Back to Journals » Infection and Drug Resistance » Volume 14
Secondary Infections in Hospitalized COVID-19 Patients: Indian Experience
Authors Vijay S, Bansal N, Rao BK, Veeraraghavan B, Rodrigues C, Wattal C, Goyal JP, Tadepalli K, Mathur P, Venkateswaran R , Venkatasubramanian R, Khadanga S, Bhattacharya S, Mukherjee S , Baveja S, Sistla S, Panda S, Walia K
Received 20 January 2021
Accepted for publication 10 April 2021
Published 24 May 2021 Volume 2021:14 Pages 1893—1903
DOI https://doi.org/10.2147/IDR.S299774
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Sonam Vijay,1 Nitin Bansal,1 Brijendra Kumar Rao,2 Balaji Veeraraghavan,3 Camilla Rodrigues,4 Chand Wattal,5 Jagdish Prasad Goyal,6 Karuna Tadepalli,7 Purva Mathur,8 Ramanathan Venkateswaran,9 Ramasubramanian Venkatasubramanian,10 Sagar Khadanga,11 Sanjay Bhattacharya,12 Sudipta Mukherjee,13 Sujata Baveja,14 Sujatha Sistla,15 Samiran Panda,1 Kamini Walia1
1Division of Epidemiology and Communicable Diseases, Indian Council of Medical Research, New Delhi, India; 2Institute of Critical Medicine, Sir Ganga Ram Hospital, New Delhi, India; 3Department of Clinical Microbiology, Christian Medical College, Vellore, India; 4Department of Microbiology, PD Hinduja Hospital, Mumbai, Maharashtra, India; 5Department of Clinical Microbiology, Sir Ganga Ram Hospital, New Delhi, India; 6Department of Pediatrics, All India Institute of Medical Sciences, Jodhpur, India; 7Department of Microbiology, All India Institute of Medical Sciences, Bhopal, India; 8Department of Laboratory Medicine, Jai Prakash Narayan Apex Trauma Center, All India Institute of Medical Sciences, New Delhi, India; 9Department of Medicine, JIPMER, Pondicherry, India; 10Department of Infectious Diseases, Apollo Hospital, Chennai, Tamil Nadu, India; 11Department of Medicine, All India Institute of Medical Sciences, Bhopal, India; 12Department of Microbiology, Tata Medical Center, Kolkata, West Bengal, India; 13Department of Critical Care Medicine, Tata Medical Center, Kolkata, West Bengal, India; 14Department of Microbiology, Lokmanya Tilak Municipal General Hospital, Mumbai, India; 15Department of Microbiology, JIPMER, Pondicherry, India
Correspondence: Kamini Walia
Division of Epidemiology and Communicable Diseases, Indian Council of Medical Research, Ansari Nagar, New Delhi, 110029, India
Email [email protected]
Purpose: Critically ill coronavirus disease 2019 (COVID-19) patients need hospitalization which increases their risk of acquiring secondary bacterial and fungal infections. The practice of empiric antimicrobial prescription, due to limited diagnostic capabilities of many hospitals, has the potential to escalate an already worrisome antimicrobial resistance (AMR) situation in India. This study reports the prevalence and profiles of secondary infections (SIs) and clinical outcomes in hospitalized COVID-19 patients in India.
Patients and Methods: A retrospective study of secondary infections in patients admitted in intensive care units (ICUs) and wards of ten hospitals of the Indian Council of Medical Research (ICMR) AMR surveillance network, between June and August 2020, was undertaken. The demographic data, time of infection after admission, microbiological and antimicrobial resistance data of secondary infections, and clinical outcome data of the admitted COVID-19 patients were collated.
Results: Out of 17,534 admitted patients, 3.6% of patients developed secondary bacterial or fungal infections. The mortality among patients who developed secondary infections was 56.7% against an overall mortality of 10.6% in total admitted COVID-19 patients. Gram-negative bacteria were isolated from 78% of patients. Klebsiella pneumoniae (29%) was the predominant pathogen, followed by Acinetobacter baumannii (21%). Thirty-five percent of patients reported polymicrobial infections, including fungal infections. High levels of carbapenem resistance was seen in A. baumannii (92.6%) followed by K. pneumoniae (72.8%).
