Back to Journals » International Journal of Nanomedicine » Volume 12
Self-assembled peptide nanomaterials for biomedical applications: promises and pitfalls
Authors Sun L, Zheng C, Webster TJ
Received 18 July 2016
Accepted for publication 3 August 2016
Published 20 December 2016 Volume 2017:12 Pages 73—86
DOI https://doi.org/10.2147/IJN.S117501
Checked for plagiarism Yes
Editor who approved publication: Professor Lei Yang
Linlin Sun,1,2 Chunli Zheng,3 Thomas J Webster1,2,4
1Wenzhou Institute of Biomaterials and Engineering, Wenzhou Medical University, Wenzhou, Zhejiang, People’s Republic of China; 2Department of Chemical Engineering, Northeastern University, Boston, MA, USA; 3Pharmaceutical Research Institute, China Pharmaceutical University, Nanjing, People’s Republic of China; 4Center of Excellence for Advanced Materials Research, King Abdulaziz University, Jeddah, Saudi Arabia
Abstract: Over the last several decades, a great number of advances have been made in the area of self-assembled supramolecules for regenerative medicine. Such advances have involved the design, preparation, and characterization of brand new self-assembled peptide nanomaterials for a variety of applications. Among all biomolecules considered for self-assembly applications, peptides have attracted a great deal of attention as building blocks for bottom-up fabrication, due to their versatility, ease of manufacturing, low costs, tunable structures, and versatile properties. Herein, some of the more exciting new designs of self-assembled peptides and their associated unique features are reviewed and several promising applications of how self-assembled peptides are advancing drug delivery, tissue engineering, antibacterial therapy, and biosensor device applications are highlighted.
Keywords: self-assembly, peptides, biomedical applications, drug delivery, antibacterial therapy, biosensor devices
Introduction
Grounded in nature, self-assembled molecules, involving lipids, peptides, proteins, sugars, and nucleic acids, are the fundamental building blocks of life as they comprise cell membranes, cell cytoskeletal structures, and extracellular matrices.1–3 In 1959, Feynman,4 who described a process to form well-organized supramolecules by combining individual units through a bottom-up approach, first proposed the concept of self-assembled molecules. In the 1960s, Lehn5,6 was the first one to introduce the term and concepts of supramolecular chemistry, and he had been awarded a Nobel Prize for his contribution to this area in 1987. Since then, efforts spanning a multitude of disciplines, involving mostly chemistry, physics, materials, mathematics, engineering, and biology, have contributed to this rapidly growing field to understand building block monomer structures, associated molecular assembly mechanisms, programmed architectures, controllable properties, and advanced functions.1,7
Just like higher hierarchical biomolecules in nature (such as proteins, DNA, and polysaccharides), most self-assembled molecules are formed by the interaction of individual monomers through weak, noncovalent interactions, including electrostatic interactions, hydrophobic interactions, hydrogen bonds, van der Waals interactions, and π–π stacking forces.3,8 Even though individual noncovalent forces are very weak (eg, hydrophobic forces <10 kcal/mol, electrostatic forces =1–20 kcal/mol, hydrogen bonds =2–30 kcal/mol, and π–π aromatic stacking =0–10 kcal/mol), the combination of several noncovalent forces together can generate very stable and well-organized structures.8 There are several advantages of developing self-assembled materials through noncovalent bonds compared to covalent forces. For example, due to strong covalent bonds prevalent in materials such as polymers, covalent bonds (eg, C–C bond =86 kcal/mol, C–H bond =103 kcal/mol, C–O bond =81 kcal/mol) are not reversible without adding catalysts or conducting extensive material processing. Compared with covalent bonding, these noncovalent forces present in numerous self-assembled materials are reversible and dynamic, providing for advanced functions, such as high synthetic convergence, error correction, programmed design, and self-organization.9
To maximize the formation of as many noncovalent forces as possible, the major component of self-assembled molecules has an amphiphilic structure with both hydrophobic and hydrophilic portions.10 When responding to an aqueous environment, such molecules display hydrophobic or hydrophilic domains on the surface to match that of the solvent properties. Among all the self-assembling molecules designed today, peptides are particularly attractive building blocks in this respect since peptide sequences can be formulated to have both hydrophilic and hydrophobic domains in the same building block. Owing to such unique properties in self-assembled peptides, they have been used for numerous applications, including storing bioinformation via their various sequences, peptide folding, fast and easy synthesis, a variety of functionalization methods, and their programmed responses to external stimulations (such as pH value, ion concentration, and hydrophobicity).8,11
In terms of the higher hierarchical structure of self-assembled peptides, their stability depends on the monomer structure in terms of the length of the amino acid chain and the shape of the functional group.12 At the same time, the external environment, including temperature, pH, ionic strength, and mechanical force, can affect or reverse the self-assembly process.13 Therefore, based on these properties, scientists can design self-assembled molecules with an “intelligence” to be triggered by the changing environment, such as specific targeting, controlled release, and improved efficacy.9
To synthesize peptides and their hybrid structures, there are three main methods: solid-phase chemical synthesis, protein engineering, and ring-opening polymerization strategy.14 Generally, solid-phase peptide synthesis is used for short-to-medium-length peptide sequences of highly precise structures. Although the yield of solid-phase peptide synthesis is >98% per step, the synthetic limitation of this method is in the range of 70 amino acids. To synthesize a long peptide sequence (>50 amino acids) with a defined structure such as silk and collagen molecules, researchers can utilize protein engineering to control genetic expression in bacteria. For the large-scale production of polypeptides, it is recommended to use ring-opening polymerization, in which cyclic monomers are added to the chain end to form a long peptide. However, this method produces a peptide primary structure with a lower accuracy than that obtained during solid-phase synthesis.14
In this review, we would like to introduce self-assembled nanostructures (nanospheres, nanotubes, nanofibers, and other ordered nanostructures) with linear peptide monomers (amphiphilic peptides, ionic-complementary peptides, etc) and nonlinear peptide monomers (cyclic peptides and hybrid peptides) as building blocks and discuss some relevant recent applications of such molecules involved during drug delivery, tissue engineering, antimicrobial control, and electronic devices (Figure 1).
Self-assembled peptide types and structures
Amphiphilic peptides
Amphiphilic peptides involve pure linear peptides, ionic-complementary peptides, long-chain alkylated peptides, peptide phospholipids, and peptide-based block copolymers.10
Inspired by lipid molecules in cell membranes, pure peptides with hydrophobic tails and hydrophilic heads can self-assemble into nanotubes, nanovesicles, and micelles, depending both on their chemical properties (eg, peptide sequences, charges, concentration, etc) and physical properties (eg, size, shape, etc).15–17 For the hydrophobic tail (which normally contains six residues), amino acids such as G, A, V, L, I, and F are good candidates; meanwhile, D, K, E, R, and H are used in the hydrophilic domain.10,18,19 For example, similar to surfactants, lipid-like peptides, such as A6D, V6D, G4DD, G6DD, G8DD, A6K, and KA6 sequences, spontaneously form self-organized nanostructures (such as micelles, vesicles, and fibers) once reaching the critical aggregation concentration (CAC) (eg, a CAC of A6D ≈1.6 mM and a CAC of A6K ≈1.5 mM).9,20,21 Moreover, due to their similarity to phospholipids, these peptides could stabilize membrane proteins, which denature and aggregate easily.
