Back to Journals » Infection and Drug Resistance » Volume 11
Second-line anti-tuberculosis drug resistance testing in Ghana identifies the first extensively drug-resistant tuberculosis case
Authors Osei-Wusu S, Amo Omari M, Asante-Poku A, Darko Otchere I , Asare P, Forson A, Otu J, Antonio M, Yeboah-Manu D
Received 27 September 2017
Accepted for publication 30 November 2017
Published 22 February 2018 Volume 2018:11 Pages 239—246
DOI https://doi.org/10.2147/IDR.S152720
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sahil Khanna
Stephen Osei-Wusu,1,2 Michael Amo Omari,3 Adwoa Asante-Poku,1 Isaac Darko Otchere,1 Prince Asare,1 Audrey Forson,3 Jacob Otu,4 Martin Antonio,4 Dorothy Yeboah-Manu1
1Noguchi Memorial Institute for Medical Research, University of Ghana, Legon, Ghana; 2West Africa Centre for Cell Biology of Infectious Pathogens, University of Ghana, Legon, Ghana; 3Department of Chest Diseases, Korle-Bu Teaching Hospital, Accra, Ghana; 4Medical Research Council Unit, Fajara, The Gambia
Background: Drug resistance surveillance is crucial for tuberculosis (TB) control. Therefore, our goal was to determine the prevalence of second-line anti-TB drug resistance among diverse primary drug-resistant Mycobacterium tuberculosis complex (MTBC) isolates in Ghana.
Materials and methods: One hundred and seventeen MTBC isolates with varying first-line drug resistance were analyzed. Additional resistance to second-line anti-TB drugs (streptomycin [STR], amikacin [AMK] and moxifloxacin [MOX]) was profiled using the Etest and GenoType MTBDRsl version 2.0. Genes associated with resistance to AMK and MOX (gyrA, gyrB, eis, rrs, tap, whiB7 and tlyA) were then analyzed for mutation.
Results: Thirty-seven (31.9%) isolates had minimum inhibitory concentration (MIC) values ≥2 µg/mL against STR while 12 (10.3%) isolates had MIC values ≥1 µg/mL for AMK. Only one multidrug-resistant (MDR) isolate (Isolate ID: TB/Nm 919) had an MIC value of ≥0.125 µg/mL for MOX (MIC = 3 µg/mL). This isolate also had the highest MIC value for AMK (MIC = 16 µg/mL) and was confirmed as resistant to AMK and MOX by the line probe assay GenoType MTBDRsl version 2.0. Mutations associated with the resistance were: gyrA (G88C) and rrs (A514C and A1401G).
Conclusion: Our findings suggest the need to include routine second-line anti-TB drug susceptibility testing of MDR/rifampicin-resistant isolates in our diagnostic algorithm.
Keywords: tuberculosis, drug resistance, diagnosis, Ghana, XDR
Introduction
Tuberculosis (TB), which has killed more humans than any infectious disease, remains a global health challenge. Over 10 million people got sick and 1.4 million individuals died of TB in 2015, with most of the affected individuals living in resource-constrained countries.1 The burden of TB is worsened by the emergence of strains that are resistant to standard TB drugs, threatening to make a treatable disease untreatable. Individuals infected with resistant strains have to take anti-TB drugs for longer periods and cure rates are lower compared to infection with susceptible strains.1,2
The standard World Health Organization (WHO)-approved treatment greatly relies on the use of two potent drugs, that is, isoniazid (INH) and rifampicin (RIF).3,4 TB strains that are resistant to these two drugs are classified as multidrug-resistant TB (MDR-TB). More severe forms of TB drug resistance are pre-extensively drug-resistant TB (pre-XDR-TB) and extensively drug-resistant TB (XDR-TB).1,5–9 XDR-TB has been reported in 117 countries worldwide with treatment success of 28%.1
The first key intervention for controlling drug resistance is early detection, for appropriate treatment.1,2 The increased use of new and rapid diagnostic tools such as Xpert MTB/RIF and line probe assays (GenoType MTBDRplus and GenoType MTBDRsl) is ensuring that significantly more TB patients are correctly diagnosed on time.
The National Tuberculosis Control Programme (NTP), Ghana, is now rolling out some of these rapid diagnostic methods to support case management: GenoType MTBDRplus for rapid detection of MDR-TB, which has been evaluated for use in Ghana,10 Xpert MTB/RIF for RIF resistance and GenoType MTBDRsl for detection of XDR-TB.
