Back to Journals » International Journal of General Medicine » Volume 16
Role of RNA Splicing Mutations in Diffuse Large B Cell Lymphoma
Authors Berta D , Girma M , Melku M , Adane T , Birke B , Yalew A
Received 1 April 2023
Accepted for publication 8 June 2023
Published 15 June 2023 Volume 2023:16 Pages 2469—2480
DOI https://doi.org/10.2147/IJGM.S414106
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Scott Fraser
Dereje Berta,1 Mekonnen Girma,2 Mulugeta Melku,1,3 Tiruneh Adane,1 Bisrat Birke,1 Aregawi Yalew1
1Department of Hematology and Immunohematology, School of Biomedical and Laboratory Sciences, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia; 2Department of Quality Assurance and Laboratory Management, School of Biomedical and Laboratory Sciences, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia; 3College of Medicine and Public Health, Flinders University, Adelaide, SA, Australia
Correspondence: Dereje Berta, Email [email protected]
Abstract: Ribonucleic acid splicing is a crucial process to create a mature mRNA molecule by removing introns and ligating exons. This is a highly regulated process, but any alteration in splicing factors, splicing sites, or auxiliary components affects the final products of the gene. In diffuse large B-cell lymphoma, splicing mutations such as mutant splice sites, aberrant alternative splicing, exon skipping, and intron retention are detected. The alteration affects tumor suppression, DNA repair, cell cycle, cell differentiation, cell proliferation, and apoptosis. As a result, malignant transformation, cancer progression, and metastasis occurred in B cells at the germinal center. B-cell lymphoma 7 protein family member A (BCL7A), cluster of differentiation 79B (CD79B), myeloid differentiation primary response gene 88 (MYD88), tumor protein P53 (TP53), signal transducer and activator of transcription (STAT), serum- and glucose-regulated kinase 1 (SGK1), Pou class 2 associating factor 1 (POU2AF1), and neurogenic locus notch homolog protein 1 (NOTCH) are the most common genes affected by splicing mutations in diffuse large B cell lymphoma.
Keywords: RNA-splicing, aberrant splicing, alternative splicing, diffuse large B-cell lymphoma
Introduction
Non-Hodgkin lymphoma is a heterogeneous group of cancers that starts during immune system differentiation.1 Recent reports show that more than 100 different lymphoma types have been identified.2 These lymphomas originate from cells of the immune system such as B cells, T cells, and dendritic cells. B-cell lymphomas are another type of cancer that develops from clonal expansion and subsequent B-cell invasion of immune organs.3 The heterogeneity is derived from different stages of mature B-cell differentiation. Diffuse large B-cell lymphoma (DLBCL) is the most common lymphoid malignancy in adults that starts from the germinal center.4,5
Genetic alterations such as chromosomal translocations, sporadic somatic mutations, and copy number alterations, deletions, and amplifications of genes have been reported in the pathogenesis of DLBCL.5–8 In addition, missense mutations such as immune-related epigenetic modifications that affect the function oncogenes have been involved in DLBCL genesis.9–11 Furthermore, current evidence reveals that mutations during RNA splicing play a role in DLBCL pathogenesis. Thus, the identification of mutations in RNA splicing is important for the diagnosis, treatment, and prognosis of DLBCL.12,13
Human genes are highly complex; it contains coding sequence exons and non-coding sequence intron. Non-coding sequence should be removed from primary transcript through RNA splicing process.14 RNA splicing is the process by which a precursor mRNA transcript is transformed into a mature mRNA. It is a crucial process to create an mRNA molecule by removing introns and subsequently ligating exons that direct the synthesis of the protein during translation.13 The introns are removed from the transcript by cleavage at splice site (the 5′ and 3′ ends of intron). This process most commonly begins at dinucleotide GU and AG site at 5′ end and 3′ end of primary transcript, respectively (Figure 1).
