Back to Journals » International Journal of General Medicine » Volume 17
SEPT3 as a Potential Molecular Target of Triple-Negative Breast Cancer
Authors Yang LH, Wang GZ , Gao C
Received 2 February 2024
Accepted for publication 22 April 2024
Published 25 April 2024 Volume 2024:17 Pages 1605—1613
DOI https://doi.org/10.2147/IJGM.S462541
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Li-Hua Yang,1,* Guo-Zhou Wang,1,* Chao Gao2
1Department of Breast Tumor Surgery, Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University, Edong Healthcare Group, Huangshi, Hubei, People’s Republic of China; 2Department of General Practitioner, Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University, Edong Healthcare Group, Huangshi, Hubei, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chao Gao, Department of General Practitioner, Huangshi Central Hospital, Affiliated Hospital of Hubei Polytechnic University, Edong Healthcare Group, Huangshi, Hubei, 435000, People’s Republic of China, Email [email protected]
Background: The survival rate for triple-negative breast cancer (TNBC) is very low due to its advanced metastatic and aggressive nature, and there is no specific target to improve the survival rate. The expression and clinical signature of neuronal-specific septin-3 (Septin3, SEPT3) in TNBC remain undetermined.
Methods: SEPT3 differential expression in TNBC was detected with the use of bioinformatic approaches based on TCGA and GEO database, which was verified with immunohistochemistry in TNBC tissues. Next, the effect of SEPT3 on survival and the association between SEPT3 expression and clinical characteristics were assessed for TNBC patients. We performed Cox analysis to evaluate whether SEPT3 is an independent predictor for TNBC patients.
Results: SEPT3 was identified as a key differentially expressed gene. SEPT3 was observed to be elevated in 112 TNBC significantly. Increased expression of SEPT3 contributed to an unfavorable prognosis in patients with TNBC. Additionally, SEPT3 was associated with several factors including TNM stage, lymph node metastasis, Ki67 level and histological grade. SEPT3 was determined to be an independent risk factor for TNBC patients through Cox regression analysis.
Conclusion: This study demonstrated that SEPT3 could be a potential disease marker for TNBC patients by bioinformatics analysis and validation in clinical samples.
Keywords: triple-negative breast cancer, STEP3, prognosis, biomarker
Introduction
As a heterogeneous disease, the absence or presence status of hormone receptors and human epidermal growth factor receptor-2 (HER2) is used to classify breast cancer subtypes.1 Triple-negative breast cancer (TNBC) is the one that lacks detectable expression of three index: estrogen and progesterone receptors (ER, PR) and HER2. Given these properties, endocrine therapy and targeted therapy have poor efficacy for patients with TNBC.2 Recently, the clinical treatment for TNBC is mainly chemotherapy combining paclitaxel and platinum with little benefit.3 Therefore, the poor prognosis of TNBC patients requires identifying specific molecular targets.
Septins, a highly conserved guanosine triphosphate (GTP) binding protein family, can be divided into four homology subgroups.4 In addition to their GTPase function, septins can take part in initiating homo- or heterooligomeric complexes, which are essential for maintaining the cytoskeleton.5 Recently, the role of septins in cancer was gradually excavated. Septin9 was found to be overexpressed in human breast cancer.6 In colorectal cancer, Septin9 could serve as a specific blood marker.7 Besides, Septin9 could affect cell invasion and metastasis in melanoma.8 Cancer-associated fibroblasts (CAFs) tend to facilitate cancer invasion and angiogenesis via remodeling the tumor matrix. It has been reported that Septin2 was involved in the regulation of matrix remodeling and tumor-growth promoting mediated by CAFs.9 A growing body of evidence demonstrated that septins play a key role in cancer.
Neuron-specific septin-3 (Septin3, SEPT3), of the SEPT3 subgroup, is present in the brain and testes. In previous studies, STEP3 was identified as a component of presynaptic nerve terminals with lipid association and GTP-regulated filament assembly properties.10 Conversely, accumulating evidence indicated that the effects of SEPT3 on neuronal development and synaptic function are negligible.11–13 The existing body of research on SEPT3 suggested that SEPT3 correlates with autophagy in synaptic/neuronal tissue. In primary neuronal cell cultures, SEPT3 binds the autophagy-related protein LC3B, and colocalization of these proteins can be enhanced by autophagy inducers.14 Current studies have focused on the biological effect of SEPT3 in the nervous system, while its expression pattern and prognostic value in cancer have not been reported.