There is limited data on drug resistance in Ghana, especially, resistance to second-line anti-TB drugs. The study objective was to determine the prevalence of second-line anti-TB drug resistance in Ghana.
Materials and methods
Ethics statement
The procedure for sample collection, diagnosis and treatment was done in accordance with NTP guidelines,11 and ethical clearance for this study was obtained from the Noguchi Memorial Institute for Medical Research Institutional Review Board (FWA 00001824, IRB 00001276).
Mycobacterial isolates
Isolates analyzed in this study were obtained from pulmonary patients attending various health facilities in the Accra Metro and the East Mamprusi District in the Greater-Accra and Northern regions of Ghana, respectively. The isolates included new TB cases, which form part of a prospective molecular epidemiology study, and those obtained from difficult-to-treat cases attending the Department of Chest Diseases, Korle-Bu Teaching Hospital. The drug susceptibility pattern of the isolates to the first-line anti-TB drugs was determined using the proportion method and GenoType MTBDRplus. In total, 71 (60.7%) MDR-TB, 33 (28.2%) INH monoresistant TB and 13 (11.1%) RIF monoresistant TB isolates were analyzed.
Isolation of genomic DNA
A loop full of mycobacterial isolate growing at the log phase was suspended in sterile distilled water and inactivated by heat killing at 95oC for 1 h. Genomic DNA was then extracted using the cetyl-trimethyl-ammonium bromide (CTAB) extraction method developed by Doyle and Doyle with modifications by Käser et al.12,13 Briefly, the initial cell lysis was done by resuspending the pellets in lysis buffer containing lysozyme and then incubating overnight at 37°C. Then, 20% sodium dodecyl sulphate (SDS) and 20 mg/mL proteinase K (PK) were added, respectively and mixed gently. The cell debris was removed by adding sodium chloride (NaCl) and prewarmed using CTAB and incubated at 65oC. After incubation with chloroform-isoamyl alcohol at a 24:1 ratio to remove protein debris, the DNA in the aqueous phase was purified with ice-cold isopropanol. The DNA was dried and resuspended in 1X Tris-EDTA buffer.12,13
Anti-TB drug susceptibility testing
The Etest (bioMérieux, Marcy-l’Étoil, France) method, which determines the minimum inhibitory concentration (MIC), was used to screen 117 resistant isolates against amikacin (AMK), streptomycin (STR) and moxifloxacin (MOX). McFarland Standard 4 bacteria suspension, corresponding to an approximate cell density of 12.0 ×108 CFU/mL,14 was prepared using Middlebrook 7H9 with 10% Tween 80 (7H9G-Tween) and inoculated on Middlebrook 7H11 agar plates with oleic albumin dextrose catalase media supplement. Etest strips of distinct drugs were placed aseptically on the plates using forceps and incubated at 37oC.15,16 The MIC was determined between 5 and 10 days.17 The laboratory reference strain, H37Rv, was used as the control for all the assays.
The GenoType MTBDRsl version 2.0 (Hain Lifesciences, Germany), a line probe assay, was used to screen 93 of the 117 phenotypically screened isolates according to the manufacturer’s instruction.18 The isolates used for this assay included all the MDR-TB (71) and RIF monoresistant (13) isolates and an additional nine INH monoresistant isolates.
Mutation analysis of drug targets
The isolates with high MICs by the Etest method (i.e., ≥2 µg/mL for STR, ≥1 µg/mL for AMK and ≥0.125 µg/mL for MOX) and resistant by the line probe assay were used for targeted DNA sequence analyses. In total, 26 isolates were sequenced including 14 control isolates that were susceptible to all the drugs. DNA segments of the respective resistance conferring genes gyrA, gyrB, rrs, eis, tlyA, tap and whiB7 were amplified using polymerase chain reaction (PCR) for direct DNA sequencing. The PCR for gyrA, gyrB, tlyA and whiB7 contained 5 µL of 10× buffer, 10 µL of Q-solution, 2.5 µL of 15 mM of MgCl2, 1 µL of dNTPs, 21.2 µL of nuclease-free water, 2.5 µL each of forward and reverse primers, 0.3 µL of HotStar Taq polymerase and 5 µL of DNA. Cycling conditions were as follows: initial activation step at 95°C for 5 min and 35 cycles of denaturation at 96°C for 1 min, annealing for 1 min at primer-specific temperature Tm (Table 1), extension at 68°C for 1 min and final extension at 72°C for 10 min. The PCR mixtures for rrs, eis and tap genes contained 25 µL of fast cycling master mix, 10 µL of Q-solution, 2.5 µL of each of the primers, 5 µL of nuclease-free water and 5 µL of DNA template. The cycling conditions included initial denaturation step at 95°C for 5 min and 40 cycles of denaturation at 96°C for 5 s, annealing for 5 s at primer-specific temperature Tm (Table 1), extension at 68°C for 48 s and final extension at 72°C for 1 min.