For example, Yeh et al22 and Zhao et al23 utilized lipid-like peptides (eg, A6D and I6K2) to stabilize the functional forms of membrane proteins Escherichia coli glycerol-3-phosphate dehydrogenase and G protein-coupled receptor bovine rhodopsin, respectively. Compared to the common surfactants (eg, n-dodecyl-β-D-maltoside and octyl-D-glucoside) used for protein stabilization, these peptides have a higher efficiency to stabilize membrane proteins.23
Ionic-complementary self-assembling peptides
Inspired from the Z-DNA binding protein zuotin, in 1993, Zhang et al24 discovered the first self-assembling peptide EAK16 (n-AEAEAKAKAEAEAKAK-c) that formed into nanofibers. Since then, the rapid development of this type of self-assembling peptide has fostered numerous applications, including three-dimensional (3D) cell culture, tissue engineering, regenerative medicine, and sensory devices.25–27 This peptide has one side group with charged side chains and another side group with hydrophobic chains. In water, the charged side is exposed on the outside and the hydrophobic side forms a double sheet inside the nanofiber.24,28 Meanwhile, the peptide sequences with periodically repeated positive and negative charges form the stable structure by ionic-complementary forces in a checkerboard-like pattern and then assemble typical beta-sheet structures and eventually form a hydrogel network of nanofibers.13,29
Owing to the combination of ionic force and hydrogen bonds inside the beta-sheet structures, these nanofibers are stable under a wide range of temperatures, pH values, and high concentrations of denaturing chemicals.26 Since the hydrophobic interactions are not specific, these nanofibers may diffuse to minimize equilibrium energy.13,30
There are different types of ionic-complementary self-assembled peptides (Figure 2): 1) −+−+−+−+ (eg, peptide RADA16-I: Ac-RADARADARADARADA); 2) −−++−−++ (eg, peptide RADA16-II: Ac-RARADADARARADADA); 3) −−−+++; and 4) −−−−++++.27,31,32 Among these peptides, the RADA16-I peptide can promote cell growth and tissue regeneration; therefore, it has been commercialized as a product termed PuraMatrix.26,32
  | Figure 2 Different types of ionic-complementary peptides. |
Once mixed with physiological fluids, amphiphilic peptides form a transparent hydrogel network within seconds. The hydrogel of these ionic-complementary self-assembled peptides is composed of 0.5%–1% peptides with 99.0%–99.5% water, and therefore, there are a large number of spaces between the nanofibers, which are enough for cells to grow and differentiate.31,33 When used in 3D cell culture systems, this type of the self-assembled peptide scaffold provides mechanical strength to encapsulate cells in a desired location; has excellent nutrient, growth factor, and oxygen diffusion for cell growth; and can release therapeutic agents in a controlled or programmed fashion.34,35 The ionic-complementary self-assembled peptides have been used to enhance various types of cell growth and differentiation, including bone, cartilage, vessel, heart, and neural systems.34,36 For example, the self-assembled peptide scaffolds made of RADA16-I and RADA16-II peptides have been used to enhance neural cell attachment, differentiation, and neurite outgrowth during in vitro studies, as well as to promote active synapse formation during in vivo studies.33 For example, Gelain et al studied the 3D cultures of mouse neural stem cells using the self-assembling peptide RADA16 with the functionalized sequences derived from neural cell adhesion motifs, bone marrow homing proteins, and collagen molecules. After 7 days of culturing, the bone marrow homing peptides (SKPPGTSS and PFSSTKT)-conjugated RADA16 peptide scaffolds promoted the most neural stem cell growth and differentiation.33
Long-chain alkylated peptides
Long-chain alkylated peptides are composed of hydrophilic peptide sequences and hydrophobic alkyl chains. The alkyl chain can be conjugated on both the N- and C-termini of the peptide sequences. A representative amphiphilic peptide molecule has four domains: hydrophobic tail, beta-sheet forming segment, charged groups, and bioactive epitopes (Figure 3A).37 In the aqueous solution, individual alkylated peptides could aggregate together to form cylinder nanofibers (Figure 3B and C).11 Since the most of bioactive epitopes are hydrophilic, these functionalized peptides can be exposed on the surface of nanofibers to interact with cellular receptors, which consequently influence cell signaling pathways, gene expression, and cell functions.11,37,38
  | Figure 3 (A) Molecular structure of an alkylated peptide with four regions: hydrophobic tail, beta-sheet forming segment, charged groups, and bioactive epitope. Adapted from Cui H, Webber MJ, Stupp SI. Self-assembly of peptide amphiphiles: from molecules to nanostructures to biomaterials. Biopolymers. 2010;94(1):1–18. Copyright © 2010 Wiley Periodicals, Inc.37 (B) Schematic illustration of an alkylated peptide (IKVAV) molecule and the nanofiber formed by IKVAV peptides. (C) TEM image of IKVAV nanofibers. From Silva GA, Czeisler C, Niece KL, et al. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science. 2004;303(5662):1352–1355. Reprinted with permission from AAAS.11 |
Yu et al39 and Forns et al40 demonstrated that the hydrocarbon chains can be used to stabilize peptide sequences and studied the folded structures. Self-assembled alkylated peptides have been utilized to functionalize carbon tubes via noncovalent bonds to enhance their solubility in water.41
To mimic cell-membrane proteins, peptides have been modified with phospholipids to form peptide–phospholipid conjugates. For example, Musiol et al42 have synthesized lipopeptides with the sequence derived from human prion proteins. HeLa cell experiments using confocal fluorescence microscopy proved that the self-assembling lipid peptide micelles were transported into cells very quickly.
Cyclic peptide
As early as 1974, De Santis et al43 determined theoretically that cyclic peptides with alternating D- and L-amino acids could self-assemble into nanotubes. However, it was not until 1993 that Ghadiri et al44 synthesized the first type of self-assembling cyclic peptide, cyclo-(L-Gln-D-Ala-L-Glu-D-Ala)2, based on De Santis’ theory.