The expression pattern, clinical significance and prognostic value of SEPT3 in TNBC have not been investigated. This study is the first to deeply uncover the role of SEPT3 in TNBC according to the bioinformatics analysis and clinical sample validation. Next, we explored the clinical significance of SEPT3 in TNBC. The study provided a pipeline for exploration of disease markers and prognostic risk factors in TNBC.
Methods
Bioinformatics Analysis
TCGA RNA data of breast cancer and TNBC were downloaded from the Cancer Genome Atlas website (http://cancergenome.nih.gov). Next, the validation cohort of TNBC was extracted from Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/; GES76250, GSE38959). Differentially expressed genes (DEGs) were identified with the following screening conditions: |log2 Fold Change| and q-value. The Venn diagram was applied to produce the common DEGs. Next, DEGs were assigned for prognostic analysis by Log rank tests and univariate cox regression, which were presented as forest plots. We divided patients into two groups according to the expression of SEPT3. The overall survival distributions between two groups were compared via the Kaplan–Meier plotter database (www.kmplot.com). Cox regression analysis was performed univariately and multivariately to determine whether SEPT3 was a prognostic factor.
TNBC Patients’ Specimens
There were 112 paired TNBC tissues and adjacent normal samples collected from patients with resection of cancer who had been treated without receiving chemotherapy beforehand at Huangshi Central Hospital. The patients provided written consent and all aspects of this study were approved by the ethical committee of Huangshi Central Hospital (NO. 2022–14). Overall survival is defined as the time from the date of surgery to the time of death.
Immunohistochemistry (IHC)
Hematoxylin and eosin (HE) staining was conducted according to routine protocols. The expression level of SEPT3 was detected by IHC with anti-STEP3 (1:500, Abcam, UK). A gradient of xylenes and ethanol was used to deparaffinize and rehydrate all sections, respectively. All sections were submerged into citrate buffer, followed by 3% hydrogen peroxide. The next step was to block nonspecific binding by using goat serum. The primary antibodies were added with sections overnight at 4 °C. For visualization, the sections were stained with diaminobenzidine after being incubated for 60 minutes with a secondary antibody. In the cohort of 112 samples, SEPT3 expression levels were categorized by a cutoff score into high- and low-expression groups.
Statistical Analysis
R v4.0.2 and SPSS 22.0 software (SPSS Inc., Chicago, IL, USA) were used to conduct statistical analysis and graph visualization. Clinicopathologic parameters in different groups were statistically analyzed with a chi-squared test. Survival analysis was performed by the Kaplan–Meier method, and the Log rank test was used to compare the survival rates between the two groups. The Cox proportional hazards model was used to analyze the joint effect of covariates. P-value less than 0.05 was considered statistically significant.
Results
SEPT3 is Identified as a Prognostic Marker in TNBC
According to TCGA and GEO database, DEGs between TNBC and normal tissues were presented in the form of volcano (Figure 1A–C). As shown in Figure 1D, the common candidate genes among three datasets were displayed. The above genes were prepared for the following cox regression analysis. A forest map identified that DEGs were significantly associated with TNBC prognosis (Figure 1E). Based on above selection, SEPT3 was included in the following analysis.
We next assessed the expression level of SEPT3 between TNBC and normal tissues using TCGA and GEO database. We observed a significantly increased expression level of SEPT3 in TNBC compared to normal tissues (P < 0.001, Figure 2A). There was a significant correlation between higher expression of SEPT3 and a worse prognosis on survival curves (Figure 2B). The model of univariate cox proportional regression suggested that SEPT3 is related to survival (Figure 2C). In Cox multivariate analysis, SEPT3 was shown to be an independent risk factor for TNBC (Figure 2D).
