Back to Journals » International Journal of Nanomedicine » Volume 16
Self-Assembled DNA Nanostructure as a Carrier for Targeted siRNA Delivery in Glioma Cells
Authors Zhou Y , Yang Q , Wang F, Zhou Z, Xu J, Cheng S, Cheng Y
Received 7 December 2020
Accepted for publication 13 February 2021
Published 3 March 2021 Volume 2021:16 Pages 1805—1817
DOI https://doi.org/10.2147/IJN.S295598
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Farooq A. Shiekh
Yanghao Zhou,1 Qiang Yang,1 Feng Wang,1 Zunjie Zhou,1 Jing Xu,1 Si Cheng,2 Yuan Cheng1
1Department of Neurosurgery, The Second Affiliated Hospital of Chongqing Medical University, Chongqing, 400010, People’s Republic of China; 2Department of Orthopedics, The Second Affiliated Hospital of Chongqing Medical University, Chongqing, 400010, People’s Republic of China
Correspondence: Yuan Cheng
Department of Neurosurgery, The Second Affiliated Hospital of Chongqing Medical University, Chongqing, 400016, People’s Republic of China
Email [email protected]
Si Cheng
Department of Orthopedics, The Second Affiliated Hospital of Chongqing Medical University, Chongqing, 400010, People’s Republic of China
Email [email protected]
Introduction: RNA interference is a promising therapy in glioma treatment. However, the application of RNA interference has been limited in glioma therapy by RNA instability and the lack of tumor targeting. Here, we report a novel DNA tetrahedron, which can effectively deliver small interfering RNA to glioma cells and induce apoptosis.
Methods: siRNA, a small interfering RNA that can suppress the expression of survivin in glioma, was loaded into the DNA tetrahedron (TDN). To enhance the ability of active targeting of this nanoparticle, we modified one side of the DNA nanostructure with aptamer as1411 (As-TDN-R), which can selectively recognize the nucleolin in the cytomembrane of tumor cells. The modified nanoparticles were characterized by agarose gel electrophoresis, dynamic light scattering, and transmission electron microscopy. The serum stability was evaluated by agarose gel electrophoresis. Nucleolin was detected by Western blot and immunofluorescence, and targeted cellular uptake was examined by flow cytometry. The TUNEL assay, flow cytometry, and Western Blot were used to detect apoptosis in U87 cells. The gene silencing of survivin was examined by qPCR, Western Blot, and immunofluorescence.
Results: As-TDN-R alone showed better stability towards siRNA, indicating that TDN was a good siRNA protector. Compared with TDN alone, there was increased intercellular uptake of As-TDN-R by U87 cells, evidenced by overexpressed nucleolin in glioma cell lines. TUNEL assay, flow cytometry, and Western Blot revealed increased apoptosis in the As-TDN-R group. The downregulation of survivin protein and mRNA expression levels indicated that As-TDN-R effectively silenced the target gene.
Conclusion: The novel nanoparticle can serve as a good carrier for targeting siRNA delivery in glioma. Further exploration of the DNA nanostructure can greatly promote the application of DNA-based drug systems in glioma.
Keywords: DNA tetrahedron, nanomedicine, tumor targeting, aptamer, apoptosis, RNA interference
Introduction
Glioma is the most malignant brain tumor that originates from glial cells. Besides, it is the most deadly type of brain tumor, with a five-year survival rate of less than 35%.1,2 The majority of glioma patients are treated with conventional surgery, combined with radiation, chemotherapy, or other drugs, due to the poor efficacy of traditional drug formulations.1,3 Although progressive therapy has been clinically used, scholars continue to explore more effective approaches to improve the survival rates of brain cancer patients.