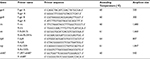  | Table 1 Primer sequences with the optimized annealing temperature for the targets Abbreviations: F, forward; R, reverse. |
Data analysis
Fisher’s exact test and chi-squared test were carried out using Stata 14.3 to determine the differences between the patient categories (new TB cases and treatment failure cases) and the frequency of second-line drug-resistant isolates. Also, the correlation coefficient was determined between the year of isolation of the MTB isolates from the patients and the prevalence of anti-TB drug resistance.
Drug resistance according to the line probe assay was indicated by the absence of wild-type band, presence of mutation band, or both. For interpretation of the results, only bands with intensities as strong as or stronger than that of the universal control band were considered.18
DNA reads obtained from sequencing were edited and cleaned to remove background noise. The cleaned sequences were screened for mutations by comparing with homologous sequences of Mycobacterium tuberculosis strain H37Rv downloaded from the Tuberculist database version 2.6 using the Staden package for DNA analysis.19,20 The results from this mutation analysis were compared with the identified mutations from the line probe assay.
Results
Biographical data of study participants
Most of the participants were males: 83 (74.8%) males, 28 (25.2%) females and six participants had no record of gender. The average age was 37 years, median age was 34 years and the range was between 13 and 79 years (Table 2). Majority of the participants, 69 (59.0%), were newly diagnosed cases; TB patients with treatment failure were 47 (40.2%) and one relapse case was also included in the study. The residential address of the study participants indicated they were from eight different administrative regions: 72 (61.5%) from Greater Accra region, 21 (17.9%) from Central region, nine (7.7%) from Northern region, seven (6.0%) from Volta region, four (3.4%) from Upper East region and two (1.7%) from Brong/Ahafo region. The participants also included one participant each from Eastern and Western region. The study participants were recruited between 2008 and 2015.
  | Table 2 Characteristics of study participants |
Second-line anti-TB drug profiles
We determined the MIC values of 117 isolates for STR, AMK and MOX (Table 3). However, one isolate was eliminated from the analysis of the results since it had a result discrepant with another laboratory.
The time for the Etest ellipse to become visible for the MIC determination was recorded for each isolate and 114 (97.4%) of the isolates were read between 5 and 10 days. Out of the total isolates screened, 37 (31.9%) had MICs ≥2 µg/mL for STR and 12 (10.3%) isolates presented with MICs ≥1 µg/mL for AMK. Only one MDR isolate (Isolate ID: TB/Nm 919) had an MIC value ≥0.125 µg/mL for MOX (MIC = 3µg/mL); this isolate also had the highest MIC value for AMK (16 µg/mL) and a very high MIC for STR (32 µg/mL) (Table 4).
Out of 69 isolates from new TB cases, eight (11.59%) were resistant to AMK, while four (8.51%) of the 47 treatment failures were AMK resistant. However, there was no significant difference between these two patient categories (p = 0.759). Also, we identified no significant difference between the proportion of isolates resistant to STR among the new TB cases (31.88%) compared to the treatment failures (31.91%, p = 0.997). There was only one MOX-resistant isolate which was a treatment failure case. There was no significant difference between the patient categories for MOX resistance (p = 0.405).
There was also no correlation between the year of isolation of the MTB isolates from the patients and the prevalence of anti-TB drug resistance (correlation coefficient, r = 0).
We screened 93 (including all MDRs) of the 117 isolates by GenoType MTBDRsl version 2.0. As summarized in Table 3, the isolate TB/Nm 919 was cross-resistant to AMK/kanamycin (KAN)/capreomycin (CAP) and also fluoroquinolone resistant. It had a mutation band MT 1, which corresponded to substitution A1401G in the rrs gene. Furthermore, there was an absence of a wild-type band (WT 1) in the gyrA of this isolate that corresponded to the G88C single-nucleotide polymorphism (SNP), which is associated with resistance to fluoroquinolones. This XDR-TB isolate was cultured from the sputum of a 42-year-old fisherman who had failed TB treatment. He died 2 years after he had reported to the hospital with persistent cough in January 2013. The first drug susceptibility testing conducted showed he was resistant to ethambutol, STR, RIF and INH.