Characteristics of TNBC Patients from a Clinicopathological Perspective
A total of 112 TNBC cases were enrolled in this study. The patient’s clinicopathological characteristics are shown in Table 1. The majorities had a moderate histological grade (58.3%). Seventy-eight patients (69.6%) had negative lymph node metastases in surgical specimen; 72 patients (64.3%) presented TNM stage (I–II). The pathologic types were predominantly of invasive ductal carcinoma (IDC).
 |
Table 1 Association Between SEPT3 Expression and Clinical Features of Patients |
Verification of SEPT3 Expression in TNBC Tissues
Clinical samples were analyzed for SEPT3 expression to provide further validation. In the first step, a HE staining was utilized to identify the tumor cells and tissues (Figure 3A). Next, we quantified protein expression levels of SEPT3 by IHC. As shown in Figure 3B, SEPT3 staining was intense in TNBC tissues, while more weak staining was observed in normal tissues. SEPT3 was mainly found in the cytoplasm. The high expression rate of SEPT3 in TNBC samples was significantly higher than that in normal samples, which indicates that SEPT3 exhibits a higher expression pattern in TNBC.
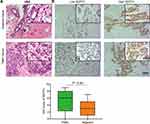 |
Figure 3 Validation expression value of SEPT3 in clinical specimen. (A) HE staining of TNBC tissue and the adjacent normal tissues. (B) IHC demonstrates immuno-expression of SEPT3 in TNBC tissues. |
The association between SEPT3 and clinical pathological features was evaluated based on the IHC score. The patients were divided into two groups, one with low levels of SEPT3 expression (n=50) and the other with high levels of SEPT3 expression (n=62). As shown in Table 1, increased expression of SEPT3 positively correlated with higher TNM stage, lymph node metastasis, histological grade and Ki67 expression (P=0.018, P=0.005, P=0.049, P=0.035). A relationship between SEPT3 expression and age, tumor size, menstrual status, or pathologic types was not found. The above findings suggested that SEPT3 may contribute to the tumor malignant progression and promotion of cancer cell proliferation.
STEP3 is an Independent Risk Factor for TNBC Patients
We next assess the effect of SEPT3 expression on prognosis. Results of the Kaplan–Meier survival curve indicated that patients with high SEPT3 expression levels had a less favorable survival than those with low SEPT3 expression levels (P=0.032) (Figure 4A–C). Univariate and multivariate Cox regression analysis showed that SEPT3 is an independent predictor for TNBC patients after adjusting for TNM stage, lymph node metastasis and Ki67 expression level (P=0.047, HR=1.212, Figure 4D). According to these results, patients with TNBC have a worse prognosis with higher SEPT3 expression.
Discussion
TNBC remains a devastating malignancy and the number of new deaths related to TNBC is increasing.15 Due to the heterogeneity of TNBC, the demand for TNBC prognostic and therapeutic markers is intensifying. Increasing evidence supported that bioinformatics technologies is a strong tool to discover novel targets for cancer.16,17 Therefore, the combination of bioinformatic approaches and experimental validation was an attractive strategy for TNBC patients. Recently, most studies focused on the screening of DEGs for prognostic prediction and treatment strategy.18,19 In this study, we identified STEP3 as a potential target for TNBC patient with the cox regression analysis based on multiple data sets.
The septin-3 protein is a member of the septin protein family that forms multimers and facilitates the formation of cytoskeletons. Previous studies mainly investigated the effect of SEPT3 on neurological disorders, such as Alzheimer’s disease (AD). AD is characterized by synaptic dysfunction. The accumulation of SEPTIN3 and SEPTIN5 might compromise synaptic transmission based on animal models.20 Using shotgun mass spectrometry, Sravani et al reported that SEPT3 might be a potential disease marker for AD.21 The expression pattern and prognostic value of septin-3 has yet to be described in cancer. From the review of literature, only one report has been published. In brain tumors, SEPT3 was observed in medulloblastoma tissues and cell lines, but not in astrocytoma tissues and cell lines.22 In this study, SEPT3 was found to be elevated in TNBC tissues from both databases and TNBC specimen. SEPT3 expression contributed to an adverse prognosis in TNBC through survival analysis, suggesting SEPT3 expression is predictive of poor prognosis. Interestingly, according to the correlation analysis between SEPT3 expression and clinical features, we found that SEPT3 may affect the progression of TNBC, especially lymph node metastasis. SEPT9, the same member of septins, has been reported to be related to the tumor growth and survival. The underlying mechanism was that SEPT9 expression could inhibit epithelial–mesenchymal transition induced by TGF-B1.23 A recent study concluded that SEPT3 might be involved in regulation of cellular migration mediated by extracellular vesicles in mesenchymal stem cells.24 Taken together, SEPT3 belongs to cytoskeleton proteins that may promote TNBC cell metastasis.