One promising approach to glioma treatment is the inactivation of proteins critical to tumor survival or progression through RNA interference (RNAi), which ultimately results in the death of tumor cells. The main targets of RNAi therapy include oncogenes and genes involved in survival, anti-apoptosis, angiogenesis, metastasis, and chemotherapeutic resistance.4,5 Among the apoptotic inhibitor proteins, survivin has attracted increasing attention because of its high expression in tumor tissues and tumor cell lines.6 Survivin is also overexpressed in human gliomas, and its high protein expression level is associated with the degree of tumor malignancy.7–9 The higher expression level of survivin is also associated with a worse prognosis. Previous studies have shown that survivin-targeted RNA interference can inhibit cancer cell growth and prolong the survival of tumor-bearing mice.10 However, due to poor RNA stability, survivin cannot precisely enter tumor cells, thus, its application remains a challenge. Current RNA delivery methods include liposomes, cationic colloid, and carbon quantum dots, however, these methods have issues related to the material itself.5
Recently, a novel nanomaterial, DNA tetrahedron, has emerged as a multifunctional drug due to its anti-inflammatory, anti-oxidative, and neuroprotective effects.11 Besides, it has been used as a multifunctional drug carrier due to its high drug delivery capacity, good biocompatibility, and biodegradability.12 DNA nanomaterials are composed of only purines and pyrimidines, which means high editability. Therefore, a variety of tumor-targeting ligands and antitumor drugs are designed and modified on the tetrahedron. AS1411 is a guanine-rich DNA oligonucleotide produced by SELEX technology. It recognizes the nucleolin protein expressed on the tumor cell membrane through the high affinity of the guanine domain.13 Studies have shown that AS1411 inhibits the formation of new blood vessels and it is applied to ophthalmology and tumor-related diseases. Based on these facts, we modified aptamer AS1411 onto one side of the DNA tetrahedron to form a drug carrier with tumor-targeted function. Moreover, cy3 (a kind of anthocyanin dye), which emit red fluorescence under excitation, was attached to the DNA oligonucleotide. Based on the location of the fluorescence signal, the biological behavior of the nanomaterial in the cell can be determined.
In this study, we constructed a tetrahedral DNA nanostructure loaded survivin interfering RNA (As-TDN-R), to selectively identify tumors cells over-expressing nucleolin protein. Through the cy3 fluorescence signal, the biological behavior of nanomaterials in the cells was determined by flow cytometry and fluorescence staining. The results showed that the nanomaterials had good structural stability and biocompatibility. Besides, compared with the tetrahedron alone, As-TDN-R was found to be absorbed more by tumor cells and concentrated around the nucleus to achieve effective concentration. By interfering with survivin expression, As-TDN-R can inhibit the anti-apoptotic pathways of gliomas and induce tumor cell death.
Materials and Methods
All oligonucleotides were synthesized and purified by Sangon Biotech Company (Shanghai, China). Survivin siRNA (5ʹ-AUUCACCAAGGGUUAAUUCdTdT-3ʹ) was synthesized by GenePharma Company (Shanghai, China). Deionized water used in all aqueous solutions was produced using a Millipore Ultrapure water machine (Massachusetts, USA). GelRed DNA gel stain solution was purchased from Biosharp (Beijing, China). Tris, magnesium chloride (MgCl2), and Triton X-100 were obtained from Macklin (Shanghai, China). Agarose was purchased from Biowest (Barcelona, Spain). U87, U251, and HUVEC cells were purchased from ATCC. Fetal bovine serum (FBS) and high-glucose Dulbecco’s modified Eagle’s medium (DMEM/high glucose) were bought from Gibco (NY, USA). A cell counting kit (CCK-8) was purchased from Bimake (Texas, USA). 4′6-Diamidino-2-phenylindole (DAPI) were obtained from Solarbio (Beijing, China). Primary antibodies β-actin (20536-1-AP), NCL (10556-1-AP), and Caspase-3 (19677-1-AP) were obtained from Proteintech (IL, USA). Survivin (YT4472) was purchased from Immunoway (TX, USA), and horseradish peroxidase-conjugated or CoraLite594-conjugated secondary antibodies were purchased from Proteintech (IL, USA).
Preparation of As-TDN-R
The purchased oligonucleotides were diluted in deionized water. The oligonucleotides (AS-s1, Si-s2, s3, s4, and siRNA-R) were mixed using the same molar ratio in TM (10mM Tris base, 5mM MgCl2, PH=8.0) buffer to assemble the target nanostructure.14 Briefly, the four single-strand DNA were heated to 95°C for 10min and slowly annealed to 4°C in 2 hours.15,16 All the oligonucleotides are listed in Table 1.
 |
Table 1 Sequence of Each Oligonucleotide |
Characterization and Stability of As-TDN-R
Agarose gel electrophoresis (3%) was used to determine the accuracy of As-TDN-R synthesis. To stain the single-strand DNA, GelRed was added to the agarose gel. The samples were run in the 1X Tris-acetate-EDTA electrophoresis buffer at a voltage of 80v for 40min, and the gel captured using the Bio-Rad ChemiDoc MP imaging system (California, USA).