There was no mutation or missing wild-type band in the eis and gyrB genes among all the isolates screened using the GenoType MTBDRsl version 2.0.
Mutations in aminoglycoside and fluoroquinolone resistance associated target genes
To identify the mutations associated with resistance to fluoroquinolones and aminoglycosides, 26 isolates were tested. Of the 26, 12 were isolates with high MICs for AMK and MOX or resistant by the line probe assay, GenoType MTBDRsl version 2.0. The other 14 were susceptible to all the second-line anti-TB drugs. A non-synonymous mutation, G263T that translates as G88C, was identified in the gyrA gene of isolate TB/Nm 919. This same isolate had the SNPs A1401G and A514C in the rrs gene (Table 4). We also detected a polymorphism N236K in tlyA gene in a drug-susceptible isolate. We did not find any mutation in gyrB, tap, eis and whiB7 genes.
Discussion
This study sought to determine the prevalence of second-line anti-TB drug resistance in Ghana. Prior to this study, only a few drug resistance surveys had been carried out on new TB cases and treatment failure cases in Ghana.17,21,22 The high STR resistance levels observed in this study is even lower than that achieved by Forson et al and Kato et al that recorded 79% and 81%, respectively, among treatment failures in Ghana.22,23 The high level of resistance to STR strengthens the WHO’s recommendation not to use STR for the treatment of MDR-TB cases.24 The high prevalence of STR resistance in Ghana can be attributed to its previous use as first-line anti-TB drug and also for treating other ailments and in veterinary medicine.21,25,26
The first second-line anti-TB drug testing in Ghana by Kato et al was carried out on 5 MDR-TB and 19 monoresistant isolates from four new cases and 17 treated cases. They did not record any resistance to AMK and ofloxacin.22 However, a later study conducted between 2009 and 2013 in eight West African countries by the West African Network of Excellence for Tuberculosis, AIDS and Malaria (WANETAM), identified resistance to KAN, CAP and ofloxacin for the first time in Ghana. They also observed a higher prevalence of MDR-TB in new and retreatment cases (6% and 35%, respectively) compared to the WHO estimates of 2% for new cases and 17% for retreatment cases.27 The study by Kato et al was actually carried out between 2008 and 2009 and the reporting of resistance by the WANETAM indicates the emergence of resistance.22,27 The resistance reported by this study also suggests the circulation of resistant strains in the country.
Mutations associated with resistance to fluoroquinolones and aminoglycosides was analyzed by GenoType MTBDRsl version 2.0 and drug target gene sequencing. The G88C mutation observed in the gyrA gene by targeted sequencing corresponded with the missing of gyrA wild-type band of the GenoType MTBDRsl version 2.0 assay. This mutation has been shown to be associated with resistance to fluoroquinolones.28,29 Although a review by Avalos et al observed geographic differences in the gyrA mutations across the globe, the use of GenoType MTBDRsl version 2.0 in Ghana was useful in the detection of this resistance-associated mutation.30
The target gene sequencing showed three SNPs (A1401G and A514C in rrs and N236K in tlyA) that are associated with aminoglycoside resistance. The isolate that had rrs MUT 1 band of the GenoType MTBDRsl version 2.0 assay also had a corresponding rrs mutation (A1401G). This is a common mutation reported in several studies to be associated with resistance to AMK, KAN and CAP.31–34 The tlyA missense mutation observed is an infrequent mutation that is associated with CAP resistance;35,36 however, the respective isolates were not tested phenotypically against CAP. There was no mutation in the eis gene by both molecular methods. The GenoType MTBDRsl version 2.0 included the eis gene probe which was missing in the GenoType MTBDRsl version 1.0 and a study by Tagliani et al showed that the inclusion of the probe for the eis promoter gene in the version 2.0 had increased the sensitivity for the detection of resistance to the aminoglycosides. However, this study did not detect any mutation band or missing wild-type band in the eis promoter region.37
Our analysis, to the best of our knowledge, identified one XDR-TB isolate for the first time in Ghana. The progressive identification of resistant isolates in Ghana if not controlled will pose a great challenge to the control of TB in Ghana. This calls for immediate action, including continuous surveillance, patient counseling/support to improve adherence to treatment and drug supply management to prevent the spread of XDR-TB in Ghana. The findings have been reported to the NTP to take the necessary actions. The contacts of the presumed XDR-TB patient have been screened for TB and are being monitored.