The self-assembling cyclic peptides are composed of stacking cyclic peptide monomers with a flat conformation structure, in which the carbonyl and amino groups are pointed perpendicular to the ring and the side chains present a pseudo-equatorial outward-pointing orientation.45,46 Then the nanotubes were stabilized by hydrogen bonds between amide groups in neighboring peptide cycles (Figure 4). Typical cyclic peptide sequences include alternating D,L-α-amino acids, β-amino acids; alternating α,β-amino acids; and alternating α,γ-amino acids, δ-amino acids, and oligoureas.46–49
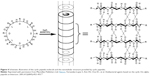  | Figure 4 Schematic illustration of the cyclic peptide molecule and the nanotubular structure provided by cyclic peptides. |
Compared to the other self-assembled peptide structures, cyclic peptide nanotubes have unique properties: 1) their precise diameter depends on the number of amino acids, chemical structure, and the side-chain size and 2) the functions of the nanotubes can be tailored by varying the side chains.46 For example, when the number of amino acids in the cyclic peptide ring increases from 4 to 12, the internal diameter increases from 2 Å to 13 Å.46
Applications in medicine
Drug delivery
The conventional administration of hydrophobic drugs suffers several disadvantages, including low water solubility, poor oral availability, quick biodegradation, nonspecific delivery, and serious side effects.50,51 Therefore, drug delivery systems could encapsulate these therapeutic agents to increase drug efficacy.52 Similarly, delivery vehicles can also improve the efficacy of certain hydrophilic drugs, which have unspecific targeting or short half-lives.53 In order to protect highly sensitive biomolecules (such as peptides, proteins, DNAs, and RNAs) from biodegradation and concentrate and retain them in the target organ, it is necessary to use a delivery vehicle, for which self-assembled peptides are a good option.54,55
For drug delivery applications, the basic requirements for self-assembled nanomaterials are biocompatibility, noncytotoxicity, and biodegradablility.56,57 For drug loading and release, the nanocarriers should encapsulate molecules at a high concentration, protect them from dilution and degradation, and release them in a controlled and prolonged manner (Figure 5).58–61 For advanced functions, the self-assembled systems should have domains specific for targeting or sensory capability to be utilized for the intended application.56,62
  | Figure 5 Schematic illustration of cell-membrane interactions and drug release of the drug-loaded self-assembling peptides. |
As an example of the aforementioned application, Webber et al63 conjugated the anti-inflammatory drug dexamethasone with the peptide amphiphile (C16-V2A2E2) via a hydrazine linkage to achieve long-term drug release in both in vitro and in vivo tests. In vitro studies showed that dexamethasone could be released over several weeks at physiological pH and temperature. Furthermore, the peptide nanofibers were mixed with polystyrene microparticles and injected into mice, and finally, a luminescence assay and histological staining were used to evaluate the inflammation after 3 days or 9 days. The results demonstrated that the localized drug-loaded nanofiber gel could promote immune suppression.63
Zhang et al64 utilized the self-assembling Tat peptide to encapsulate the hydrophobic drug paclitaxel to facilitate drug delivery and efficacy. First, four sequences with different numbers of hydrophobic tails were tested, and only the four-tailed sequence qC8-Tat self-assembled into nanotubes under aqueous solutions at a pH of 7.4 (Figure 6). After the coumarin-6-loaded nanofibers were delivered into KB-3-1 cervical cancer cells, confocal microscopy indicated that hydrophobic drugs were transported into cancer cells at high efficacy through the adsorptive-mediated pathway.64
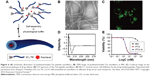  | Figure 6 (A) Schematic illustration of paclitaxel-loaded Tat peptide nanofibers. (B) TEM image of paclitaxel-loaded Tat nanofibers in PBS. (C) Confocal image of the endocytosis pathway of drug release. (D) CD spectrum of the Tat peptide nanofibers. (E) KB-3-1 cervical cancer cell inhibition by the drug-loaded peptides. Reprinted with permission from Zhang P, Cheetham AG, Lin YA, Cui H. Self-assembled tat nanofibers as effective drug carrier and transporter. Acs Nano. 2013;7(7):5965–5977. Copyright 2013 American Chemical Society.64 |
For controlled drug releases, peptide hydrogels could encapsulate small molecules or proteins such as enzymes and antibodies.65,66 The release profiles are determined not only by the molecule size, charge, and hydrophobicity but also by the characteristics of the peptide hydrogel, including peptide sequences, concentration, and the hydrogel pore size.59
Tissue engineering
For tissue regeneration, self-assembled peptides with nanotubular or nanofibered structures could mimic the natural extracellular matrix of many tissues and localize drug release at the desired site.11,58,67 Similar to the spherical shape of nanoparticles, nanotubes or fibers could not only display a high density of signals on the surface but also can carry multifunctional molecules to target specific cellular receptors, such as G protein-coupled receptors and kinase-linked receptors.58,68,69 Different from micelles, nanofibers have a precise geometry that retains signals over long distances on the cell surface to benefit many applications in medicine that require directional growth, such as spinal cord regeneration, vessel growth, and cartilage and bone repair.11,29,35,70,71 Moreover, several types of peptides could form a rigid network or filaments with good mechanical properties to serve as substrates or 3D scaffolds to support the growth, proliferation, differentiation, and function of cells.34,72–74 To introduce some specific bioactive functions, it is generally a good idea to incorporate the functional peptide motifs on the C-termini of the peptide sequence with a GG spacer between the motifs and ionic-complementary sequence.27 The functional motifs can then provide cells with more external stimuli than the 2D coating in terms of bioactive molecule concentration.27,75
Conventionally, a variety of polymers, such as poly(L-lactic acid) and poly(glycolic acid), and biopolymers, such as collagen and alginate, have been used as artificial scaffolds for various tissue engineering applications. Although these polymers are biodegradable and easy to fabricate, most of them lack bioactivity to direct cell growth and functions.76 On the other hand, with their ability to self-assemble in the physiological environment, the peptide solution could be injected into the injured location and forms a scaffold to promote tissue regeneration.34,72,77 With peptides derived from growth factor, nanofibers can expose these peptides on their surfaces to bind and activate cellular receptor for cell signaling and/or protein synthesis.78,79 Furthermore, the scaffolds composed of peptides can encapsulate stem cells and induce their differentiation into normal cells through bioactive peptides at a high density on the scaffold surface.76,78,80
In a previous study, to improve cartilage regeneration, the hydrogels formed by the peptide KLD12 (KLDLKLDLKLDL) were cocultured with primary bovine chondrocytes, and the results showed that peptide hydrogel retained the chondrocyte phenotype to produce a cartilage-like extracellular matrix and type II collagen.35 After 4 weeks of culturing, the material stiffness of the extracellular matrix increased, and therefore, it was suggested that the KLD12 self-assembling peptide scaffold was promising for cartilage tissue repair.35
Moreover, Stroumpoulis et al81 studied cell responses on vesicles formed by (C16)2-Glu-C2-GRGDSP and (C16)2-Glu-PEO-GRGDSP sequences. The sequence with PEO promoted the highest fibroblast cell adhesion and growth, which could be used to develop a membrane system to screen biological probes for cell adhesion and growth. Later on, Lin et al synthesized pH-sensitive ampiphilic peptides C16GSH and C16EOSH composed of histidine and serine with a single fatty acid tail and branched structure.77 Using histidine as the switch, these peptides were viscoelastic liquids at a pH value <5.5. In a neutral solution, these peptides formed micelles. While in pH >6.5, the amphiphilic peptides assembled into nanofibers to form a hydrogel. Once cultured with cells, the hydrogel kept fibroblasts proliferating for 96 hours. This type of nanofibers is promising to develop a pH-sensitive cell scaffold to transfer the peptide solution into a hydrogel under physiological environment.77
In addition, mimicking the ligand–receptor interactions of extracellular matrix components, Lee et al82 synthesized amphiphilic peptides with sequences specifically binding to heparin sulfate. Then the hybrid scaffold localized with the bone morphogenetic protein-2 to modulate the ligand–receptor signaling to promote bone regeneration.
