limitations. For example, the molecular mechanisms underlying abnormal SEPTIN3 expression in TNBC remain unclear. Second, we did not construct in vivo tumor metastasis models, which limited the level of evidence. Molecular inhibitors targeting SEPTIN3 require further investigation.
Conclusions
This study suggests that SEPTIN3 facilitates the malignant progression of TNBC by activating the Wnt signaling pathway. This indicates the importance of SEPTIN3 and Wnt signaling pathways in TNBC. The specific molecular regulatory mechanisms of SEPTIN3 require further investigation. Our findings may have indirect therapeutic implications, as the direct modulation of SEPTIN3 may alter TNBC cell fate and unlock additional treatment options. These findings provide new insights for the development of novel therapeutic targets.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval
All procedures performed were in accordance with the declaration of the ethical standards of the institutional research committee and with the Helsinki Declaration and its later amendments. The ethics committee has approved this study of the Huangshi Central Hospital.
Acknowledgments
We are very grateful for Huangshi Central Hospital for its help in this research.
Funding
No funding was received.
Disclosure
All authors declare no conflicts of interest for this work.
References
1. Gradishar WJ, Moran MS, Abraham J, et al. Breast cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2022;20(6):691–722. doi:10.6004/jnccn.2022.0030
2. Wang C, Xu K, Wang R, Han X, Tang J, Guan X. Heterogeneity of BCSCs contributes to the metastatic organotropism of breast cancer. J Exp Clin Cancer Res. 2021;40(1):370. doi:10.1186/s13046-021-02164-6
3. Altundag K. Pembrolizumab in triple-negative breast cancer. N Engl J Med. 2022;387(15):1435–1436. doi:10.1056/NEJMc2210919
4. Li Y, Zhang H, Merkher Y, et al. Recent advances in therapeutic strategies for triple-negative breast cancer. J Hematol Oncol. 2022;15(1):121. doi:10.1186/s13045-022-01341-0
5. Denkert C, Liedtke C, Tutt A, von Minckwitz G. Molecular alterations in triple-negative breast cancer-The road to new treatment strategies. Lancet. 2017;389(10087):2430–2442. doi:10.1016/S0140-6736(16)32454-0
6. Schneider BP, Winer EP, Foulkes WD, et al. Triple-negative breast cancer: risk factors to potential targets. Clin Cancer Res. 2008;14(24):8010–8018. doi:10.1158/1078-0432.CCR-08-1208
7. Zhou L, Rueda M, Alkhateeb A. Classification of breast cancer Nottingham prognostic index using high-dimensional embedding and residual neural network. Cancers. 2022;14(4). doi:10.3390/cancers14040934
8. Parsons MJ, Tammela T, Dow LE. WNT as a driver and dependency in cancer. Cancer Discov. 2021;11(10):2413–2429. doi:10.1158/2159-8290.CD-21-0190
9. Galluzzi L, Spranger S, Fuchs E, Lopez-Soto A. WNT signaling in cancer immunosurveillance. Trends Cell Biol. 2019;29(1):44–65. doi:10.1016/j.tcb.2018.08.005
10. Xu X, Zhang M, Xu F, Jiang S. Wnt signaling in breast cancer: biological mechanisms, challenges and opportunities. Mol Cancer. 2020;19(1):165. doi:10.1186/s12943-020-01276-5
11. Freeman J, Smith D, Latinkic B, et al. A functional connectome: regulation of Wnt/TCF-dependent transcription by pairs of pathway activators. Mol Cancer. 2015;14:206. doi:10.1186/s12943-015-0475-1
12. Liu X, Klein PS. Glycogen synthase kinase-3 and alternative splicing. Wiley Interdiscip Rev RNA. 2018;9(6):e1501. doi:10.1002/wrna.1501
13. Bernemann C, Hulsewig C, Ruckert C, et al. Influence of secreted frizzled receptor protein 1 (SFRP1) on neoadjuvant chemotherapy in triple negative breast cancer does not rely on WNT signaling. Mol Cancer. 2014;13:174. doi:10.1186/1476-4598-13-174
14. Rodgers SJ, Ooms LM, Oorschot VMJ, et al. INPP4B promotes PI3Kalpha-dependent late endosome formation and Wnt/beta-catenin signaling in breast cancer. Nat Commun. 2021;12(1):3140. doi:10.1038/s41467-021-23241-6
15. Rosa HVD, Leonardo DA, Brognara G, et al. Molecular recognition at septin interfaces: the switches hold the key. J Mol Biol. 2020;432(21):5784–5801. doi:10.1016/j.jmb.2020.09.001
16. Fujishima K, Kiyonari H, Kurisu J, Hirano T, Kengaku M. Targeted disruption of Sept3, a heteromeric assembly partner of Sept5 and Sept7 in axons, has no effect on developing CNS neurons. J Neurochem. 2007;102(1):77–92. doi:10.1111/j.1471-4159.2007.04478.x
17. Xue J, Tsang CW, Gai WP, et al. Septin 3 (G-septin) is a developmentally regulated phosphoprotein enriched in presynaptic nerve terminals. J Neurochem. 2004;91(3):579–590. doi:10.1111/j.1471-4159.2004.02755.x
18. Toth V, Vadaszi H, Ravasz L, et al. Neuronal-specific septin-3 binds Atg8/LC3B, accumulates and localizes to autophagosomes during induced autophagy. Cell Mol Life Sci. 2022;79(9):471. doi:10.1007/s00018-022-04488-8
19. Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi:10.1016/j.cels.2015.12.004
20. Wu SY, Wang H, Shao ZM, Jiang YZ. Triple-negative breast cancer: new treatment strategies in the era of precision medicine. Sci China Life Sci. 2021;64(3):372–388. doi:10.1007/s11427-020-1714-8
21. Calvo F, Ranftl R, Hooper S, et al. Cdc42EP3/BORG2 and septin network enables mechano-transduction and the emergence of cancer-associated fibroblasts. Cell Rep. 2015;13(12):2699–2714. doi:10.1016/j.celrep.2015.11.052
22. Montagna C, Lyu MS, Hunter K, et al. The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 2003;63(9):2179–2187.
23. Targa B, Klipfel L, Cantaloube I, et al. Septin filament coalignment with microtubules depends on SEPT9_i1 and tubulin polyglutamylation, and is an early feature of acquired cell resistance to paclitaxel. Cell Death Dis. 2019;10(2):54. doi:10.1038/s41419-019-1318-6
24. Kamatari YO, Ohta S, Inoshima Y, et al. Identification and characterization of a multispecific monoclonal antibody G2 against chicken prion protein. Protein Sci. 2014;23(8):1050–1059. doi:10.1002/pro.2491
25. Zhang G, Feng W, Wu J. Down-regulation of SEPT9 inhibits glioma progression through suppressing TGF-beta-induced epithelial-mesenchymal transition (EMT). Biomed Pharmacother. 2020;125:109768. doi:10.1016/j.biopha.2019.109768
26. Yu J, Zhang W, Tang H, et al. Septin 2 accelerates the progression of biliary tract cancer and is negatively regulated by mir-140-5p. Gene. 2016;589(1):20–26. doi:10.1016/j.gene.2016.05.005
27. Koch S, Acebron SP, Herbst J, Hatiboglu G, Niehrs C. Post-transcriptional Wnt signaling governs epididymal sperm maturation. Cell. 2015;163(5):1225–1236. doi:10.1016/j.cell.2015.10.029
28. Marquardt J, Chen X, Bi E. Septin assembly and remodeling at the cell division site during the cell cycle. Front Cell Dev Biol. 2021;9:793920. doi:10.3389/fcell.2021.793920
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.