 |
Figure 1 A schematic representative pre-mRNA, coding and non-coding sequence and splicing site. |
The RNA splicing process contains more than five small nuclear ribonucleoproteins (snRNPs) and trans and cis-splicing elements such as more than 200 proteins (at the splice acceptor, donor, and splice site), branch points (BP), splice enhancers, splice silencers, and splicing reaction catalysts. A large RNA-protein complex called the spliceosome recognizes auxiliary elements to promote the excision of introns from the nuclear pre-mRNA.12,13
Based on their involvement, spliceosome is classified as major and minor. About 99.5% of introns are recognized and excised by major spliceosome, which is known as U2 dependent spliceosome (U1, U2, U4, U6, and U5 snRNPs).15,16 The major splicing reaction occurs in order to recognized relatively poorly conserved sequence of pre-mRNA at the 5’ and 3’ ends. On the other hand, remaining about 05% of introns are excised by minor spliceosome, it is called U12 dependent (U11, U12, U5, U4atac, and U6atac).16,17 They target a rare group of introns and highly conserved sequences at the 5’ and 3’ ends. Both of splicing reactions are important to formation of mature mRNA.15,18
The alteration during the RNA splicing could potentially alter mRNA maturation and the subsequent production of protein.13,19 Recurrent genetic abnormalities due to splicing mutations have been reported in all forms of myeloid neoplasms, several lymphoid neoplasms, and solid tumors. These abnormalities are implicated during pre-RNA splicing.19 Furthermore, epigenetic changes can sometimes be linked to RNA splicing.17
The abnormalities detected in DLBCL due to RNA splicing are mutant splice sites, aberrant alternative splicing site, exon skipping, and intron retention.3,4,20 Alternative splicing is physiologic process, which occurs in most human gene and creates transcript variability. It is also allowing formation of various proteins from single transcript.21 However, sometimes it leading to pathologic conditions and induces human diseases such DLBCL.20
RNA splicing helps to regulate gene expression for cellular proliferation, survival, and differentiation.22 Nevertheless, mutations in the RNA splicing could potentially affect the maturation of mRNA for most genes and the subsequent production of protein.13 Most of these mutations affect the tumor suppressor activities and DNA repair activities of different genes.23–26 Currently, many splicing site gene mutations in B-cell lymphomas are reported.13 Mutated B cell has a high proliferation rate, a low differentiation rate, and a long lifetime, which leads it to malignant transformation.27
Even though, different scholars published original articles related with splicing site mutations in DLBCL, there is scarcity of a comprehensive review which shows splicing mutation as a cause of DLBCL. Thus, this review tried to discuss the general mechanisms of splicing mutation and splicing regulation. In addition, the review elucidates the role of different splicing mutation in DLBCL pathogenesis. In order to achieve the objective, the articles were identified through searching of the literature published in English using National Library of Medicine, PubMed, Google Scholar, and Google databases. Articles already identified in the references were also manually searched and included. After article identified, imported to end note version 8.1 and exported to Microsoft word for citation.
RNA Splicing Regulation
Ribonucleic acid splicing is a nuclear process catalyzed by the spliceosomes. It consists of snRNPs (U1, U2, U4, U5, and U6), over 200 related proteins, and other auxiliary components.28 The first step in the process of RNA splicing is the binding of U1 snRNPs to the 5’ splice site. Splicing factor 1 then binds the BP. After that, the U2 auxiliary factor (U2AF) complex binds the polypyrimidine tract and 3’ splice site. Binding of U2AF either strengthens or repels the recruitment of the spliceosome complex to the splice site.19,28 These U2AF include members of the serine/arginine (SR) protein family that promote splicing by recognizing specific splicing sequences in pre-mRNA called exonic and intronic splicing enhancers. Conversely, heterogeneous nuclear ribonucleoproteins (hnRNPs) suppress splicing by interacting with exonic and intronic splicing silencers.3 The splicing process consists of two sequential enzymatic steps named transesterification reactions. The BP nucleotide helps form the intron lariat through nucleophilic attack. After this, a ubiquitin-specific peptidase 59 (Usp59)-mediated attack on the ubiquitin-specific peptidase 39 (Usp39) occurred and led to the removal of the intron lariat from RNA for the formation of the spliced RNA product (Figure 2).3,19,28