Conclusion
We thus recommend the inclusion of routine testing for second-line drug resistance among RIF and MDR-TB cases in our diagnostic algorithm.
Acknowledgments
We thank Dr Frank Bonsu, the Director of the NTP Ghana and Department of Chest Diseases, Korle-Bu Teaching Hospital, Ghana, for his support during this study. This study was funded by Wellcome Trust (Grant Number: 097134/Z/11/Z). Stephen Osei-Wusu, a graduate student, was supported by the Holger Pöhlmann Foundation and the West Africa Centre for Cell Biology of Infectious Pathogens (WACCBIP; ACE02-WACCBIP: Awandare), University of Ghana. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Disclosure
The authors report no conflicts of interest in this work.
References
World Health Organization. Global tuberculosis report. Geneva: WHO; 2016. Available from: http://apps.who.int/iris/bitstream/10665/250441/1/9789241565394-eng.pdf?ua=1&ua=1. Accessed February 9, 2017. | ||
Cegielski JP, Chin DP, Espinal MA, et al. The global tuberculosis situation. Progress and problems in the 20th century, prospects for the 21st century. Infect Dis Clin North Am. 2002;16(1):1–58. | ||
World Health Organization. Treatment of tuberculosis: guidelines 4th ed. Geneva: WHO; 2010. Available from: http://apps.who.int/iris/bitstream/10665/44165/1/9789241547833_eng.pdf?ua=1&ua=1. Accessed April 25, 2017. | ||
Horsburgh CR, Jr., Barry CE, 3rd, Lange C. Treatment of tuberculosis. N Engl J Med. 2015;373(22):2149–2160. | ||
World Health Organization. Multidrug and extensively drug-resistant TB (M/XDR-TB): 2010 global report on surveillance and response. Geneva: WHO; 2010. Available from: http://apps.who.int/iris/bitstream/10665/44286/1/9789241599191_eng.pdf. Accessed April 25, 2017. | ||
Centers for Disease Control Prevention. Emergence of Mycobacterium tuberculosis with extensive resistance to second-line drugs--worldwide, 2000-2004. MMWR Morbid Mortal Wkly Rep. 2006;55(11):301. | ||
Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–281. | ||
Fisher JF, Mobashery S. Enzymology of bacterial resistance. In: Mander L. Liu HW, editors. Comprehensive Natural Products II. Vol 8. Oxford: Elsevier; 2010:443–487. | ||
Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. Tuberculosis drug resistance mutation database. PLoS Med. 2009;6(2):e1000002. | ||
Asante-Poku A, Otchere ID, Danso E, et al. Evaluation of GenoType® MTBDRplus for the rapid detection of drug-resistant tuberculosis in Ghana. Int J Tuberc Lung Dis. 2015;19(8):954–959. | ||
National Tuberculosis Programme (NTP) Ghana. NTP comprehensive review report. Accra: Ghana Health Service; 2007. | ||
Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. | ||
Käser M, Ruf M-T, Hauser J, Marsollier L, Pluschke G. Optimized method for preparation of DNA from pathogenic and environmental mycobacteria. Appl Environ Microbiol. 2009;75(2):414–418. | ||
McFarland J. The nephelometer: an instrument for estimating the number of bacteria in suspensions used for calculating the opsonic index and for vaccines. J Am Med Assoc. 1907;49(14):1176–1178. | ||
Joloba ML, Bajaksouzian S, Jacobs MR. Evaluation of Etest for susceptibility testing of Mycobacterium tuberculosis. J Clin Microbiol. 2000;38(10):3834–3836. | ||
Sader HS, Pignatari ACC. E test: a novel technique for antimicrobial susceptibility testing. Sao Paulo Med J. 1994;112(4):635–638. | ||
Newman MJ, Frimpong E, Donkor ES, Opintan JA, Asamoah-Adu A. Resistance to antimicrobial drugs in Ghana. Infect Drug Resist. 2011;4:215–220. | ||
Hain Lifesciences. GenoType MTBDRsl VER 2.0 Manual. Molecular Genetic Assay for Identification of the M. tuberculosis Complex and its Resistance to Fluoroquinolones and Aminoglycosides/Cyclic Peptides from Sputum Specimens or Cultivated Samples. 2015. Available from: https://www.immunodiagnostic.fi/wp-content/uploads/MTBDRsl-V2_kit-insert.pdf. Accessed February 9, 2017. | ||