In our group, anticancer drugs (eg, curcumin) were encapsulated into the self-assembled peptide C18GR7RGDS to inhibit selectively bone cancer cells (MG-63 osteosarcoma).83 Fourier transform infrared and X-ray diffraction analyses proved that curcumin was encapsulated in the hydrophobic core of peptide nanoparticles. The cell studies showed that self-assembled peptides significantly improved the delivery efficiency. Moreover, compared to healthy bone cells, curcumin-loaded peptide nanoparticles demonstrated selective cytoxicity against bone cancer cells.83
Zhang et al84 designed temperature-sensitive nanofibers (V3A3E3) to form aligned cellular wires. Figure 7 demonstrates that the amphiphilic peptides could form a string-like hydrogel in a phosphate-buffered saline solution or can be mixed with a CaCl2 solution after a temperature treatment (heated to 80°C for 30 minutes and then cooled down to 20°C). Accordingly, mechanical tests and scanning electron microscopy proved that the nanofibers organized along the length of the string. When incubated in the hydrogel, human mesenchymal stem cells grew both cell bodies and filopodia along the direction of the string formed by the peptides.84
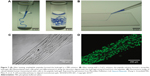  | Figure 7 (A) After heating, amphiphilic peptides formed the hydrogel in a PBS solution. (B) After mixing with a CaCl2 solution, the peptide solution formed a string-like hydrogel. (C) Mesenchymal stem cells grew and differentiated at along the direction aligned with the hydrogel string formed from the peptides. (D) Fluorescence image of calcein-labeled human mesenchymal stem cells in the peptide hydrogel. Reprinted by permission from Macmillan Publishers Ltd: Nature Materials. Zhang S, Greenfield MA, Mata A, et al. A self-assembly pathway to aligned monodomain gels. 2010;9(7):594–601., Copyright 2010.84 |
Antibacterial agents
Over the past 3 decades, the antibacterial effects of various bioactive peptides have attracted extensive attention.85,86 As a part of our innate defense systems, antimicrobial peptides were widely found in different types of organisms. With lengths of 10–40 amino acids, most antimicrobial peptides are capable of interacting with negatively charged bacterial membranes to show a broad spectrum of ability. The mechanism of antimicrobial peptides is not only determined by the size, hydrophobicity, amphipathicity, net charge, and secondary structure of the peptide sequence but also affected by their interaction with bacterial membranes.87–90
For example, Veiga et al synthesized an antibacterial hydrogel using arginine-rich self-assembled peptides with different arginine contents (two, four, six, or eight Arg residues in the sequence). They investigated the viability of E. coli and Staphylococcus aureus (105 CFU/dm2) on the peptide hydrogel surface with varying weight percentages (0.5, 1, 1.5, and 2) after 24 hours of culturing. Both increases in the number of arginine groups and the weight percent could enhance the antibacterial capability of the self-assembled hydrogels. For gels containing four, six, or eight arginine groups, both E. coli and S. aureus were completely inhibited after 24 hours, irrespective of the weight percentage of the peptides. However, the hydrogels with two arginine groups and containing 2 wt% peptides inhibited only 50% of E. coli growth. The antibacterial results indicated that the hydrogel formed by the self-assembled PEP6R peptides (VKVRVRVRVDPPTRVRVRVKV) had the greatest antibacterial ability with a low cytotoxicity toward human erythrocytes and mesenchymal stem cells.86
Chu-Kung et al91,92 studied the effect of the length of fatty acids of peptides on antimicrobial properties. In their study, the peptides YGAAKKAAKAAKKAAKAA (AKK) and LKKLLKLLKLLKL (LKK) were conjugated with fatty acids, and the results indicated that the increased length of the fatty tails conjugated to AKK peptides enhanced interactions between the peptides and membranes, which consequently increased the antibacterial activity (for E. coli and S. epidermidis).91 However, once conjugated with a longer fatty acid tail and peptides start to assemble into nanofibers, the antibacterial activity was reduced.
As another example on the use of self-assembled peptides for antibacterial applications, bone fracture healing, and antimicrobial applications, KLD12 peptides (KLDLKLDLKLDL) with variable numbers of arginine residues at the N-terminus have been synthesized to introduce the antimicrobial properties while maintaining the β-sheet and self-assembled structures.93 The results of propidium iodide staining and scanning electron microscopy demonstrated that the increased arginine caused leaking of the E. coli membrane and death.
Chen et al94 also assembled antimicrobial peptides (surfactin) on photoluminescent gold nanodots (SFT/DT-Au) to inhibit both normal and multidrug-resistant bacteria. In vitro studies indicated that the synergistic effect of peptides and gold dots enhanced anti-infection properties dramatically by the disruption of the membrane. Animal studies also demonstrated that SFT/DT-Au not only inhibited methicillin-resistant S. aureus infection but also promoted collagen production in wounded skin of rats infected by methicillin-resistant S. aureus.94
In previous studies, cyclic peptide nanotubes were mainly used for antibacterial applications.45,95,96 With stacking peptide cycles, nanotubes are able to form transmembrane pores to mimic ion channels in membranes.45,97,98 Mechanistic studies showed that the antibacterial activity is due to the enhancement of the membrane permeability.96,97 Moreover, these cyclic peptides could also inhibit virus infections by blocking virus entry or endosome escape in mammalian cells. To inhibit methicillin-resistant bacteria, it is significant to choose a specific peptide sequence against bacterial selectively rather than mammalian cells.99
Khalfa and Tarek98 simulated the interaction of self-assembling cyclic peptides with cytoplasmic membrane models using coarse-grained molecular dynamic simulations to study the mechanism of the antibacterial process. First, the cyclic peptide ([RRKWLWLW]) self-assembled on the top of the membrane formed by palmitoyl-oleoyl-phosphatidylethanolamine (POPE) and palmitoyl-oleoyl-phosphatidylglycerol (POPG) lipid bilayers. With the increased concentration of peptides on the surface, the lipids released from the POPE–POPG bilayers. Moreover, the mechanical properties of the membrane were affected by cyclic peptides. Figure 8 indicates that in the beginning of the interaction, cyclic peptides with a low concentration started to form short nanotubes and did not penetrate the membrane. At the 1/10 cyclic peptide/lipid ratio, lipid protrusion appeared. Once the cyclic peptide/lipid ratio increased to 1/5, the peptides aggregated with negative lipid molecules and caused large perturbations of the membrane.98
  | Figure 8 CG-MD images of the interaction of self-assembling cyclic peptides ([RRKWLWLW]) with POPE–POPG lipid membranes. |
Nanosensors
Once targeting biomolecules or metallic nanoparticles into self-assembled peptides were incorporated, peptide nanotubes could act as biosensors with enhanced sensitivity and selectivity for mineralization, imaging probes, and electronic devices.100–103
Inspired from peptide sequences with a biomineralized capability in nature, peptide nanotubes can be conjugated with these sequences (such as peptide amphiphiles containing the RGD sequence) to nucleate metal/semiconductor materials on the surface. Several groups have utilized peptide nanomaterials to control mineralization for bone tissue regeneration.104,105 This method could facilitate bone cell growth along the peptide fibers with a desired direction in the scale of nanometers.