Conclusion: Predominance of Gram-negative pathogens in COVID-19 patients coupled with high rates of resistance to higher generation antimicrobials is an alarming finding. A high rate of mortality in patients with secondary infections warrants extra caution to improve the infection control practices and practice of antimicrobial stewardship interventions not only to save patient lives but also prevent selection of drug-resistant infections, to which the current situation is very conducive.
Keywords: COVID-19, secondary infections, antimicrobial resistance, hospital acquired infections, antibiotics
Introduction
COVID-19, caused by severe acute respiratory syndrome coronavirus 2 (SARS Cov2), has resulted in 119.2 million infections and 2.64 million deaths by 14 March 2021, globally.1 As of March 14, 2021, India has seen 11.35 million infections and 0.15 million deaths.2 Just like other viral pneumonias, bacterial and fungal infections are common complications seen in hospitalized COVID-19 patients. A proportion of COVID-19 patients also require prolonged stay in hospitals and are at a higher risk of developing hospital-acquired bacterial and fungal infections. This is due to the impaired ability of the host to clear bacterial pathogens owing to release of specific cytokines like IL-10, IL-6, IL-17 and IL-23; reduced dendritic cells function, macrophages, natural killer cells, CD4+ and CD8+ T-cells; and also certain phagocyte-independent mechanisms by which virus infection may facilitate secondary bacterial infection.3,4
Secondary infections in COVID-19 patients are known to be associated with negative health outcomes. As per recent studies, bacterial co-infection upon admission has been reported in 3.1–3.5% of COVID-19 patients, while secondary bacterial infections, following hospitalization, occurred in up to 15% of patients.5–8
Higher risk of mortality in COVID-19 patients with bacterial super-infection has been previously reported, and several recommendations encourage empirical use of antibiotics in severely ill patients.9,10 The prevalence and profile of secondary infections (SIs) in COVID-19 Indian patients is not very well understood. The present study was carried out with an objective to understand the etiology and antimicrobial resistance profile of secondary bacterial and fungal infections and subsequent clinical outcomes in hospitalized COVID-19 Indian patients. The overarching aim is to use this evidence to guide the optimal antimicrobial use in COVID-19 patients.
Patients and Methods
The Indian Council of Medical Research, as part of the antimicrobial resistance (AMR) surveillance and research initiative, has a network of tertiary care hospitals which undertake AMR surveillance, infection control and antimicrobial stewardship activities.11,12 These hospitals regularly capture AMR data from patients admitted in ICUs, wards and visiting OPD and information on antibiotics prescribed in hospitalized patients.
Study Design
We analyzed retrospective data of hospitalized COVID-19 patients from ten ICMR AMR network hospitals from June 2020 to August 2020. Ethical clearance, for reporting AMR data for all patients admitted to these hospitals to ICMR, is in place in all the hospitals who reported COVID-19 data. The hospitals collected the following information on COVID-19 patients: demographic details of admitted patients, presence of comorbidities (if any), date of admission, date of culture-positive results, antimicrobial susceptibility profile of isolates, antibiotics administered, day of hospitalization on which bacterial or fungal culture was sent, length of hospital stay and patient outcome.
Pathogens Identification and Antimicrobial Susceptibility Testing (AST)
All clinical samples including respiratory specimens, blood, urine, pus and other samples like deep wound were collected and processed. All these hospitals performed routine microbiological investigations including collection and culture of specimens. The pathogen identification and AST were performed using standard operating procedures (SOPs) formulated by the ICMR in which AST data were classified as per Clinical and Laboratory Standards Institute (CLSI) guidelines 2019.13 Isolates having resistance to at least one agent in three or more different classes of antibiotics (1. Cephalosporins-Aminoglycosides-Fluoroquinolones; 2. Carbapenem- Cephalosporins-Aminoglycosides-Fluoroquinolones; 3. Carbapenem- Cephalosporins-Fluoroquinolones) were classified as multidrug-resistant pathogens (MDROs).
Antibiotics Usage
The antibiotics prescribed to each hospitalized patient were recorded, and total prescriptions were analyzed and classified as per the Access, Watch and Reserve (AWaRe) classification of WHO.
Data Analysis
All data were collated in Microsoft Excel for analysis. Here, n represented the number of patients, organism and site of infection. Patient’s age was mentioned as mean ± standard deviation (SD). The comparison of ward and ICU data were analyzed using GraphPad 8, and p-value of P<0.05 was considered to be statistically significant.