The morphological characteristics of the nanoparticles were detected using a transmission electron microscope (TEM). A drop of nanoparticles was dried on the copper mesh overnight. The sample was imaged at an accelerating voltage of 80kV using the Hitachi-7500 (Tokyo, Japan). Moreover, the size and distribution of the nanoparticle were detected using the Malvern Zetasizer ZS90 (Malvern, UK). The As-TDN-R stability in serum was determined by agarose gel electrophoresis. Briefly, As-TDN-R were incubated with 10% FBS at various periods (6 h, 12 h, 24 h, and 48 h), and agarose gel electrophoresis was used to confirm the As-TDN-R structure.
Cell Culture
The human glioma cell lines (U87 and U251) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) High Glucose containing 1% penicillin-streptomycin solution, and 10% fetal bovine serum. HUVEC cell line was incubated in DMEM/F12 mixed with 0.1mg/mL heparin, 0.03mg/mL ECGs, 1% penicillin-streptomycin solution and 10% fetal bovine serum. All cell lines were cultured in a humidified incubator with 5% CO2 at 37°C. The culture medium was replaced every 2–3 days.
Cellular Uptake of Nanoparticles
cy3-s3 was used to synthesize cy3-TDN as previously reported.17 U87 cells were digested, placed on the coverslips in 24-well culture plates and grown for 24 h until the cells reached at least 75% confluence. The cells were treated with PBS, cy3-TDN (100nM), and cy3-As-TDN-R (100nM) at 37°C in a humidified atmosphere with 5% CO2 for 6 h. The cells were rinsed three times using 0.01% PBS and fixed with cold 4% paraformaldehyde for 15 minutes in the dark. After rinsing three times with PBS, 4′,6-diamidino-2-phenylindole (DAPI) was used to stain the nucleus for 10 min. The coverslips were sealed with glycerin, and a fluorescence microscope (Olympus, Tokyo, Japan) was used to observe cellular uptake and capture images. Moreover, cellular uptake state was also detected by flow cytometry.
U87 cells were digested and cultured in 6-well plates at a density of 5×105 for 24 h until the cells reached confluence. The cells were then treated with PBS, cy3-TDN (100nM), and cy3-As-TDN-R (100nM) at 37°C for 6 h (as described above). The treated cells were digested using 0.25% trypsin and collected by centrifugation. The harvested cells were subsequently counted using a Beckman flow cytometer (California, USA). To further confirm the selectivity of nanoparticles to cells, HUVEC cells were also treated as described above.
CCK-8 Assays
U87 cells were digested and grown in 96-well plates at a density of 5000 cells per well. After pretreatment with a low FBS culture medium for 2 h, an equivalent concentration of PBS, TDN (150nM), or As-TDN-R (150nM) were added and incubated for 48 h. To each well, 10 μL of CCK-8 (Bimake, USA) was added and incubated for 1 h. The absorbance was determined at 450 nm using a microplate reader. To further determine the effect of As-TDN-R on cell cytotoxicity, different concentrations (0nM, 50nM, 150nM, and 250nM) were added to U87 cells, and all the other steps were performed as previously described.
TUNEL Assays
Apoptosis is accompanied by DNA damage, which can be detected using the apoptosis detection kit. U87 cells at a density of 5000 per well were placed in a 24-well climbing slice and cultured overnight. Subsequently, the media was replaced with a complete medium containing different nanoparticles (PBS, 150nM TDN, or 150nM As-TDN-R) and incubated for another 48 h. The cells were fixed with 4% paraformaldehyde for 20 min, blocked with 5% goat serum for 30 min, and treated with 0.3% Triton X-100 for 10 min at room temperature. The terminal deoxynucleotidyl transferase (TdT) reaction reagent (Beyotime, China) was added, incubated for 60 minutes, and DAPI was used to stain the cell nucleus. A fluorescence microscope (Olympus, Tokyo, Japan) was used to capture the images.
Flow Cytometry of Apoptosis
U87 cells were seeded into 6-well plates at a density of 5×105 per well and incubated at 37 °C. An equivalent amount of PBS, TDN (150nM), and As-TDN-R (150nM) was added and the cells incubated at 37 °C for 48 h. The cells were harvested and incubated with reagents from the Annexin V-FITC apoptosis kit (Beyotime, China) according to the manufacturer’s instructions, and the cells were counted using a Beckman flow cytometer (California, USA).