For diagnostic imaging, peptide nanomaterials have been also conjugated with radiometals for in vivo tracking using the near-infrared fluorescence106 or MRI.107,108 Bull et al synthesized two Gd(III) conjugated amphiphilic peptides (DOTA-KK(K)K-LL-CCC-K-C16 and DOTA-KGRGDS(K)K-LLL-AAA-K-C16) to form spherical and tubular structures. Since the self-assembling structure could increase the molecular weight of Gd, the rotational correlation time and consequently the relaxivity were enhanced. MRI tests demonstrated that these Gd functionalized peptides could increase T1 relaxation time for medical applications.108
With advantages of one-dimensional direction and a controllable size, peptide nanotubes are suitable to work as building blocks for nanoelectronic devices.109 After nucleation, the electronic properties of the peptide nanotubes can be tuned by the size, shape, arrangement, and density of a mineralized coating. Additionally, the conditions (eg, pH, ion concentration, etc) of the extra environment also affected the mineralization process and the final conductivity.109,110 Previous studies utilized such nanotubes to modify membranes to detect the conductivity change from the antigen–antibody interactions.111
For example, the self-assembled histidine-rich HGGGHGHGGGHG (HG12) peptides with linear or branched alkyl chains were labeled by pyrene to detect metal ions (Figure 9).112 Since the HG12 sequence bonded Cu2+ specifically, the fluorescence could be blocked. On the other hand, the Ag+ could “light up” the fluorescence response of the nanofibers with branched alkyl chains (Figure 9). Moreover, the nanofibers could work as templates for Ag nanoparticles for cell imaging and antibacterial effects.112 Table 1 summarizes the highlighted medical applications of self-assembled peptides in this review.
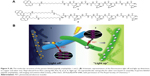  | Figure 9 (A) The molecular structure of the pyrene-labeled peptide amphiphiles 1 and 2. (B) Schematic representation of the fluorescence light off and light up detections to copper and silver ions. Reproduced from Kim I, Jeong HH, Kim YJ, et al. A “light-up” 1D supramolecular nanoprobe for silver ions based on assembly of pyrene-labeled peptide amphiphiles: cell-imaging and antimicrobial activity. J Mat Chem. 2014;2(38):6478–6486, with permission of The Royal Society of Chemistry.112 |
Future directions and conclusion
Based on noncovalent forces, self-assembled molecules could construct ordered structures of defined shapes, various properties, and multiple functions. Examples from recent studies have shown that self-assembled peptides with different structures have been used for a wide range of medical applications. However, in this field, scientists and researchers are still facing challenges to predict higher hierarchical structures, properties, and functions from the structure of the individual building molecule. With multidisciplinary efforts, self-assembled peptides will help us not only to study complex biological phenomena and create various applications but also to conquer the diseases and improve human health.
Acknowledgments
The authors would like to thank Wenzhou Institute of Biomaterials and Engineering and Northeastern University for funding.
Disclosure
The authors report no conflicts of interest in this work.
References
Whitesides GM, Boncheva M. Beyond molecules: self-assembly of mesoscopic and macroscopic components. Proc Natl Acad Sci U S A. 2002;99(8):4769–4774. | ||
Whitesides GM, Grzybowski B. Self-assembly at all scales. Science. 2002;295(5564):2418–2421. | ||
Tu RS, Tirrell M. Bottom-up design of biomimetic assemblies. Adv Drug Deliv Rev. 2004;56(11):1537–1563. | ||
Feynman RP. There’s plenty of room at the bottom. Caltech Eng Sci. 1959;23(5):22–36. | ||
Lehn JM. Supramolecular chemistry – scope and perspectives molecules, supermolecules, and molecular devices. Angew Chem Int Ed Engl. 1988;27(1):89–112. | ||
Lehn JM. Perspectives in chemistry-aspects of adaptive chemistry and materials. Angew Chem Int Ed Engl. 2015;54(11):3276–3289. | ||
Hagstrom JE. Self-assembling complexes for in vivo gene delivery. Curr Opin Mol Ther. 2000;2(2):143–149. | ||
Shi JF, Xu B. Nanoscale assemblies of small molecules control the fate of cells. Nano Today. 2015;10(5):615–630. | ||
Hosseinkhani H, Hong PD, Yu DS. Self-assembled proteins and peptides for regenerative medicine. Chem Rev. 2013;113(7):4837–4861. | ||
Lowik D, van Hest JCM. Peptide based amphiphiles. Chem Soc Rev. 2004;33(4):234–245. | ||
Silva GA, Czeisler C, Niece KL, et al. Selective differentiation of neural progenitor cells by high-epitope density nanofibers. Science. 2004;303(5662):1352–1355. | ||
Schneider JP, Pochan DJ, Ozbas B, Rajagopal K, Pakstis L, Kretsinger J. Responsive hydrogels from the intramolecular folding and self-assembly of a designed peptide. J Am Chem Soc. 2002;124(50):15030–15037. | ||
Kim S, Kim JH, Lee JS, Park CB. Beta-sheet-forming, self-assembled peptide nanomaterials towards optical, energy, and healthcare applications. Small. 2015;11(30):3623–3640. | ||
Klok HA. Protein-inspired materials: synthetic concepts and potential applications. Angew Chem Int Ed. 2002;41(9):1509–1513. | ||
Antonietti M, Forster S. Vesicles and liposomes: a self-assembly principle beyond lipids. Adv Mater Deerfield. 2003;15(16):1323–1333. | ||
Huang RL, Qi W, Su RX, Zhao J, He ZM. Solvent and surface controlled self-assembly of diphenylalanine peptide: from microtubes to nanofibers. Soft Matter. 2011;7(14):6418–6421. | ||