Results
A total of 17,534 COVID-19 patients were admitted in the 10 hospitals of ICMR AMR surveillance between 1 June 2020 and 30 August 2020. Among these, overall incidence of secondary infections was 3.6% (640 patients) with the range of 1.7%–28% across the hospitals. From the overall pooled proportion of COVID-19 patients with SIs, the mean age of admitted patients was 53.3 ± 9.36 (SD) years (range 1–97 years); 451 patients (70.5%) were males and 189 (29.5%) females. Demographic characteristics of all patients of individual hospitals (H1 to H10) are summarized in Table 1. Among patients with confirmed secondary infections (SIs), 459 patients (71.7%) were in ICUs and 181 (28.3%) in wards at the time of sending cultures. The median days to develop a hospital-acquired infection for COVID-19 patients varied between 3 and 15 days. Out of the total patients having confirmed diagnosis of SIs, a majority (78%) of patients acquired secondary bacterial/fungal infections in the hospital (>48 hours of admission). Mortality among COVID-19 patients who acquired secondary infections was 56.7% (363/640).
 |
Table 1 Demographic Characteristics and Outcome of Hospitalized COVID-19 Patients |
Etiology of the Secondary Infections
Out of a total of 7,163 samples collected for microbiological culture, 852 (11.89%) samples were positive for bacterial or fungal growth. Of the total samples tested, 44.1% were blood, 35.1% respiratory specimens [bronchoalveolar lavage (BAL), endotracheal aspirate (ETA), pleural fluid (PF) and sputum], 9.1% urine samples and 5.6% pus. Of the pathogens isolated, Gram-negative bacteria were the predominant pathogen (78.03%, 785/1006). The most commonly isolated pathogens were Klebsiella pneumoniae (29.3%, 295/1006), Acinetobacter baumannii (21.07%, 212/1006), Pseudomonas aeruginosa (9.6%, 97/1006) and E. coli (8.2%, 83/1006). Other Gram-negative bacteria (9.70%) causing SIs were Stenotrophomonas maltophilia, Enterobacter cloacae, Proteus mirabilis and Serratia spp. Enterococcus was isolated in 7.25% (73/1006) of patients, and methicillin-resistant Staphylococcus aureus (MRSA) was isolated only in 1.29% (13/1006) of patients. Candida spp. were isolated from 6% (61/1006) of admitted COVID-19 patients, and, among these, 13 isolates (1.3%) of Candida auris were found (Figure 1). The most common site of isolation of a pathogen was blood (n=444) followed by respiratory specimen (n=354) and urine (n=92). K. pneumoniae was the most commonly isolated bacterium from respiratory (35%) and blood samples (29.7%). Other common bacteria in respiratory samples were A. baumannii (27%) and Pseudomonas aeruginosa (14%). A. baumannii (27%), Enterococcus spp. (8.7%) and Candida spp. (9.2%) were other isolated pathogens from blood. Among urine samples, E. coli (27.17%), K. pneumoniae (18.4%) and Candida spp. (18.4%) had the highest isolation rates. Isolates from pus samples were predominately E. coli (28%). The site of infection and distribution of bacteria are shown in Figure 1 and Table 2.
 |
Table 2 Site-Specific Etiological Distribution of Pathogens |
 |
Figure 1 Distribution of bacterial and fungal pathogens isolated from COVID-19 patients. |
Drug Resistance Profile of Isolated Pathogens
Of the total hospitalized patients with secondary infections, 47.1% (302/640) were infected with MDROs (Supplementary Table S1). Highest resistance was seen in K. pneumoniae isolates against third-generation cephalosporin, ceftriaxone (91.7%), fluoroquinolones (82%) and β-lactam-β-lactamase inhibitor combinations, piperacillin/tazobactam (79.2%) and cefoperazone-sulbactam (76.4%). Among carbapenems, ertapenem, meropenem and imipenem resistance in K. pneumoniae was 79%, 72% and 66.8%, respectively (Figure 2A). A. baumannii isolates showed high resistance to nearly all antibiotics tested (Figure 2B), with highest resistance against ceftazidime (96%), followed by meropenem (94%), fluoroquinolone (93.5%), imipenem (92%) and piperacillin/tazobactam (91%). In total, 76.8% of K. pneumoniae isolates and 90.3% of A. baumannii isolates were intermediate to colistin. Among the Enterobacteriaceae, extended spectrum beta-lactamases (ESBL) were detected in 83.2% (284/341), reflected by 91.7% of K. pneumoniae showing resistance to ceftriaxone (Table 3). Overall, 74.2% (484/652) of Gram-negative bacteria were resistant to carbapenems (92.6% of A. baumannii isolates and 72.8% of K. pneumoniae isolates) (Table 4). All Staphylococcus aureus and 68% of Enterococcus spp. isolates were susceptible to vancomycin.