Western Blot
HUVEC, U87, and U251 cells were lysed with RIPA lysate containing 1mm of phenylmethane sulfonyl fluoride (Beyotime, China). After centrifugation, the total protein concentration was determined using a Pierce bicinchoninic add (BCA) protein assay kit (Beyotime, China). The equivalent amount of protein (20ug) was separated by electrophoresis on 12.5% sodium dodecyl sulfate-polyacrylamide gel followed by transfer to the polyvinylidene fluoride membrane (Millipore, USA). The membranes were blocked with 5% skimmed milk powder in TBST (pH=8.0) for 2 h at room temperature. The membrane was incubated with nucleolin (1:1000), caspase-3 (1:1000), and survivin (1:1000) antibody overnight at 4°C, followed by incubation with horseradish peroxidase-labeled secondary antibodies (1:5000) for 1 h. The Bio-Rad detection system was used to detect the chemiluminescence, and β-actin was used as the internal standard.
Quantitative Reverse Transcription-Polymerase Chain Reaction (qRT-PCR)
U87 cells were incubated with different DNA nanoparticles (TDN, As-TD-R) at a concentration of 150nM for 48 h. Total RNA of U87 cells was extracted using TRIzol according to the manufacturer’s instructions. The concentration and quality of nucleic acids in each sample was determined using a NanoDrop Spectrophotometer (Thermo Fisher, USA). The RNA samples were reverse-transcribed to cDNA using a Primescript RT reagent kit (TaKaRa, Japan). The following primers were used for real-time PCR analysis: survivin forward: TCAAGGACCACCGCATCTCTA, reverse: TGAAGCAGAAGAAACACTGGGC, GAPDH forward: CACCACCATGGAGAAGGCTGG, reverse: CCAAAGTTGTCATGGAT-GACC. Amplification of target genes was performed with the real-time PCR machine (Bio-rad, USA). GAPDH was used as the internal control, and the relative fold-changes in mRNA expression levels were calculated using the ΔΔCT method.18
Immunofluorescence
U87 cells were digested and seeded onto the coverslip of 24-well plates. The cells were treated with TDN and As-TDN-R for 48 h. After drug treatment, the cells were fixed with 4% paraformaldehyde for 20 min and permeabilized with 0.5% Triton X-100 in PBS for 15 min. Subsequently, the coverslips were incubated with 5% goat serum for 1 h to block the cells. The cells were then incubated with nucleolin (1:100) and survivin (1:150) overnight at 4°C, followed by incubation with CoraLite594 labeled secondary antibody (1:1000) for 1 h at 37°C. DAPI was used to stain the cell nucleus, and a fluorescence microscope (Olympus, Tokyo, Japan) was used to capture the images.
Statistical Analysis
All data were presented as mean ± standard deviation. Statistical analysis was performed using SPSS 22.0. One-way analysis of variance was used to determine any statistical differences between the groups (IBM, NY, USA). *P<0.05, **P<0.01, ***p<0.001 were considered statistically significant. Graphs were generated by GraphPad Prism 8.0.
Results
Synthesis and Characterization of As-TDN-R
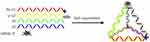
A tetrahedron (TDN) is a three-dimensional structure based on DNA material.19 It can be rapidly self-assembled from four designed single-strand DNA (ssDNA) via Watson-Crick base pairing.20 To synthesize As-TDN-R previously reported methods were used with slight modifications21 as shown in Table 1. AS1411 aptamer was strengthened on the 5ʹ end of s1, and the complementary strand of siRNA-R on s2. Each single-strand DNA formed one face of As-TDN-R, whereas the modified As1411 and siRNA were on the apex of the tetrahedron (Scheme 1).
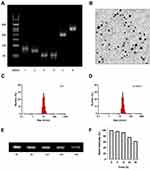
To confirm the successful synthesis of As-TDN-R, 3% agarose gel was used to separate each of the DNA samples based on their migration speed.22 As shown in Figure 1A, the first four bands represented four single-strand DNA (Lane 1: As-s1, Lane 2: si-s2, Lane 3: s3, Lane 4: s4). Due to external modification, band 1 and band 2 moved slowly compared with the other two bands. Besides, As-TDN-R migration (Lane 6) was slower compared with the single TDN (Lane 5), suggesting that As-TND-R was successfully formed.