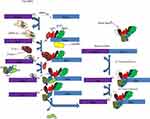 |
Figure 2 Splicing regulation mechanism. |
Mechanism of Splicing Site Mutation
The splicing process is a highly regulated one, but any alteration in splicing factors, splicing sites, or auxiliary components affects the final products of a gene.3,17 As a result of the alteration in mRNA, malignant transformation, cancer progression, and metastasis occur.21 Splicing mutations may be formed by different mechanisms.3,13,19,29,30 One of the mechanisms is related to cis-acting elements in RNA that act as an anchor for trans-acting factors to produce functional RNA transcripts. Mutations in cis-acting elements can lead to exon suppression, exon inclusion, blocking the binding of snRNPs, exon skipping, and intron retention. Consequently, the final product of mRNA is altered, which codes for tumor suppressors, oncogene inactivation, and DNA repair products.3,13
Another mechanism of splicing site mutation is related to alternative splicing. It is essential physiologically for the production of diverse proteins from a single pre-mRNA.20 However, it can sometimes result in genetic changes such as exon skipping, intron retention, altered 5’ splice, altered 3’ splice, and mutually exclusive splicing.22 These alterations may play an important role in tumor growth through tumor suppressor gene inactivation and oncogene activation, as well as inhibition of cell differentiation.31
Dysregulation of splicing factors’ expression is another mechanism related to splicing mutations. Spliceosome recognizes intron-exon boundaries and removes the intron.20 Exon skipping and intron retention are caused by mutations in the spliceosome complex.13,19 As a result of the spliceosome mutation, gene expression is dysregulated, and tumor cells proliferate (Figure 3).32,33
 |
Figure 3 Schematic representation of constitutive and alternative splicing dysregulation. |
Splicing Mutation in DLBCL Pathogenesis
B-cell lymphoma is a heterogeneous group of hematological malignancies originating from B cells and accounts for up to 35% of non-Hodgkin’s lymphomas.6,27 DLBCL has a global annual incidence of more than 100,000 cases. As a result, identification of the cause, pathogenesis, prognosis, and presentation is critical for improving the diagnostic process, classification, outcome stratification, and personalized therapy. As a result, splicing site mutation also has a contribution to DLBCL diagnosis, pathogenesis, and prognosis.5–8
Mutations during splicing affect different genes, which contribute to the inactivation of oncogenes,34 the activation of tumor suppressor genes,35 the activation of DNA repair genes, and the inhibition of apoptosis.27,30 Splicing mutations most commonly affect B-cell lymphoma 7 protein family member A (BCL7A), cluster of differentiation 79B (CD79B), myeloid differentiation primary response gene 88 (MYD88), tumor protein P53 (TP53), signal transducer and activator of transcription (STAT), serum- and glucose-regulated kinase 1 (SGK1), caspase recruitment domain-containing protein 11 (CARD11), zinc finger proteins 36L1 (ZFP36L1), PR domain zinc finger protein 1 (PRDM1), pou class 2 associating factor 1 (POU2AF1), and neurogenic locus notch homolog protein 1 (NOTCH1) (Figure 4).6,35,36
 |
Figure 4 Schematic representation of pathological mechanisms DLBCL in splicing mutation. |
Role of Splice Site Mutation on BCL7A Gene
The BCL7A gene is located on chromosome 12 and is affected most commonly by a splicing mutation.6 Its products play a role in B cell activation and tumor suppressor function. In the BCL7A gene, three different splicing site mutations, such as mutant splice site, cryptic splice site, and exon skipping, were found. So, splicing mutations in the BCL7A gene inactivate and alter its normal function.5–7 The tumor suppressor function is one of the functions of genes affected by splice site mutations. The gene’s function is altered due to splice site mutations of the portion of the BCL7A protein’s amino terminal domain, which coded by the BCL7A gene.7 As a result, binding of the BCL7A protein to the Switch/Sucrose non-fermentable complex was affected. The loss of Switch/Sucrose non-fermentable complex integrity suppresses the differentiation of B cells into plasma and memory cells. B cell activation is another function of a gene affected by splice site mutation. This is because BCL7A restoration induces transcriptomic changes in genes involved in B-cell activation. Furthermore, evidence suggests that the BCL7A gene is involved in oncogene inactivation.3,36
Role of Splice Site Mutation on MYD88, CARD11, CD79B and MYD88 Genes