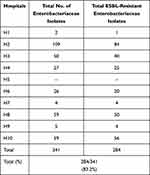 |
Table 3 Distribution of ESBL-Resistant Enterobacteriaceae Isolates |
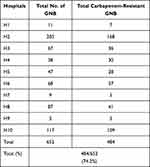 |
Table 4 Distribution of Carbapenem-Resistant Gram-Negative Bacteria (GNB) |
 |
Figure 2 Susceptibility pattern (%) of Gram-negative bacteria isolated from COVID-19 patients. (A) Klebsiella pneumoniae. (B) Acinetobacter baumannii. |
In our study 91.4% (585/640) of admitted COVID-19 patients had bacterial infections, and 5.4% (35/640) of patients had only fungal infections. Mixed infection with more than one organism isolated from different or same site was seen in 34.8% (223/640) of patients. Both bacterial and fungal organisms were reported in 8.43% (54/640) of patients.
Antibiotic Usage
Third-generation cephalosporin (16.06%), β-lactam-β-lactamase inhibitors like piperacillin-tazobactam, cefoperazone-sulbactam (57.3%) and carbapenems (43.7%) were the most commonly prescribed antibiotics in our study group. Vancomycin or teicoplanin was prescribed to 24.9% of patients. Most antibiotics were prescribed from the “watch” (52.36%) and “reserve” categories (22.05%), and relatively fewer prescriptions were from the “access“ category of WHO AWaRe classification (16.49%) (Figure 3).
 |
Figure 3 Depiction of antibiotics as per WHO Aware, Watch and Reserve (AWaRe) classification. (A) Total antibiotics usage in %. (B) Class of antibiotics usage in %. |
Patient Outcomes
Overall mortality among admitted COVID-19 patients, in these ten hospitals, was 11.6% (range 2.5%–45%), and mortality among COVID-19 patients with SIs was 56.7% (27%–78.9%). Mortality was higher in critically ill patients (in ICUs), ie 68% (313/459) as compared to patients in wards 27.6% (50/181) (p=0.0016). Mortality following incidence of SIs was 55% (248/451) in male patients and 56% (107/189) in female patients. Out of the patients who died with SIs, 72% had Gram-negative infections, 10.8% had Gram-positive infections, 8% had mixed infections with Gram-positive and Gram-negative pathogens, 4% had fungal pathogens with Gram-negative pathogens and 6% of patients had fungal infections (Figure 4). Mortality was very high among those infected with MDROs (60.5%).
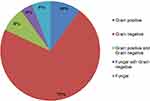 |
Figure 4 Depiction of patient outcomes linked to secondary infections. |
Discussion
There is paucity of data to make a firm estimate on the exact prevalence of bacterial or fungal co-infections in COVID-19 patients, as most studies are retrospective and suffer from selection bias. But from the emerging data, it is clear that bacterial co-infection rate is lower than seen in patients with influenza H1N1 and H3N2.14,15 Two meta-analyses published so far from COVID-19 patients documented rates of bacterial co-infections as 3.5% and 7%, respectively.6,16 We found overall prevalence of SIs in our study to be 3.6%. However, the presence of secondary infection in more than 20% of the admitted COVID-19 patients within 48 hours of admission is higher than the rates of bacterial community-acquired infections documented by other studies.5 With predominance of Gram-negative pathogens and isolation of fungal pathogens in these patients, the profile of secondary infections also did not resemble the typical picture of community-acquired infections. Hence, these patients may not represent a true picture of co-infection at the time of admission. This could be due to the fact that most of hospitals of the ICMR network are tertiary care hospitals and the patients may have reached these tertiary hospitals after previous hospitalization where they acquired these pathogens.