In addition to a relationship between tumor occurrence and metastasis, SEPT3 might be involved in tumor growth. The results of IHC demonstrated that SEPT3 expression was positively correlated with Ki67 expression. Septins has been reported to be a cell cycle-regulated protein, which facilitates the process of cytokinesis in human astrocytoma cells.25 As a consequence of septin overexpression, cytokinesis is compromised, centrosome amplification occurs, and multipolar mitoses occur, which are characteristics of cancer cells.26 However, the exact mechanism of tumor growth mediated by SEPT3 has not been discovered. The biological function and mechanisms of SEPT3 in TNBC need to be further verified.
Conclusions
Our study indicated that SEPT3 could be a potential predictor for TNBC prognosis. Patients with TNBC may benefit from targeting SEPT3. In this study, we provide a foundation for identifying and treating TNBC patients at an early stage.
Data Sharing Statement
All data can be obtained in TCG, GEO and conclusions of this article will be made available by the corresponding author.
Ethics Approval
All procedures performed were in accordance with the declaration of the ethical standards of the institutional research committee and with the 1964 Helsinki 387 Declaration and its later amendments. The ethics committee has approved this study of the Huangshi Central Hospital.
Acknowledgments
We are very grateful for data provided by databases such as TCGA, GEO database and like to thank Huangshi Central Hospital for its help in this research. Lihua Yang and Guozhou Wang are the joint first authors for this study. Chao Gao is the corresponding author for this study.
Funding
No funding was received.
Disclosure
All authors declare no conflicts of interest for this work.
References
1. Gradishar WJ, Moran MS, Abraham J, et al. Breast cancer, version 3.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw. 2022;20(6):691–722. doi:10.6004/jnccn.2022.0030
2. Diana A, Carlino F, Franzese E, et al. Early triple negative breast cancer: conventional treatment and emerging therapeutic landscapes. Cancers. 2020;12(4):819. doi:10.3390/cancers12040819
3. Kim C, Gao R, Sei E, et al. Chemoresistance evolution in triple-negative breast cancer delineated by single-cell sequencing. Cell. 2018;173(4):879–893.e13. doi:10.1016/j.cell.2018.03.041
4. Mostowy S, Cossart P. Septins: the fourth component of the cytoskeleton. Nat Rev Mol Cell Biol. 2012;13(3):183–194. doi:10.1038/nrm3284
5. Spiliotis ET, Kesisova IA. Spatial regulation of microtubule-dependent transport by septin GTPases. Trends Cell Biol. 2021;31(12):979–993. doi:10.1016/j.tcb.2021.06.004
6. Montagna C, Lyu MS, Hunter K, et al. The Septin 9 (MSF) gene is amplified and overexpressed in mouse mammary gland adenocarcinomas and human breast cancer cell lines. Cancer Res. 2003;63(9):2179–2187.