Liu L, Busuttil K, Zhang S, et al. The role of self-assembling polypeptides in building nanomaterials. Phys Chem Chem Phys. 2011;13(39):17435–17444. | ||
Caplan MR, Schwartzfarb EM, Zhang SG, Kamm RD, Lauffenburger DA. Control of self-assembling oligopeptide matrix formation through systematic variation of amino acid sequence. Biomaterials. 2002;23(1):219–227. | ||
Marini DM, Hwang W, Lauffenburger DA, Zhang SG, Kamm RD. Left-handed helical ribbon intermediates in the self-assembly of a beta-sheet peptide. Nano Lett. 2002;2(4):295–299. | ||
Vauthey S, Santoso S, Gong HY, Watson N, Zhang SG. Molecular self-assembly of surfactant-like peptides to form nanotubes and nanovesicles. Proc Natl Acad Sci U S A. 2002;99(8):5355–5360. | ||
von Maltzahn G, Vauthey S, Santoso S, Zhang SU. Positively charged surfactant-like peptides self-assemble into nanostructures. Langmuir. 2003;19(10):4332–4337. | ||
Yeh JI, Du SC, Tortajada A, Paulo J, Zhang SG. Peptergents: peptide detergents that improve stability and functionality of a membrane protein, glycerol-3-phosphate dehydrogenase. Biochemistry. 2005;44(51):16912–16919. | ||
Zhao X, Nagai Y, Reeves PJ, Kiley P, Khorana HG, Zhang S. Designer short peptide surfactants stabilize G protein-coupled receptor bovine rhodopsin. Proc Natl Acad Sci U S A. 2006;103(47):17707–17712. | ||
Zhang SG, Holmes T, Lockshin C, Rich A. Spontaneous assembly of a self-complementary oligopeptide to form a stable macroscopic membrane. Proc Natl Acad Sci U S A. 1993;90(8):3334–3338. | ||
Gelain F, Horii A, Zhang S. Designer self-assembling peptide scaffolds for 3-D tissue cell cultures and regenerative medicine. Macromol Biosci. 2007;7(5):544–551. | ||
Zhao XB, Pan F, Xu H, et al. Molecular self-assembly and applications of designer peptide amphiphiles. Chem Soc Rev. 2010;39(9):3480–3498. | ||
Yang Y, Khoe U, Wang X, Horii A, Yokoi H, Zhang S. Designer self-assembling peptide nanomaterials. Nano Today. 2009;4(2):193–210. | ||
Zhang S, Lockshin C, Cook R, Rich A. Unusually stable beta-sheet formation in an ionic self-complementary oligopeptide. Biopolymers. 1994;34(5):663–672. | ||
Zhang S, Holmes TC, DiPersio CM, Hynes RO, Su X, Rich A. Self-complementary oligopeptide matrices support mammalian cell attachment. Biomaterials. 1995;16(18):1385–1393. | ||
Altman M, Lee P, Rich A, Zhang SG. Conformational behavior of ionic self-complementary peptides. Protein Science. 2000;9(6):1095–1105. | ||
Zhang SG. Emerging biological materials through molecular self-assembly. Biotechnol Adv. 2002;20(5–6):321–339. | ||
Yokoi H, Kinoshita T, Zhang SG. Dynamic reassembly of peptide RADA16 nanofiber scaffold. Proc Natl Acad Sci U S A. 2005;102(24):8414–8419. | ||
Gelain F, Bottai D, Vescovi A, Zhang S. Designer self-assembling peptide nanofiber scaffolds for adult mouse neural stem cell 3-dimensional cultures. Plos One. 2006;1(2):e119. | ||
Davis ME, Motion JPM, Narmoneva DA, et al. Injectable self-assembling peptide nanofibers create intramyocardial microenvironments for endothelial cells. Circulation. 2005;111(4):442–450. | ||
Kisiday J, Jin M, Kurz B, et al. Self-assembling peptide hydrogel fosters chondrocyte extracellular matrix production and cell division: Implications for cartilage tissue repair. Proc Natl Acad Sci U S A. 2002;99(15):9996–10001. | ||
Wen Y, Roudebush SL, Buckholtz GA, et al. Coassembly of amphiphilic peptide EAK16-II with histidinylated analogues and implications for functionalization of beta-sheet fibrils in vivo. Biomaterials. 2014;35(19):5196–5205. | ||
Cui H, Webber MJ, Stupp SI. Self-assembly of peptide amphiphiles: from molecules to nanostructures to biomaterials. Biopolymers. 2010;94(1):1–18. | ||
Newcomb CJ, Sur S, Ortony JH, et al. Cell death versus cell survival instructed by supramolecular cohesion of nanostructures. Nat Commun. 2014;5:3321. | ||
Yu YC, Tirrell M, Fields GB. Minimal lipidation stabilizes protein-like molecular architecture. J Am Chem Soc. 1998;120(39):9979–9987. | ||
Forns P, Lauer-Fields JL, Gao S, Fields GB. Induction of protein-like molecular architecture by monoalkyl hydrocarbon chains. Biopolymers. 2000;54(7):531–546. | ||
Arnold MS, Guler MO, Hersam MC, Stupp SI. Encapsulation of carbon nanotubes by self-assembling peptide amphiphiles. Langmuir. 2005;21(10):4705–4709. | ||
Musiol HJ, Dong SL, Kaiser M, et al. Toward semisynthetic lipoproteins by convergent strategies based on click and ligation chemistry. Chembiochem. 2005;6(4):625–628. | ||
De Santis P, Forni E, Rizzo R. Conformational analysis of DNA-basic polypeptide complexes: possible models of nucleoprotamines and nucleohistones. Biopolymers. 1974;13(2):313–326. | ||
Ghadiri MR, Granja JR, Milligan RA, McRee DE, Khazanovich N. Self-assembling organic nanotubes based on a cyclic peptide architecture. Nature. 1993;366(6453):324–327. | ||
Fernandez-Lopez S, Kim HS, Choi EC, et al. Antibacterial agents based on the cyclic D,L-alpha-peptide architecture. Nature. 2001;412(6845):452–455. | ||
Chapman R, Danial M, Koh ML, Jolliffe KA, Perrier S. Design and properties of functional nanotubes from the self-assembly of cyclic peptide templates. Chem Soc Rev. 2012;41(18):6023–6041. | ||
Ishihara Y, Kimura S. Nanofiber formation of amphiphilic cyclic tri-beta-peptide. J Peptide Sci. 2010;16(2):110–114. | ||
Guerra A, Brea RJ, Amorin M, Castedo L, Granja JR. Self-assembling properties of all gamma-cyclic peptides containing sugar amino acid residues. Org Biomol Chem. 2012;10(44):8762–8766. | ||
Montenegro J, Ghadiri MR, Granja JR. Ion channel models based on self-assembling cyclic peptide nanotubes. Acc Chem Res. 2013;46(12):2955–2965. | ||