Out of the total number of patients reporting secondary infections, three-quarters of infections were identified after 48 hours of hospitalization, making hospital-acquired infections (HAIs) as the likely cause of these secondary infections. While this may not be a very accurate way of calculating the exact prevalence of secondary infections, this is in line with other studies published world-wide in which authors could not make distinction between co-infections at the time of admission and secondary infections following hospitalization.17
Blood and respiratory sites were the most common sites of secondary infection in COVID-19 patients. Gram-negative pathogens were predominant in respiratory infections, with a significant proportion of Gram-positive pathogens isolated from bloodstream infections. It is well known that most hospitalized moderate or severe COVID-19 patients are prescribed steroids, undergo invasive procedures and sometimes have a prolonged hospital stay, rendering them vulnerable to HAIs. Predominance of Gram-negative pathogens could be due to the invasive device-associated infections during hospitalization due to mechanical ventilation and central venous catheter implantation in these patients.
When compared with the overall mortality rates in hospitalized COVID-19 patients, the mortality rates in this study were significantly higher (56%) among patients with secondary infections, against an overall mortality rate of 11.6% in hospitalized COVID-19 patients. Out of 640 patients with SIs, 55.7% (357) of patients were also reported to have comorbid conditions, with 30% (193) of patients having diabetes and 25.6% (164) having hypertension. We suspect that the presence of comorbid conditions could have also contributed to the higher mortality seen in these patients. In our analysis, we did not find any significant difference in mortality between males and females, suggesting that gender was not a risk factor for death in Indian patients with SIs. Studies from other countries have reported male gender to be a risk factor for disease severity status, and a risk factor related to death.18,19
Out of the total deaths because of SIs, 72% of deaths were attributable to Gram-negative pathogens and 11% due to Gram-positive pathogens. Multidrug-resistant Gram-negative pathogens (K. pneumoniae and A. baumannii) were the predominant pathogens of SIs, and the rate of carbapenem resistance among these two pathogens was very high (72% and 92%, respectively). Mortality among patients with K. pneumoniae and A. baumannii infections was 76% and 63%, respectively. These results were in line with the previously documented high mortality associated with carbapenem-resistant A. baumannii and K. pneumoniae.20
Among secondary infections in patients with COVID-19, extremely drug-resistant (XDR) K. pneumoniae and XDR A. baumannii constituted almost 50% of the isolates. This is different from ICMR data published in last few years, wherein E. coli was the most commonly isolated pathogen, except in respiratory specimens where K. pneumoniae was most commonly isolated.21,22 Successful persistence of both K. pneumoniae and A. baumannii has been mainly due to high genome plasticity that results in the acquisition, maintenance and spread of multiple mobile elements carrying AMR determinants. In our study we observe “superiority of K. pneumoniae”, over E. coli in COVID-19 patients. High isolation rates along with reduced susceptibility of K. pneumoniae to drugs like piperacillin-tazobactam, cefoperazone sulbactam and carbapenems are alarming and need urgent containment measures through appropriate infection control and antimicrobial stewardship interventions.23 Additionally, considerable numbers of other Gram-negative bacteria like Stenotrophomonas maltophilia (2.7%) were also isolated from respiratory (12.4%) and blood (10.4%) infections. The overuse of carbapenems as an empirical treatment selects for and promotes resistance in Stenotrophomonas maltophilia infections, since it is intrinsically resistant to carbapenems. Reducing such antibiotic selection pressure should be urgently considered to prevent selection of resistant isolates.
Candida spp. were isolated from 9% of blood culture samples and had more than 30% resistance to fluconazole. Aspergillosis complicating COVID-19, associated with high morbidity and mortality in critically ill patients, was reported only in 16 patients (1.5%) in our study as compared to some other published studies which report incidence of COVID-19-associated pulmonary aspergillosis as high as 26% to 33%.24–26 This could be due to lack of awareness or lack of availability/use of appropriate diagnostic tests within the hospitals.