The discrete and uniform state of nanoparticles were investigated by Transmission electron microscopy. The image showed that the size of As-TDN-R was approximately 18nm with good dispersion (Figure 1B). Meanwhile, the dynamic light scattering was used to measure the size distribution of As-TDN-R. Compared with TDN (~10 nm), As-TDN-R had a larger size of ~18 nm (Figure 1C and D), which was hypothesized to be due to the inclusion of AS1411 and siRNA.
Good stability of nanoparticles in serum is a crucial factor for efficient drug delivery.23 Serum stability of As-TDN-R was determined by placing the nanoparticles in a complete medium with 10% FBS at 37 °C for 48 h. Agarose gel electrophoresis showed that the As-TND-R band was observed after 48 h incubation (Figure 1E). Semiquantitative analysis of the relative band intensity showed that over 50% of As-TDN-R remained intact (Figure 1F), which reflected that the nanoparticle can be a good siRNA cargo.
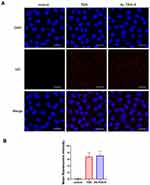
Expression of Nucleolin in Glioma Cell Lines
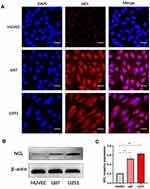
Nucleolin, a multifunctional protein that is highly expressed in a variety of tumors, promotes the development and progression of tumors.24,25 It is also the ligand of aptamer AS1411, which is a G-rich DNA oligonucleotide produced by SELEX technology.26 When combined with AS1411, nucleolin supports the cellular entry of AS1411 as a molecular chaperone.13 As shown in Figure 2B and C, compared with HUVEC cells, U87 and U251 cells highly expressed nucleolin protein. Moreover, immunofluorescence showed that nucleolin was localized in both the cytoplasm and membrane in glioma cells (Figure 2A), which can act as the ligand of active targeting.
Intercellular Uptake of As-TND-R
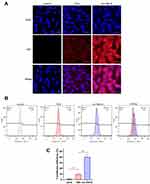
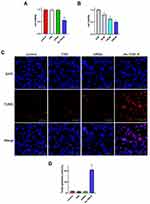
To investigate the cellular uptake of nanoparticles, U87 cells were treated with cy3-TDN or cy3-As-TDN-R. As shown in Figure 3A, both groups were stained with cy3. Earlier studies have shown that single and double DNA cannot cross the cell membrane due to their negative potentials.16 However, when single-stranded DNA forms a three-dimensional structure, it can be efficiently ingested by cells via endocytosis and the caveolin-dependent pathway.27 Compared with TDN alone, As-TDN-R showed more cellular uptake, which was largely concentrated around the nucleus, and partially gained entry into the nucleus. Moreover, flow cytometry analysis showed that 39.2% of cells were cy3 positive in the As-TDN-R group and 8.86% positive in the TDN group (Figure 3B and C). The aptamer AS1411 can combine with nucleolin, which is widely expressed in the cytoplasm and nucleus of tumor cells. The localization of the nucleolin enabled As-TND-R showed better permeability than TDN alone.
Besides, we also treated HUVEC cells with cy3-TDN or As-TDN-R. Interestingly, there was no statistical difference between the two groups (Figure 4A and B). The results indicated that the enhanced As-TDN-R permeability was driven by AS1411 aptamer.
The Effect of As-TDN-R on Cell Viability
Survivin, an apoptotic inhibitor protein, is highly expressed in human gliomas, where increased expression levels are related to the higher grade of malignancy.28,29 The inhibition of survivin can result in apoptosis and tumor growth suppression. Therefore, we treated U87 cells with siRNA, TDN, or As-TDN-R at concentrations of 150nM, respectively to detect their influence on cell viability. As shown in Figure 5A, a significant decrease was observed in the As-TDN-R group, while there was no statistical difference between the siRNA or TDN group. The results indicated that As-TDN-R can maintain the influence of siRNA on cell viability. Furthermore, when treated with a set of gradient concentrations of As-TDN-R, the cell viability of U87 cells decreased (Figure 5B). The results revealed that there was dose-dependent cytotoxicity of As-TND-R on U87 cell viability.