7. Warren JD, Xiong W, Bunker AM, et al. Septin 9 methylated DNA is a sensitive and specific blood test for colorectal cancer. BMC Med. 2011;9(1):133. doi:10.1186/1741-7015-9-133
8. Farrugia AJ, Rodríguez J, Orgaz HL, et al. CDC42EP5/BORG3 modulates SEPT9 to promote actomyosin function, migration, and invasion. J Cell Biol. 2020;219(9). doi:10.1083/jcb.201912159
9. Calvo F, Ranftl R, Hooper S, et al. Cdc42EP3/BORG2 and septin network enables mechano-transduction and the emergence of cancer-associated fibroblasts. Cell Rep. 2015;13(12):2699–2714. doi:10.1016/j.celrep.2015.11.052
10. Xue J, Tsang CW, Gai WP, et al. Septin 3 (G-septin) is a developmentally regulated phosphoprotein enriched in presynaptic nerve terminals. J Neurochem. 2004;91(3):579–590. doi:10.1111/j.1471-4159.2004.02755.x
11. Tsang CW, Fedchyshyn M, Harrison J, et al. Superfluous role of mammalian septins 3 and 5 in neuronal development and synaptic transmission. Mol Cell Biol. 2008;28(23):7012–7029. doi:10.1128/MCB.00035-08
12. Tsang CW, Estey MP, Diciccio JE, et al. Characterization of presynaptic septin complexes in mammalian hippocampal neurons. Biol Chem. 2011;392(8–9):739–749. doi:10.1515/BC.2011.077
13. Fujishima K, Kiyonari H, Kurisu J, et al. Targeted disruption of Sept3, a heteromeric assembly partner of Sept5 and Sept7 in axons, has no effect on developing CNS neurons. J Neurochem. 2007;102(1):77–92. doi:10.1111/j.1471-4159.2007.04478.x
14. Tóth V, Vadászi H, Ravasz L, et al. Neuronal-specific septin-3 binds Atg8/LC3B, accumulates and localizes to autophagosomes during induced autophagy. Cell Mol Life Sci. 2022;79(9):471. doi:10.1007/s00018-022-04488-8
15. Bianchini G, De Angelis C, Licata L, et al. Treatment landscape of triple-negative breast cancer - expanded options, evolving needs. Nat Rev Clin Oncol. 2022;19(2):91–113. doi:10.1038/s41571-021-00565-2
16. Wang Y, Wu S, Zhu X, et al. LncRNA-encoded polypeptide ASRPS inhibits triple-negative breast cancer angiogenesis. J Exp Med. 2020;217(3). doi:10.1084/jem.20190950
17. Guo B, Wu S, Zhu X, et al. Micropeptide CIP2A-BP encoded by LINC00665 inhibits triple-negative breast cancer progression. EMBO j. 2020;39(1):e102190. doi:10.15252/embj.2019102190
18. Acharya S, Yao J, Li P, et al. Sphingosine kinase 1 signaling promotes metastasis of triple-negative breast cancer. Cancer Res. 2019;79(16):4211–4226. doi:10.1158/0008-5472.CAN-18-3803
19. Zamberlan M, Boeckx A, Muller F, et al. Inhibition of the mitochondrial protein Opa1 curtails breast cancer growth. J Exp Clin Cancer Res. 2022;41(1):95. doi:10.1186/s13046-022-02304-6
20. Györffy BA, Tóth V, Török G, et al. Synaptic mitochondrial dysfunction and septin accumulation are linked to complement-mediated synapse loss in an Alzheimer’s disease animal model. Cell Mol Life Sci. 2020;77(24):5243–5258. doi:10.1007/s00018-020-03468-0
21. Musunuri S, Wetterhall M, Ingelsson M, et al. Quantification of the brain proteome in Alzheimer’s disease using multiplexed mass spectrometry. J Proteome Res. 2014;13(4):2056–2068. doi:10.1021/pr401202d
22. Kim DS, Hubbard SL, Peraud A, et al. Analysis of mammalian septin expression in human malignant brain tumors. Neoplasia. 2004;6(2):168–178. doi:10.1593/neo.03310
23. Zhang G, Feng W, Wu J. Down-regulation of SEPT9 inhibits glioma progression through suppressing TGF-β-induced epithelial-mesenchymal transition (EMT). Biomed Pharmacother. 2020;125:109768. doi:10.1016/j.biopha.2019.109768
24. Zubkova E, Evtushenko E, Beloglazova I, et al. Analysis of MicroRNA profile alterations in extracellular vesicles from mesenchymal stromal cells overexpressing stem cell factor. Front Cell Dev Biol. 2021;9:754025. doi:10.3389/fcell.2021.754025
25. Xu D, Liu A, Wang X, et al. Repression of Septin9 and Septin2 suppresses tumor growth of human glioblastoma cells. Cell Death Dis. 2018;9(5):514. doi:10.1038/s41419-018-0547-4
26. Poüs C, Klipfel L, Baillet A. Cancer-related functions and subcellular localizations of septins. Front Cell Dev Biol. 2016;4:126. doi:10.3389/fcell.2016.00126
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.