High isolation rates of carbapenem-resistant A. baumannii and Pseudomonas aeruginosa, also indicate a near-patient environmental source, pointing to poor hand hygiene and lack of compliance with device-related bundle care protocols as contributing factors. All the participating hospitals are also part of the infection control capacity-building initiative and capture HAI rates on regular basis.12 From the data collected during the same period under HAI surveillance, we understand that most of the hospitals have documented 2–3 fold increased bloodstream infection (BSI) rates from June 2020 to August 2020, as compared to data from the previous year. Compromised hand hygiene compliance due to double-gloving practiced in COVID-19 wards and use of personal protective equipment (PPE) in humid tropical environments, added to the fear factor of the SARS-CoV-2 virus amongst the health care workers, doctors and nurses, were some of the practical issues that could have led to high nosocomial infections rates.
Only three out of ten hospitals administered more than three antibiotics to hospitalized COVID-19 patients. Since these hospitals are also part of a capacity-building initiative for antimicrobial stewardship, it may have led to judicious and responsible prescribing of antimicrobials. Although there is no data to support the use of broad-spectrum antibiotics in COVID-19 patients, still these are prescribed to many hospitalized COVID-19 patients, also documented in our study. β-Lactam-β-lactamase inhibitors (piperacillin-tazobactam, cefoperazone-sulbactam) and carbapenems were the most commonly prescribed antibiotics. ESBL rates and carbapenem resistance rates among isolated pathogens in our study could be the consequence of excessive use of these antibiotics. Most antibiotics prescribed were from the “watch” and “reserve” categories of WHO AWaRe classification, which may be further adding “fuel to the fire” of the already alarming antimicrobial resistance levels in India. Since secondary infections were predominantly caused by Gram-negative pathogens (78.3%) in our study, the empirical cover for resistant Gram-positive pathogens (eg, vancomycin) may not be warranted. We found that 10% of patients (70/640) received antifungals without any evidence of fungal infection, warranting a focused intervention around rational use of antifungals. Since these data were captured at the time when there was limited understanding and experience of managing COVID-19 patients, this could have led to indiscriminate use of antimicrobials, including antifungals.
As most of the secondary infections in our study were nosocomial in origin, and that too with highly drug-resistant pathogens, it highlighted poor infection control practices and irrational antibiotic prescription practices. Hand hygiene practices are most affected during COVID-19 pandemic as all health personnels use gloves as part of PPE and do not feel the need to perform hand hygiene, and there is a lack of concern for inter-patient transmission of infections in patients. Gloves are known to get colonized by pathogens and can lead to cross-contamination, and glove hygiene practices of performing hand hygiene before and after putting on gloves, removal of gloves after care of a single patient, not using hand rubs over gloves and compliance to device care bundles need to be re-enforced in all health care settings. Fear of missing a secondary infection and lack of specific therapy for COVID-19 leads to overprescription of antibiotics. Sending appropriate cultures, use of biomarkers like procalcitonin and galactomannan and antibiotic time-out at 48 hours of prescription can help in reducing unnecessary antibiotic prescriptions. Following these simple but effective measures can lead to reduced opportunities for antimicrobial prescriptions and reduce empirical prescriptions, leading to better outcomes for containment of antimicrobial resistance during this pandemic.
There are several limitations to our study. Firstly, the protocol of sending cultures may have differed between different hospitals, and also cultures were sent as per discretion of treating physicians, so prevalence of infections calculated from our study may not be accurate. There is a possibility that many cultures may have been sent only when the initial empirical antibiotics did not help the patients, which may have led to isolation of pathogens with drug resistance. Hence, we would not like to propose any changes in policy regarding agent of choice for secondary infections in COVID-19 patients based on findings from this retrospective study. Prospective studies are needed to throw more light into this issue. There was significant difference in the number cultures sent per patient, and we found a positive correlation between the detection rate of SIs and the total number of cultures sent (Supplementary Table S2 and Figure S1). Secondly, most of these hospitals are tertiary care centers, so microbial flora and antimicrobial susceptibility patterns depicted in our study may not represent the true picture at other hospitals. Thirdly, many of the positive cultures are from non-sterile sites, like respiratory tract secretions and urine, and may represent colonization rather than true infection. Fourthly, steroids and other immune-modulatory agents like tocilizumab are used in treatment of COVID-19, and these agents are known to cause secondary infections. Although most hospitals in our network administered steroids to COVID-19 patients, we were not able to analyze the effect of these agents in patients due to incomplete data on some patients. Despite all these limitations, to our knowledge, this is the first study that gives the prevalence and spectrum of secondary bacterial and fungal infections among COVID-19 patients in India, and it highlights an urgent need to improve infection control practices in hospitals and also rationalize antibiotic prescriptions. Our findings are important when defining the role of empiric antimicrobial therapy or stewardship strategies in hospitalized patients with COVID-19 to achieve better therapeutic outcomes.