Langer R. Drug delivery and targeting. Nature. 1998;392(6679):5–10. | ||
Su CW, Chiang CS, Li WM, Hu SH, Chen SY. Multifunctional nanocarriers for simultaneous encapsulation of hydrophobic and hydrophilic drugs in cancer treatment. Nanomedicine. 2014;9(10):1499–1515. | ||
Rosler A, Vandermeulen GWM, Klok HA. Advanced drug delivery devices via self-assembly of amphiphilic block copolymers. Adv Drug Deliv Rev. 2001;53(1):95–108. | ||
Kopecek J. Smart and genetically engineered biomaterials and drug delivery systems. Eur J Pharm Sci. 2003;20(1):1–16. | ||
Maurer N, Fenske DB, Cullis PR. Developments in liposomal drug delivery systems. Expert Opin Biol Ther. 2001;1(6):923–947. | ||
Huang RL, Qi W, Feng LB, Su RX, He ZM. Self-assembling peptide-polysaccharide hybrid hydrogel as a potential carrier for drug delivery. Soft Matter. 2011;7(13):6222–6230. | ||
Pawar R, Ben-Ari A, Domb AJ. Protein and peptide parental controlled delivery. Expert Opin Biol Ther. 2004;4(8):1203–1212. | ||
Collins L, Parker AL, Gehman JD, et al. Self-assembly of peptides into spherical nanoparticles for delivery of hydrophilic moieties to the cytosol. ACS Nano. 2010;4(5):2856–2864. | ||
Rajagopal K, Schneider JP. Self-assembling peptides and proteins for nanotechnological applications. Curr Opin Struct Biol. 2004;14(4):480–486. | ||
Xu XH, Li YK, Li HP, et al. Smart nanovehicles based on pH-triggered disassembly of supramolecular peptide-amphiphiles for efficient intracellular drug delivery. Small. 2014;10(6):1133–1140. | ||
Lin BF, Missirlis D, Krogstad DV, Tirrell M. Structural effects and lipid membrane interactions of the pH-responsive GALA peptide with fatty acid acylation. Biochemistry. 2012;51(23):4658–4668. | ||
Santana H, Avila CL, Cabrera I, et al. How does growth hormone releasing hexapeptide self-assemble in nanotubes? Soft Matter. 2014;10(46):9260–9269. | ||
Boekhoven J, Zha RH, Tantakitti F, et al. Alginate-peptide amphiphile core-shell microparticles as a targeted drug delivery system. RSC Adv. 2015;5(12):8753–8756. | ||
Webber MJ, Matson JB, Tamboli VK, Stupp SI. Controlled release of dexamethasone from peptide nanofiber gels to modulate inflammatory response. Biomaterials. 2012;33(28):6823–6832. | ||
Zhang P, Cheetham AG, Lin YA, Cui H. Self-assembled tat nanofibers as effective drug carrier and transporter. Acs Nano. 2013;7(7):5965–5977. | ||
Peppas NA, Bures P, Leobandung W, Ichikawa H. Hydrogels in pharmaceutical formulations. Eur J Pharmaceutics and Biopharm. 2000;50(1):27–46. | ||
Davis ME, Hsieh PCH, Takahashi T, et al. Local myocardial insulin-like growth factor 1 (IGF-1) delivery with biotinylated peptide nanofibers improves cell therapy for myocardial infarction. Proc Natl Acad Sci U S A. 2006;103(21):8155–8160. | ||
Wan ACA, Ying JY. Nanomaterials for in situ cell delivery and tissue regeneration. Adv Drug Deliv Rev. 2010;62(7–8):731–740. | ||
Li Y, Yu SM. Targeting and mimicking collagens via triple helical peptide assembly. Curr Opin Chem Biol. 2013;17(6):968–975. | ||
Alsbaiee A, Beingessner RL, Fenniri H. Self-assembled nanomaterials for tissue-engineering applications. In: Webster TJ, editor. Nanomedicine: Technologies and Applications. Woodhead Publishing Limited, Cambridge, UK. 2012:490–533. | ||
Nah JW, Yu L, Han SO, Ahn CH, Kim SW. Artery wall binding peptide-poly (ethylene glycol)-grafted-poly(L-lysine)-based gene delivery to artery wall cells. J Controll Release. 2002;78(1–3):273–284. | ||
Semino CE, Kasahara J, Hayashi Y, Zhang SG. Entrapment of migrating hippocampal neural cells in three-dimensional peptide nanofiber scaffold. Tissue Eng. 2004;10(3–4):643–655. | ||
Kretsinger JK, Haines LA, Ozbas B, Pochan DJ, Schneider JP. Cytocompatibility of self-assembled beta-hairpin peptide hydrogel surfaces. Biomaterials. 2005;26(25):5177–5186. | ||
Kim SJ, Kim JE, Kim SH, et al. Therapeutic effects of neuropeptide substance P coupled with self-assembled peptide nanofibers on the progression of osteoarthritis in a rat model. Biomaterials. 2016;74:119–130. | ||
Kumada Y, Hammond NA, Zhang SG. Functionalized scaffolds of shorter self-assembling peptides containing MMP-2 cleavable motif promote fibroblast proliferation and significantly accelerate 3-D cell migration independent of scaffold stiffness. Soft Matter. 2010;6(20):5073–5079. | ||
Hauser CAE, Zhang S. Designer self-assembling peptide nanofiber biological materials. Chem Soc Rev. 2010;39(8):2780–2790. | ||
Zhang ZP, Hu J, Ma PX. Nanofiber-based delivery of bioactive agents and stem cells to bone sites. Adv Drug Deliv Rev. 2012;64(12):1129–1141. | ||
Lin BF, Megley KA, Viswanathan N, et al. pH-responsive branched peptide amphiphile hydrogel designed for applications in regenerative medicine with potential as injectable tissue scaffolds. J Mater Chem. 2012;22(37):19447–19454. | ||
Kim JE, Kim SH, Jung Y. In situ chondrogenic differentiation of bone marrow stromal cells in bioactive self-assembled peptide gels. J Biosci Bioeng. 2015;120(1):91–98. | ||
Wu EC, Zhang SG, Hauser CAE. Self-assembling peptides as cell-interactive scaffolds. Adv Funct Mater. 2012;22(3):456–468. | ||
Cheng TY, Chen MH, Chang WH, Huang MY, Wang TW. Neural stem cells encapsulated in a functionalized self-assembling peptide hydrogel for brain tissue engineering. Biomaterials. 2013;34(8):2005–2016. | ||
Stroumpoulis D, Zhang H, Rubalcava L, Gliem J, Tirrell M. Cell adhesion and growth to peptide-patterned supported lipid membranes. Langmuir. 2007;23(7):3849–3856. | ||
Lee SS, Huang BJ, Kaltz SR, et al. Bone regeneration with low dose BMP-2 amplified by biomimetic supramolecular nanofibers within collagen scaffolds. Biomaterials. 2013;34(2):452–459. | ||