MYD88, CARD11, and CD79B genes are among those whose functions are altered by RNA splicing mutations. Splicing mutations such as exon skipping in MYD88, intron retention in CARD11 and CD79B, and a mutant splice site in CARD1 affect the final product of the gene.23,30 Because there is participation in the inactivation of the nuclear factor kappa-light-chain-enhancer of activated B cells (NF-B) signaling pathway, alteration of the product of these genes plays a significant role in DLBCL pathogenesis.25 The NF-kB signaling pathway has negative feedback for B cell differentiation and positive feedback for B cell proliferation.6 According to the report of available literature, splicing mutations in MYD88, CARD11, and CD79B gene products are the driving force for activating the NF-κB pathway that is implicated in the pathogenesis of DLBCL.37–39 Similarly, splicing factor mutations called spliceosome mutations in pre-mRNAs trigger exon skipping and intron retention. These changes are caused by mutations in the splicing factor 3A and 3B genes.40,41 These shorten the MyD88 gene transcript, which is essential to induce toll-like receptor signaling in cells and the differentiation of B cells. Lack of differentiation may cause DLBCL.40,42 In contrast, two studies found no link between splicing mutations and the MYD88 gene in the pathogenesis of DLBCL.30,34
Role of Splice Site Mutation on STAT Gene
The splicing mutations might be linked to changes in STAT gene products.35 According to the literature, cryptic splice sites, intron retention, and exon skipping all play a role in STAT gene product alteration. Cryptic splicing leads to a lost serine codon at the acceptor sites, while exon skipping and intron retention lead to premature stop codons.43,44 These mutations affect STAT3’s normal function because it mediates the expression of various genes that play an important role in cell proliferation, survival, and differentiation. However, abnormal expression of STAT3 products promotes malignant transformation and tumor progression through oncogenic gene expression and blocking cell differentiation.43
Role of Splice Site Mutation on SGK1, PRDM1, TP53, ZFP36L1, POU2AF1 and NOTCH1 Genes
Splicing mutations such as exon skipping, mutant splice sites, intron retention, cryptic splice sites, and aberrant alternative splicing are also involved in the SGK1 gene. Available data shows that SGK1 gene products are involved in the regulation of cell apoptosis and the cell cycle. So, alteration of this gene loses its tumor suppressor role and restricts B cell differentiation at the germinal center.45,46 Similarly, splicing mutations in the PR domain containing 1 with a zinc finger domain (PRDM1) affect B cell differentiation at the germinal center. Splicing mutations detected in PRDM1 are mutant splice site and exon skipping. It cause premature translation termination and inhibition of B cell differentiation at the germinal center, respectively.6,47,48
Moreover, splicing mutations affect the products of a tumor suppressor gene called TP53. The gene mediates various stimuli for stressed cells through controlling the expression of target genes. In addition, splicing mutations altered the p53 protein, which participates in DNA repair and apoptosis.49 The reports of different publications revealed that the splicing mutations found in the TP53 gene are aberrant alternative splicing, mutant splice sites, and exon skipping. As a result, the tumor suppressor, DNA repair, and apoptotic functions of genes were altered.4,27,31,35,49–51 Similarly, splicing mutations such as exon skipping and mutant splice sites affect the product of ZFP36L1 and POU2AF1 genes that block B cell differentiation at the germinal center.52,53 Furthermore, exon skipping and mutant splice sites altered the apoptotic and tumor suppressor functions of POU2AF1 gene products.6,54
Furthermore, splicing mutations affect the NOTCH1 gene, which encodes a transmembrane protein for regulating cell differentiation, proliferation, and apoptosis. So, altered splicing of NOTCH gene transcripts may affect B-cell lineage development and cause malignant transformations.55 So, altered splicing of NOTCH genes transcripts may affect B-cell lineage development and cause malignant transformations.54 The findings of various studies show that the splicing mutations detected in the NOTCH gene transcript are aberrant alternative splicing and skipping of exon.54,55
Role of Splicing Mutation on SF1, CD58, TNFAIP3, NFKBIE, STXBP2, NFKBI, STXBP, CYLD and PTPN1 Genes