In conclusion, prevalence of bacterial and fungal superinfections in hospitalized Indian patients with COVID-19 is low; however, when such infections are present, they cause severe disease with worst outcomes, as most superinfections are nosocomial and due to XDR pathogens. These data were captured when the COVID-19 cases were on the rise, and the findings suggest that a lot of overprescribing of antimicrobials happened during that time. The practice of poor infection control and empirical over-use of broad-spectrum antimicrobials also provides fertile ground for future outbreaks with highly drug-resistant pathogens. Hence, all efforts should be made to improve infection control practices (like hand hygiene/glove hygiene, device bundle compliance and air quality). This will help in reducing incidence of secondary infections, particularly those of nosocomial origin. Strict practice of antimicrobial stewardship will help in reducing drug pressure, which is a key driver of drug resistance. This study provides an important lesson on the need to go back to the basics of diagnostic stewardship and practice culture-directed therapy using narrow-spectrum agents, from the “access” category of AWaRe classification. High usage of broad-spectrum agents from the “watch“ and “reserve” categories will not only make drugs redundant but will also create highly drug-resistant bugs which may become a clinicians’ nightmare. With increasing cases of COVID-19 in India, yet again, and fear of another wave around the corner, there is a need to reinforce the principles of infection control and antibiotic stewardship. Together, both these interventions will lead to reduction in mortality and morbidity related to not only COVID-19 patients, but will also restrain development of drug-resistant pathogens/infections.
Abbreviations
AMR, antimicrobial resistance; AST, antimicrobial susceptibility testing; AWaRe, Access Watch Reserve; BAL, bronchoalveolar lavage; BSI, bloodstream infection; CLSI, Clinical and Laboratory Standards Institute; ESBL, extended spectrum beta-lactamases; ETA, endotracheal aspirate, HAIs, hospital-acquired infections; ICMR, Indian Council of Medical Research; ICU, intensive care unit; PF, pleural fluid; PPE, personal protective equipment; SD, standard deviation; SIs, secondary infections; SOPs, standard operating procedures; XDR, extremely drug-resistant.
Data Sharing Statement
The datasets used and/or analyzed for this study are available from the corresponding author on request. For the COVID-19 patients admitted to these hospitals, all data were anonymized to maintain participants’ privacy.
Ethics Approval
This retrospective study includes data from the ten hospitals which are part of the antimicrobial resistance (AMR) surveillance and research initiative of the Indian Council of Medical Research (ICMR). These sites routinely share AMR data for clinical isolates received by clinical microbiology labs and capture antimicrobial prescriptions and clinical outcomes of patients with drug-resistant infections. The study does not require taking informed consent from all patients for which AMR data are added to ICMR data management systems, as (i) no patient is sampled exclusively for project work; (ii) all samples are collected as part of standard of care for patient management; (iii) the data are utilized maintaining full confidentiality after removing all the patient identifiers. The study has been approved by Ethics Committees of all the hospitals part of this study. The progress of the ICMR AMR project is reviewed annually by a project review committee (PRC), and the PRC has approved all technical, scientific and ethical aspects of the project. All hospitals included in this analysis gave written consent for the same, and the study was conducted in accordance with the Declaration of Helsinki.
Disclosure
Dr Camilla Rodrigues reports personal fees from Pfizer, Sanofi, B Braun, Becton Dickinson, Biomerieux, Cipla and Glenmark, outside the submitted work. The authors declare that they have no other potential conflict of interests regarding this work.
References
1. World Health Organization. Coronavirus disease (COVID-19) pandemic. Available from: https://covid19.who.int.
2. COVID-19 pandemic in India. Available from: https://covid19.who.int/region/searo/country/in.
3. Mirzaei R, Goodarzi P, Asadi M, et al. Bacterial co-infections withSARS-CoV-2. IUBMB Life. 2020;72:2097–2111.
4. Bakaletz LO. Viral-bacterial co-infections in the respiratory tract. Curr Opin Microbiol. 2017;35:30–35.
5. Garcia-Vidal C, Sanjuan G, Moreno-García E, et al. Incidence of co-infections and super infections in hospitalized patients with COVID-19: a retrospective cohort study. Clin Microbiol Infect. 2021;27(1):83–88.