Chang R, Sun L, Webster TJ. Selective inhibition of MG-63 osteosarcoma cell proliferation induced by curcumin-loaded self-assembled arginine-rich-RGD nanospheres. Int J Nanomedicine. 2015;10:3351–3365. | ||
Zhang S, Greenfield MA, Mata A, et al. A self-assembly pathway to aligned monodomain gels. Nat Mater. 2010;9(7):594–601. | ||
Chen C, Pan F, Zhang S, et al. Antibacterial activities of short designer peptides: a link between propensity for nanostructuring and capacity for membrane destabilization. Biomacromolecules. 2010;11(2):402–411. | ||
Veiga AS, Sinthuvanich C, Gaspar D, Franquelim HG, Castanho M, Schneider JP. Arginine-rich self-assembling peptides as potent antibacterial gels. Biomaterials. 2012;33(35):8907–8916. | ||
Galanth C, Abbassi F, Lequin O, et al. Mechanism of antibacterial action of dermaseptin B2: interplay between helix-hinge-helix structure and membrane curvature strain. Biochemistry. 2009;48(2):313–327. | ||
Zelezetsky I, Tossi A. Alpha-helical antimicrobial peptides – using a sequence template to guide structure-activity relationship studies. Biochim Biophys Acta. 2006;1758(9):1436–1449. | ||
Bechinger B, Lohner K. Detergent-like actions of linear amphipathic cationic antimicrobial peptides. Biochimi Biophys Acta. 2006;1758(9):1529–1539. | ||
van Kan EJM, van der Bent A, Demel RA, de Kruijff B. Membrane activity of the peptide antibiotic clavanin and the importance of its glycine residues. Biochemistry. 2001;40(21):6398–6405. | ||
Chu-Kung AF, Nguyen R, Bozzelli KN, Tirrell M. Chain length dependence of antimicrobial peptide-fatty acid conjugate activity. J Colloid Interface Sci. 2010;345(2):160–167. | ||
Chu-Kung AF, Bozzelli KN, Lockwood NA, Haseman JR, Mayo KH, Tirrell MV. Promotion of peptide antimicrobial activity by fatty acid conjugation. Bioconjug Chem. 2004;15(3):530–535. | ||
Tripathi JK, Pal S, Awasthi B, et al. Variants of self-assembling peptide, KLD-12 that show both rapid fracture healing and antimicrobial properties. Biomaterials. 2015;56:92–103. | ||
Chen WY, Chang HY, Lu JK, et al. Self-assembly of antimicrobial peptides on gold nanodots: against multidrug-resistant bacteria and wound-healing application. Adv Funct Mat. 2015;25(46):7189–7199. | ||
Bionda N, Pitteloud JP, Cudic P. Cyclic lipodepsipeptides: a new class of antibacterial agents in the battle against resistant bacteria. Future Med Chem. 2013;5(11):1311–1330. | ||
Dartois V, Sanchez-Quesada J, Cabezas E, et al. Systemic antibacterial activity of novel synthetic cyclic peptides. Antimicrob Agents Chemother. 2005;49(8):3302–3310. | ||
Brea RJ, Vazquez ME, Mosquera M, Castedo L, Granja JR. Controlling multiple fluorescent signal output in cyclic peptide-based supramolecular systems. J Am Chem Soc. 2007;129(6):1653–1657. | ||
Khalfa A, Tarek M. On the antibacterial action of cyclic peptides: insights from coarse-grained MD simulations. J Phys Chem. 2010;114(8):2676–2684. | ||
Bionda N, Stawikowski M, Stawikowska R, et al. Effects of cyclic lipodepsipeptide structural modulation on stability, antibacterial activity, and human cell toxicity. Chemmedchem. 2012;7(5):871–882. | ||
Williams RJ, Mart RJ, Ulijn RV. Exploiting biocatalysis in peptide self-assembly. Biopolymers. 2010;94(1):107–117. | ||
Mishra A, Loo YH, Deng RH, et al. Ultrasmall natural peptides self-assemble to strong temperature-resistant helical fibers in scaffolds suitable for tissue engineering. Nano Today. 2011;6(3):232–239. | ||
Mishra A, Chan KH, Reithofer MR, Hauser CAE. Influence of metal salts on the hydrogelation properties of ultrashort aliphatic peptides. RSC Adv. 2013;3(25):9985–9993. | ||
Rodriguez AL, Parish CL, Nisbet DR, Williams RJ. Tuning the amino acid sequence of minimalist peptides to present biological signals via charge neutralised self assembly. Soft Matter. 2013;9(15):3915–3919. | ||
Hartgerink JD, Beniash E, Stupp SI. Self-assembly and mineralization of peptide-amphiphile nanofibers. Science. 2001;294(5547):1684–1688. | ||
Huang Z, Sargeant TD, Hulvat JF, et al. Bioactive nanofibers instruct cells to proliferate and differentiate during enamel regeneration. J Bone Mineral Res. 2008;23(12):1995–2006. | ||
Tanisaka H, Kizaka-Kondoh S, Makino A, Tanaka S, Hiraoka M, Kimura S. Near-infrared fluorescent labeled peptosome for application to cancer imaging. Bioconjug Chem. 2008;19(1):109–117. | ||
Morisco A, Accardo A, Gianolio E, Tesauro D, Benedetti E, Morelli G. Micelles derivatized with octreotide as potential target-selective contrast agents in MRI. J Peptide Sci. 2009;15(3):242–250. | ||
Bull SR, Guler MO, Bras RE, Meade TJ, Stupp SI. Self-assembled peptide amphiphile nanofibers conjugated to MRI contrast agents. Nano Lett. 2005;5(1):1–4. | ||
Reches M, Gazit E. Casting metal nanowires within discrete self-assembled peptide nanotubes. Science. 2003;300(5619):625–627. | ||
Cavalli S, Albericio F, Kros A. Amphiphilic peptides and their cross-disciplinary role as building blocks for nanoscience. Chem Soc Rev. 2010;39(1):241–263. | ||
Krol S, Macrez R, Docagne F, et al. Therapeutic benefits from nanoparticles: the potential significance of nanoscience in diseases with compromise to the blood brain barrier. Chem Rev. 2013;113(3):1877–1903. | ||
Kim I, Jeong HH, Kim YJ, et al. A “light-up” 1D supramolecular nanoprobe for silver ions based on assembly of pyrene-labeled peptide amphiphiles: cell-imaging and antimicrobial activity. J Mat Chem. 2014;2(38):6478–6486. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