The splicing factor 1 (SF1) gene is one splicing factor gene out of 66 splicing factor coding genes. It recognizes the 3’ splice site by binding the BP of intron.56 In addition, it has played a critical role in the retention of nuclear pre-mRNA during time-alternative splicing. Recently, there has been an increase in the evidence supporting mutations of the SF1 gene related to various cancers, but less in lymphoma.57 Only a couple of studies report mutations in SF1 in DLBCL.56,58 In addition, mutant splice sites and exon skipping altered CD 58 gene products’ normal function. The loss of CD 58 receptors on B cells due to mutation allows tumor cells to avoid immune recognition.51,59 In addition, intron retention and aberrant splicing inhibit the expression of the CD 58 receptor on B cells. As a result, abnormal B cells escaped recognition by cytotoxic T cells and caused overproliferation of abnormal cells.6,59
As reported in different studies, splice site mutations in the TNFAIP3, NFKBIE, and STXBP2 genes play a significant role in DLBCL pathogenesis.3,6,36,60 The TNFAIP3 gene encodes a ubiquitin-modifying enzyme that is involved in the termination of NF-kB signaling pathways because its activation is important for malignant cell proliferation and blocks B cell differentiation.39 Splicing mutation is one of the mechanisms involved in TNFAIP3 gene alteration. As evidence shows, mutant splicing sites and exclusion of exons are splicing mutations involved in the TNFAIP3 gene mutation.8 As a result, the negative feedback for NF-Kb pathways is altered.6,30 Similarly, when the NFKBIA gene was mutated, the NF-B signaling pathway was activated because it codes for IkBs proteins that inhibit NF-kB dimers.6,38 Similarly, during mutation of NFKBIA gene, NF-κB signaling pathway activated because it codes IκBs proteins that inhibits NF-kB dimers.3,39 There are defects in the NFKBIA gene due to the exclusion of an exon that inhibits B cell differentiation at the germinal center, according to the literature.6,8,27 Studies show that the STXBP2 gene is mutated due to exon skipping, but the details of the mechanisms need to be investigated further.44,61
Splicing mutations such as exon skipping, cryptic splices at acceptor sites of exons, and mutant splicing sites were detected in CYLD gene transcripts. These splicing mutations inhibit the tumor suppressor role of the gene through inactivation of NF-Kb signaling pathways. The pathway’s activation may contribute to malignant cells’ ability to proliferate.39,62 Splicing mutations like exon skipping are found in protein tyrosine phosphatase, nonreceptor type 1 (PTPN1) gene transcripts. This gene has crucial negative or positive regulators of JAK/STAT signaling. These functions of genes contribute to preventing the proliferation, differentiation, and survival of tumor cells.63,64 Splicing mutations altered the normal function of the PTPNI gene, leading to enhanced proliferation and survival of malignant B cells and blocking the differentiation of B cells.63
Role of Splicing Mutation on ZEB2, CD70, FOXP1, EBF1 and CD37 Genes
Recently, mutant splice sites were detected in zinc finger E-box-binding homeobox 2 (ZEB2) transcripts. Consequently, the effects of ZEB2 on the growth, migration, invasion, cell cycle distribution, and apoptosis of B cells were explored.6 As hypothesised by different scholars, alteration of ZEB2’s role may inhibit the differentiation of B cells and promote malignant transformation and metastasis.6,51,60 Similarly, malignant transformation and metastasis are detected in the alteration of cluster of differentiation 70 (CD70). This is because, as evidence shows, its tumor suppressor and anti-apoptotic roles are affected due to exon skipping. This may lead to DLBCL formation.6,36 In contrast, one study found no significant link between splicing mutations and changes in CD70 expression.65 It is convincing that CD70 only serves as a co-stimulator during the activation of a signal transduction pathway, and that their function is compensated for by the function of other co-stimulators.3,51
Forkhead box protein P1 (FOXP1) is a master regulator of stem and progenitor cells.65 However, aberrant expression of the oncogenic transcription factor is due to a splicing mutation in FOXP1 detected commonly in DLBCL.6,39 This contributes to tumor cell survival because intron retention during RNA splicing leads to sphingosine-1-phosphate receptor 2 being repressed in the germinal center B-cell. It is important to activate downstream signaling pathways to induce apoptosis, restrict tumor growth, and inactivate oncogenes.65 Similarly, early B-cell factor 1 (EBF1) functions as a regulator of committed B cell progenitors and B cell-specific gene expression programs. Despite the fact that splicing mutations known as exon skipping altered its function, recent studies revealed that splicing mutations affect efficient B cell commitment and differentiation.6,66,67