The Effect of As-TDN-R on Cell Apoptosis
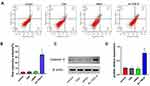
We performed a terminal deoxynucleotidyl Transferase (TdT) mediated dUTP nick end labeling (TUNEL) assay and flow cytometry analysis to evaluate the effect of inhibition of survivin on apoptosis. In TUNEL assay, the 3ʹ-OH on the fragments of broken DNA is added with cy3-labeled dUPT through the catalysis of TdT.30 Quantification of dUPT+ cells showed nearly 40% of the cells were labeled with dUPT after As-TDN-R treatment. However, dUPT+ cells treated with TDN or siRNA as the control group showed similar results, with approximately 6% of the cells labeled, which was much lower than the As-TDN-R group (Figure 5C and D). Flow cytometry analysis revealed that the As-TDN-R group showed the highest apoptosis rate among the four groups (Figure 6A and B), with an apoptosis rate of 34.3% compared with the other three groups 3.3%, 3.2%, 3.1%, respectively. Furthermore, the expression of the apoptotic effector protein also showed a similar trend in the four groups (Figure 6C). The protein level of caspase-3 was about 2.8-fold higher in the As-TDN-R group than that of the other three groups (Figure 6D). These results indicated that As-TND-R can induce the apoptosis of U87 cells.
Downregulation of Survivin by As-TDN-R in U87 Cells
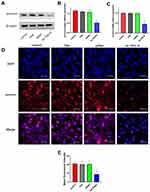
To further investigate the effect of As-TND-R on the target genes, we detected the expression levels of survivin in U87 cells in three different aspects. qPCR results (Figure 7C), showed that survivin was considerably downregulated in the As-TND-R group, with a mean value of 0.45-fold. Besides, Western blot analysis showed that the protein levels of survivin was decreased in the corresponding group, and was 0.41-times lower than that in the control and TDN alone groups (Figure 7A and B). Moreover, immunofluorescence images showed a decrease in survivin expression in the As-TDN-R group (Figure 7D and E), whereas the survivin expression in the cytoplasm and nucleus was decreased. Decreased expression of survivin at the gene and protein levels indicated a successful reduction in the translation of the target gene in the cytoplasm and nucleus.
Discussion
The main objective of this study was to evaluate the effective transportation of siRNA into gliomas cells by aptamer-modified DNA tetrahedrons. First, we successfully constructed siRNA loaded, aptamer modified DNA tetrahedron (As-TDN-R) in a one-step synthesis. TEM and DLS analysis revealed that the size of As-TDN-R was approximately 18nm. Subsequently, flow cytometry and fluorescence microscopy analysis revealed that the nanoparticle was effectively absorbed by U87 cells, but not HUVEC cells, indicating that the nanostructure was selective to glioma cells. Survivin expression analysis showed that the nanostructure inhibited survivin expression level and induced apoptosis in gliomas.
RNA interference technology has great potential in the treatment of gliomas. However, RNA instability limits its application in clinical research. In this study, the nanomaterials had a protective effect on RNA, thus, enhancing RNA stability.
Due to the lack of tissue selectivity, traditional drugs not only have anti-tumor effects but also cause systemic toxicity.31 In this study, differential expression of nucleolin in glioma cells and normal cells was observed, meaning that the receptor can be useful glioma markers.32 After attachment with the aptamer AS1411, the novel nanostructure showed differential intercellular uptake between glioma cells and normal cells. However, the exact mechanism of this differential uptake remains unclear. Le reported that the DNA tetrahedron with negative potential enters the cell via endocytosis and the caveolin-dependent pathway,27 while Reyes-Reyes showed that AS1411 increased the permeability of the linked-material through nucleolin-dependent endocytosis.33 Therefore, we hypothesized that these two mechanisms work together to allow As-TDN-R to enter the cell.
Overexpression of survivin is common in tumor cells, which enables them to grow rapidly. In this study, Western blot, qPCR, and immunofluorescence showed decreased expression of surviving, which was associated with efficient gene silencing by As-TDN-R. Studies on the mechanism of survivin found that it can induce the inactivation of apoptotic effector caspase-3,34 which is consistent with our experimental results. The protein level of caspase-3 increased after the As-TDN-R treatment. Moreover, As-TDN-R induced apoptosis and efficient gene silencing by AS-TDN-R.
Conclusion
In this study, we present a successfully synthesize novel nanostructure (As-TDN-R) to deliver survivin siRNA for efficient regulation of gene expression. In contrast to traditional strategies, this siRNA delivery platform is safe, efficient, and easily manufactured. In this study, As-TND-R is found to efficiently enter U87 cells. Through overexpression of nucleolin in glioma, aptamer AS1411 enhances the cellular uptake by glioma cells, thus playing a role in active targeted delivery. Besides, the As-TDN-R effectively induces apoptosis in glioma cells which are activated by inhibiting survivin expression. These findings suggested that As-DNA -R is a promising and efficient system for siRNA delivery into glioma. Due to the high editability of the DNA tetrahedron, further exploration of the nanostructure can greatly promote the application of DNA-based drug systems in glioma.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (No.81771961).