6. Langford BJ, So M, Raybardhan S, et al. Bacterial co-infection and secondary infection in patients with COVID-19: a living rapid review and meta-analysis. Clin Microbiol Infect. 2020;26(12):1622–1629.
7. Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult in patients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395(10229):1054–1062.
8. Li J, Wang J, Yang Y, et al. Etiology and antimicrobial resistance of secondary bacterial infections in patients hospitalized with COVID-19 in Wuhan, China: a retrospective analysis. Antimicrob Resist Infect Control. 2020;9:153.
9. World health organization. Clinical Management of COVID-19 Interim Guidance. Geneva: WHO; 2020. Available from:: https://www.who.int/publications/i/item/clinical-management-of-covid-19.
10. NICE. Managing suspected or confirmed pneumonia COVID-19 rapid guideline: managing suspected or confirmed pneumonia in adults in the community. Guidance 2020. Available from: https://www.nice.org.uk/guidance/ng165/chapter/4-Managing-suspected-or-confirmed-pneumonia.
11. Walia K, Madhumathi J, Veeraraghavan B, et al. Establishing Antimicrobial resistance surveillance & research network in India: journey so far. Indian J Med Res. 2019;149:164–179.
12. Swaminathan S, Prasad J, Dhariwal AC, et al. Strengthening infection prevention and control and systematic surveillance of healthcare associated infections in India. BMJ. 2017;358:j3768.
13. Indian Council of Medical Research. Standard Operating Procedures: Bacteriology. Antimicrobial Resistance Surveillance and Research Network. New Delhi: ICMR; 2019. Available from:: https://main.icmr.nic.in/sites/default/files/guidelines/Bacteriology_SOP_2nd_Ed_2019.pdf.
14. Morris DE, Cleary DW, Clarke SC. Secondary bacterial infections associated with influenza pandemics. Front Microbiol. 2017;8:1041.
15. Martin-Loeches I, Schultz JM, Vincent JL, et al. Increased incidence of co-infection in critically ill patients with influenza. Intensive Care Med. 2017;43(1):48–58.
16. Lansbury L, Lim B, Baskaran V, Shen lim W. Co-infections in people with COVID-19: a systematic review and meta-analysis. J Infect. 2020;81(2):266–275.
17. Rawson TM, Moore LSP, Zhu N, et al. Bacterial and Fungal coinfection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2020;71(9):2459–2468.
18. Yanez ND, Weiss NS, Romand JA, Treggiari MM. COVID-19 mortality risk for older men and women. BMC Public Health. 2020;20:1742.
19. Jin JM, Bai P, He W, et al. Gender differences in patients with COVID-19: focus on severity and mortality. Front Public Health. 2020;8:152.
20. Balkhair A, Al-Muharrmi Z, Al’Adawi B, et al. Prevalence and 30-day all-cause mortality of carbapenem-and colistin-resistant bacteraemia caused by Acinetobacter baumannii, Pseudomonas aeruginosa, and Klebsiella pneumoniae: description of a decade-long trend. Int J Infect Dis. 2019;85:10–15.
21. Indian Council of Medical Research. Annual Report Antimicrobial Resistance Surveillance and Research Network. New Delhi. January 2019- December 2019; 2019. Available from: https://main.icmr.nic.in/sites/default/files/upload_documents/Final_AMRSN_Annual_Report_2019_29072020.pdf.
22. Indian Council of Medical Research. Annual Report Antimicrobial Resistance Surveillance Network. New Delhi. January 2018- December 2018. 2018. Available from: http://iamrsn.icmr.org.in/index.php/resources/amr-icmr-data.
23. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275.
24. Koehler P, Cornely OA, Bottiger BW, et al. COVID-19 associated pulmonary aspergillosis. Mycoses. 2020;63(6):528–534.
25. Verweij PE, Gangneux JP, Bassetti M, et al. Diagnosing COVID-19-associated pulmonary aspergillosis. Lancet Microbe. 2020;1(2):e53–e55.
26. Wu CP, Adhi F, Highland K. Recognition and management of respiratory coinfection and secondary bacterial pneumonia in patients with COVID-19. Cleve Clin J Med. 2020;87(11):659–663.
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