Splicing mutations such as exon skipping affect cell cycle regulation and the apoptosis role in the proto-oncogene serine/threonine-protein kinase 1. This promotes malignant cell growth and survival through cell cycle dysregulation and inhibition of apoptosis in B cells.68,69 Similarly, due to exon skipping in pre-RNA, the apoptotic role of cluster of differentiation 37 (CD37) is affected. It causes malignant B cells to evade immune cells.70
Role of Splicing Mutation on Others Minor Genes
According to recent findings, B cells in DLBCL fail to express cell-surface molecules required for tumor cell recognition by immune effector cells.5,6,71 It may be due to splicing mutations (exon skipping) that inactivate the β2-microglobulin (B2M) gene. Inactivation of the gene prevents the expression of the cell-surface human leukocyte antigen 1 complex that is necessary for the recognition of tumor cells by CD8+ cytotoxic T cells. As a result, malignant B cells escape from immune surveillance and progress to DLBCL.59,71
In addition, ETS translocation variant 6 (ETV6), cluster of differentiation 83 (CD83), tumor necrosis factor receptor superfamily member 14 (TNFRSF14), myosin-Ie (MYO1E), dual specificity protein phosphatase 2 (DUSP2), lysosomal-associated transmembrane protein 5 (LAPTM5), histone-lysine N-methyltransferase 2D (KMT2D), and T-cell leukemia/lymphoma protein 1A (TCL1A) are less frequently detected genes in DLBCL. To identify the detailed mechanisms involved in the pathogenesis of DLBCL, they require further investigation4,6,27,36,69,70 (Table 1).
 |
Table 1 Summarizes the Role of Splicing Mutation in Different Genes in DLBCL as Well as Their Mechanisms |
Splicing Mutation and Targeted Drugs for the Treatment of DLBCL
Diffuse large B-cell lymphoma is a heterogeneous group of aggressive lymphoma that has specific clinical course and response to therapy.75,76 As a result, DLBCL is still remained challenging for physicians in developing specific targeted treatments for patients care. Recurrent mutations such as splicing mutation associated with poor clinical outcomes in DLBCL.77 The drugs targeted to RNA binding regulatory proteins have predictive prognostic values to treat patients with DLBCL.76,78
As evidences show in the past decades, immunochemotheraphy such as rituximab plus cyclophosphamide, vincristine, etoposide, brentuximab, bleomycin, doxorubicin, and prednisone have a significant role in DLBCL patients to extend survival rate and cure rate as well as decrease relapse rate.77,79–81 Those drugs target RNA binding protein during splicing process. RNAs splicing is crucial process for the formation of mature mRNA which codes specific proteins. The proteins formed from translation regulate cell apoptosis, differentiation, DNA repair and inactivate oncogene.35 Hence, the immunochemotheraphy drugs may target RNA binding factors and prevent the occurrence of mutation. However, prognosis of some patients still refractory to standard treatment. Thus, in order to improve prognostic outcomes, new treatment option is under the preclinical trials.56,77,79
Furthermore, currently some drugs such as inhibitor of splice factor kinases (dasatinib), inhibitor of NF-kB pathway (ibrutinib), inhibitor of JAK-STAT pathway (niacinamide) and inhibitors bruton tyrosine kinase pathway (calabrutinib, tirabrutinib, and spebrutinib) on pre-clinical stage are reported as novel treatment option to treat patient with DLBCL.75,76,82 The pathways targeted by drugs are important for malignant B cell proliferation and differentiation. Consequently, the drugs inactivate the malignant B cells proliferation, differentiation and induce malignant B cell apoptosis at germinal center during RNA splicing. Based on subtypes of DLBCL, providing and applying specific therapeutic strategies is important to treat patients with DLBCL.76,83
Conclusion
In conclusion, different types of splicing mutations were detected in DLBCL. These mutations are mutant splice site, aberrant alternative splicing, exon skipping and intron retention. Most of these RNA splicing mutations affect normal function of different genes. The genes most frequently affected by splicing mutation in DLBCL are BCL7A, CD79B, SF1, MYD88, TP53, STAT, SGK1, ZFP36L1 and POU2AF1. As a result of alteration in different gene by RNA splicing mutation, malignant transformation, cancer progression, and metastasis occur in DLBCL. Although, most of the literature published is focused on only the role of splicing mutation for DLBCL; risk factors, prevention mechanisms, and treatment options for these mutations were not well discussed. As a recommendation, it is important for researchers to find risk factors, prevention mechanisms, prognostic role and treatment options of RNA splicing mutation in DLBCL.