Disclosure
The authors declare no conflicts of interest.
References
1. Lapointe S, Perry A, Butowski NA. Primary brain tumours in adults. Lancet. 2018;392(10145):432–446. doi:10.1016/s0140-6736(18)30990-5
2. Laug D, Glasgow SM, Deneen B. A glial blueprint for gliomagenesis. Nat Rev Neurosci. 2018;19(7):393–403. doi:10.1038/s41583-018-0014-3
3. Brandner S, Jaunmuktane Z. Neurological update: gliomas and other primary brain tumours in adults. J Neurol. 2018;265(3):717–727. doi:10.1007/s00415-017-8652-3
4. Zheng M, Tao W, Zou Y, Farokhzad OC, Shi B. Nanotechnology-based strategies for siRNA brain delivery for disease therapy. Trends Biotechnol. 2018;36(5):562–575. doi:10.1016/j.tibtech.2018.01.006
5. Xin Y, Huang M, Guo WW, Huang Q, Zhang LZ, Jiang G. Nano-based delivery of RNAi in cancer therapy. Mol Cancer. 2017;16(1):134. doi:10.1186/s12943-017-0683-y
6. Garg H, Suri P, Gupta JC, Talwar GP, Dubey S. Survivin: a unique target for tumor therapy. Cancer Cell Int. 2016;16(1):49. doi:10.1186/s12935-016-0326-1
7. Andersen MH, Svane IM, Becker JC, Straten PT. The universal character of the tumor-associated antigen survivin. Clin Cancer Res. 2007;13(20):5991–5994. doi:10.1158/1078-0432.CCR-07-0686
8. Dahan P, Martinez Gala J, Delmas C, et al. Ionizing radiations sustain glioblastoma cell dedifferentiation to a stem-like phenotype through survivin: possible involvement in radioresistance. Cell Death Dis. 2014;5:e1543. doi:10.1038/cddis.2014.509
9. Zhang S, Zhang C, Song Y, Zhang J, Xu J. Prognostic role of survivin in patients with glioma. Medicine (Baltimore). 2018;97(17):e0571. doi:10.1097/MD.0000000000010571
10. Colombo M, Figueiro F, de Fraga Dias A, Teixeira HF, Battastini AMO, Koester LS. Kaempferol-loaded mucoadhesive nanoemulsion for intranasal administration reduces glioma growth in vitro. Int J Pharm. 2018;543(1–2):214–223. doi:10.1016/j.ijpharm.2018.03.055
11. Zhang Q, Lin S, Shi S, et al. Anti-inflammatory and antioxidative effects of tetrahedral DNA nanostructures via the modulation of macrophage responses. ACS Appl Mater Interfaces. 2018;10(4):3421–3430. doi:10.1021/acsami.7b17928
12. Xie N, Liu S, Yang X, He X, Huang J, Wang K. DNA tetrahedron nanostructures for biological applications: biosensors and drug delivery. Analyst. 2017;142(18):3322–3332. doi:10.1039/c7an01154g
13. Bates PJ, Reyes-Reyes EM, Malik MT, Murphy EM, O’Toole MG, Trent JO. G-quadruplex oligonucleotide AS1411 as a cancer-targeting agent: uses and mechanisms. Biochim Biophys Acta Gen Subj. 2017;1861(5 Pt B):1414–1428. doi:10.1016/j.bbagen.2016.12.015
14. Fu W, You C, Ma L, et al. Enhanced efficacy of temozolomide loaded by a tetrahedral framework DNA nanoparticle in the therapy for glioblastoma. ACS Appl Mater Interfaces. 2019;11(43):39525–39533. doi:10.1021/acsami.9b13829
15. Liu X, Wang G, You Z, et al. Inhibition of hypoxia-induced proliferation of pulmonary arterial smooth muscle cells by a mTOR siRNA-loaded cyclodextrin nanovector. Biomaterials. 2014;35(14):4401–4416. doi:10.1016/j.biomaterials.2014.02.009
16. Yang J, Jiang Q, He L, et al. Self-assembled double-bundle DNA tetrahedron for efficient antisense delivery. ACS Appl Mater Interfaces. 2018;10(28):23693–23699. doi:10.1021/acsami.8b07889
17. Tian T, Li J, Xie C, et al. Targeted imaging of brain tumors with a framework nucleic acid probe. ACS Appl Mater Interfaces. 2018;10(4):3414–3420. doi:10.1021/acsami.7b17927