Lew JM, Kapopoulou A, Jones LM, Cole ST. TubercuList – 10 years after. Tuberculosis (Edinb). 2011;91(1):1–7. | ||
Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5(3):233–241. | ||
Owusu-Dabo E, Adjei O, Meyer CG, et al. Mycobacterium tuberculosis drug resistance, Ghana. Emerg Infect Dis. 2006;12(7):1170. | ||
Kato T, Addo KK, Nartey N, Nyarko AK, Bonsu FA, Mitarai S. First susceptibility testing of Mycobacterium tuberculosis for second-line anti-tuberculosis drugs in Ghana. Trop Med Health. 2014;42(1):53–55. | ||
Forson A, Kudzawu S, Kwara A, Flanigan T. High frequency of first-line anti-tuberculosis drug resistance among persons with chronic pulmonary tuberculosis at a teaching hospital chest clinic. Ghana Med J. 2010;44(2):42–46. | ||
World Health Organization. Guidelines for the programmatic management of drug-resistant tuberculosis – 2011 update. Geneva: WHO; 2011. Available from: http://apps.who.int/iris/bitstream/10665/44597/1/9789241501583_eng.pdf. Accessed April 25, 2017. | ||
Ministry of Health. Standard treatment guidelines. Ghana: Ministry of Health; 2004. Available from: http://www.moh.gov.gh/wp-content/uploads/2016/02/Standard-Treatment-Guideline-2010.pdf. Accessed February 9, 2017. | ||
Osei Sekyere J. Antibiotic types and handling practices in disease management among pig farms in Ashanti Region, Ghana. J Vet Med. 2014;2014:531952. | ||
Gehre F, Otu J, Kendall L, et al. The emerging threat of pre-extensively drug-resistant tuberculosis in West Africa: preparing for large-scale tuberculosis research and drug resistance surveillance. BMC Med. 2016;14(1):160. | ||
Xu C, Kreiswirth BN, Sreevatsan S, Musser JM, Drlica K. Fluoroquinolone resistance associated with specific gyrase mutations in clinical isolates of multidrug-resistant Mycobacterium tuberculosis. J Infect Dis. 1996;174(5):1127–1130. | ||
Sun Z, Zhang J, Zhang X, Wang S, Zhang Y, Li C. Comparison of gyrA gene mutations between laboratory-selected ofloxacin-resistant Mycobacterium tuberculosis strains and clinical isolates. Int J Antimicrob Agents. 2008;31(2):115–121. | ||
Avalos E, Catanzaro D, Catanzaro A, et al. Frequency and geographic distribution of gyrA and gyrB mutations associated with fluoroquinolone resistance in clinical Mycobacterium tuberculosis isolates: a systematic review. PLoS One. 2015;10(3):e0120470. | ||
Evans J, Segal H. Novel multiplex allele-specific PCR assays for the detection of resistance to second-line drugs in Mycobacterium tuberculosis. J Antimicrob Chemother. 2010;65(5):897–900. | ||
Perdigão J, Macedo R, Malaquias A, Ferreira A, Brum L, Portugal I. Genetic analysis of extensively drug-resistant Mycobacterium tuberculosis strains in Lisbon, Portugal. J Antimicrob Chemother. 2009;65(2):224–227. | ||
Ajbani K, Rodrigues C, Shenai S, Mehta A. Mutation detection and accurate diagnosis of extensively drug-resistant tuberculosis: report from a tertiary care center in India. J Clin Microbiol. 2011;49(4):1588–1590. | ||
Yuan X, Zhang T, Kawakami K, et al. Molecular characterization of multidrug and extensively drug-resistant Mycobacterium tuberculosis strains in Jiangxi, China. J Clin Microbiol. 2012;50(7):2404–2413. | ||
Maus CE, Plikaytis BB, Shinnick TM. Mutation of tlyA confers capreomycin resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2005;49(2):571–577. | ||
Jnawali HN, Yoo H, Ryoo S, et al. Molecular genetics of Mycobacterium tuberculosis resistant to aminoglycosides and cyclic peptide capreomycin antibiotics in Korea. World J Microbiol Biotechnol. 2013;29(6):975–982. | ||
Tagliani E, Cabibbe AM, Miotto P, et al. Diagnostic performance of the new version (v2. 0) of GenoType MTBDRsl assay for detection of resistance to fluoroquinolones and second-line injectable drugs: a multicenter study. J Clin Microbiol. 2015;53(9):2961–2969. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