Abbreviations
BCL7A, B-cell Lymphoma 7 Protein Family Member; BP, branch points; CARD11, caspase recruitment domain-containing protein 11; CD, cluster of differentiation; CD79B, cluster of differentiation 79B; cHL, classical Hodgkin lymphoma; CYLD, cylindromatosis lysine differentiation gene; DLBCL: diffuse large B cell lymphoma; MYD88, myeloid differentiation primary response gene 88; NF-κB, nuclear factor kappa B cells; NOTCH1, neurogenic locus notch homolog protein 1; Pre-RNA, premature ribonucleic acid; SF1, splicing factor 1; snRNPS, small nuclear ribonucleoproteins; STAT, signal transducer and activator of transcription; TP53, tumor protein P53; U2AF, U2 auxiliary Factor.
Acknowledgment
We would like to acknowledge the authors of the studies included in this review.
Disclosure
The authors report no conflicts of interest in this work.
References
1. de Leval L, Jaffe ES. Lymphoma classification. Cancer J. 2020;26(3):176–185. doi:10.1097/PPO.0000000000000451
2. Swerdlow SH, Campo E, Pileri SA, et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127(20):2375–2390. doi:10.1182/blood-2016-01-643569
3. Moghanibashi M, Mohamadynejad P. Splicing in Cancer. hematologica. 2022;14(23):214.
4. Asmar F, Punj V, Christensen J, et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica. 2013;98(12):1912. doi:10.3324/haematol.2013.088740
5. Chapuy B, Stewart C, Dunford AJ, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat med. 2018;24(5):679–690. doi:10.1038/s41591-018-0016-8
6. Andrades A, Álvarez-Pérez JC, Patiño-Mercau JR, Cuadros M, Baliñas-Gavira C, Medina PP. Recurrent splice site mutations affect key diffuse large B-cell lymphoma genes. Blood. 2022;139(15):2406–2410. doi:10.1182/blood.2021011708
7. Reddy A, Zhang J, Davis NS, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171(2):481–94.e15. doi:10.1016/j.cell.2017.09.027
8. Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396–1407. doi:10.1056/NEJMoa1801445
9. Wang X, Hong Y, Meng S, et al. A novel immune-related epigenetic signature based on the transcriptome for predicting the prognosis and therapeutic response of patients with diffuse large B-cell lymphoma. Clin Immun. 2022;243:109105. doi:10.1016/j.clim.2022.109105
10. Zhang S, Zhang T, Liu H, et al. Tracking the evolution of untreated high‐intermediate/high‐risk diffuse large B‐cell lymphoma by circulating tumour DNA. Br J Haematol. 2022;196(3):617–628. doi:10.1111/bjh.17894
11. Zhang H, Lu Y, Zhang T, et al. PIM1 genetic alterations associated with distinct molecular profiles, phenotypes and drug responses in diffuse large B‐cell lymphoma. Clin Transl Med. 2022;12(4). doi:10.1002/ctm2.808
12. Hahn CN, Venugopal P, Scott HS, Hiwase DK. Splice factor mutations and alternative splicing as drivers of hematopoietic malignancy. Immunol Rev. 2015;263(1):257–278. doi:10.1111/imr.12241
13. Ebert B, Bernard OA. Mutations in RNA splicing machinery in human cancers. N Engl J Med. 2011;365(26):2534–2535. doi:10.1056/NEJMe1111584