18. Angileri FF, Aguennouz M, Conti A, et al. Nuclear factor-kappaB activation and differential expression of survivin and Bcl-2 in human grade 2-4 astrocytomas. Cancer. 2008;112(10):2258–2266. doi:10.1002/cncr.23407
19. Goodman RP, Berry RM, Turberfield AJ. The single-step synthesis of a DNA tetrahedron. Chem Commun. 2004;(12):1372–1373. doi:10.1039/B402293A
20. Sun W, Ji W, Hall JM, et al. Self-assembled DNA nanoclews for the efficient delivery of CRISPR-Cas9 for genome editing. Angew Chem Int Ed Engl. 2015;54(41):12029–12033. doi:10.1002/anie.201506030
21. Wang F, Zhou Y, Cheng S, et al. Gint4.T-modified DNA tetrahedrons loaded with doxorubicin inhibits glioma cell proliferation by targeting PDGFRbeta. Nanoscale Res Lett. 2020;15(1):150. doi:10.1186/s11671-020-03377-y
22. Ma W, Xie X, Shao X, et al. Tetrahedral DNA nanostructures facilitate neural stem cell migration via activating RHOA/ROCK 2 signalling pathway. Cell Prolif. 2018;51(6):e12503. doi:10.1111/cpr.12503
23. Jiang D, England CG, Cai W. DNA nanomaterials for preclinical imaging and drug delivery. J Control Release. 2016;239:27–38. doi:10.1016/j.jconrel.2016.08.013
24. Li F, Lu J, Liu J, et al. A water-soluble nucleolin aptamer-paclitaxel conjugate for tumor-specific targeting in ovarian cancer. Nat Commun. 2017;8(1):1390. doi:10.1038/s41467-017-01565-6
25. Xu Z, Joshi N, Agarwal A, et al. Knocking down nucleolin expression in gliomas inhibits tumor growth and induces cell cycle arrest. J Neurooncol. 2012;108(1):59–67. doi:10.1007/s11060-012-0827-2
26. Bates PJ, Laber DA, Miller DM, Thomas SD, Trent JO. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp Mol Pathol. 2009;86(3):151–164. doi:10.1016/j.yexmp.2009.01.004
27. Liang L, Li J, Li Q, et al. Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells. Angew Chem Int Ed Engl. 2014;53(30):7745–7750. doi:10.1002/anie.201403236
28. Fenstermaker RA, Ciesielski MJ, Qiu J, et al. Clinical study of a survivin long peptide vaccine (SurVaxM) in patients with recurrent malignant glioma. Cancer Immunol Immunother. 2016;65(11):1339–1352. doi:10.1007/s00262-016-1890-x
29. Zhang F, Chu J, Wang F. Expression and clinical significance of cyclooxygenase 2 and survivin in human gliomas. Oncol Lett. 2017;14(2):1303–1308. doi:10.3892/ol.2017.6281
30. Kyrylkova K, Kyryachenko S, Leid M, Kioussi C. Detection of apoptosis by TUNEL assay. Odontogenesis. 2012;41–47. doi:10.1007/978-1-61779-860-3_5
31. Wang H, Yu J, Lu X, He X. Nanoparticle systems reduce systemic toxicity in cancer treatment. Future Med. 2016. doi:10.2217/nnm.15.166
32. Balça-Silva J, Do Carmo A, Tão H. Nucleolin is expressed in patient-derived samples and glioblastoma cells, enabling improved intracellular drug delivery and cytotoxicity. Exp Cell Res. 2018;370(1):68–77. doi:10.1016/j.yexcr.2018.06.005
33. Reyes-Reyes EM, Teng Y, Bates PJ. A new paradigm for aptamer therapeutic AS1411 action: uptake by macropinocytosis and its stimulation by a nucleolin-dependent mechanism. Cancer Res. 2010;70(21):8617–8629. doi:10.1158/0008-5472.CAN-10-0920
34. Shin S, Sung B-J, Cho Y-S, et al. An anti-apoptotic protein human survivin is a direct inhibitor of caspase-3 and-7. Biochemistry. 2001;40(4):1117–1123. doi:10.1021/bi001603q
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.