14. Saez B, Walter MJ, Graubert TA. Splicing factor gene mutations in hematologic malignancies. Blood. 2017;129(10):1260–1269. doi:10.1182/blood-2016-10-692400
15. Akinyi MV, Frilander MJ. At the intersection of major and minor spliceosomes: crosstalk mechanisms and their impact on gene expression. Front Genet. 2021;12:700744. doi:10.3389/fgene.2021.700744
16. Chen W, Moore MJ. Spliceosomes. Curr. 2015;25(5):R181–R183. doi:10.1016/j.cub.2014.11.059
17. Kitamura K, Nimura K. Regulation of RNA splicing: aberrant splicing regulation and therapeutic targets in cancer. Cells. 2021;10(4):923. doi:10.3390/cells10040923
18. Olthof AM, White AK, Mieruszynski S, et al. Disruption of exon-bridging interactions between the minor and major spliceosomes results in alternative splicing around minor introns. Nucleic Acids Res. 2021;49(6):3524–3545. doi:10.1093/nar/gkab118
19. Chen S, Benbarche S, Abdel-Wahab O. Splicing factor mutations in hematologic malignancies. Blood. 2021;138(8):599–612. doi:10.1182/blood.2019004260
20. Gurnari C, Pagliuca S, Visconte V. Alternative splicing in myeloid malignancies. Biomedicine. 2021;9(12):1844.
21. Leivonen S, Taskinen M, Cervera A, et al. Alternative splicing discriminates molecular subtypes and has prognostic impact in diffuse large B-cell lymphoma. Blood Cancer j. 2017;7(8):596. doi:10.1038/bcj.2017.71
22. Taylor J, Lee SC. Mutations in spliceosome genes and therapeutic opportunities in myeloid malignancies. Genes Chromosom Cancer. 2019;58(12):889–902. doi:10.1002/gcc.22784
23. Ngo VN, Young RM, Schmitz R, et al. Oncogenically active MYD88 mutations in human lymphoma. Nature. 2011;470(7332):115–119. doi:10.1038/nature09671
24. Schneider M, Schneider S, Zühlke‐Jenisch R, et al. Alterations of the CD58 gene in classical Hodgkin lymphoma. Gene Chromosom Cancer. 2015;54(10):638–645. doi:10.1002/gcc.22276
25. Takeuchi T, Yamaguchi M, Kobayashi K, et al. MYD88, CD79B, and CARD11 gene mutations in CD5‐positive diffuse large B‐cell lymphoma. Cancer. 2017;123(7):1166–1173. doi:10.1002/cncr.30404
26. Voropaeva EN, Pospelova TI, Voevoda MI, Maksimov VN, Orlov YL, Seregina OB. Clinical aspects of TP53 gene inactivation in diffuse large B-cell lymphoma. BMC Genom. 2019;12(2):35–44. doi:10.1186/s12920-019-0484-9
27. Chapuy B, Stewart C, Dunford AJ, et al. Author Correction: molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat med. 2018;24(8):1290–1291. doi:10.1038/s41591-018-0097-4
28. Wilkinson ME, Charenton C, Nagai K. RNA splicing by the spliceosome. Annu Rev Biochem. 2020;89(1):359–388. doi:10.1146/annurev-biochem-091719-064225
29. Zhou J, Chng WJ. Aberrant RNA splicing and mutations in spliceosome complex in acute myeloid leukemia. Stem Cell Investig. 2017;4:6. doi:10.21037/sci.2017.01.06
30. Dobashi A. Molecular pathogenesis of diffuse large B-cell lymphoma. JCEH. 2016;56(2):71–78. doi:10.3960/jslrt.56.71
31. Jung H, Lee KS, Choi JK. Comprehensive characterisation of intronic mis-splicing mutations in human cancers. Oncogene. 2021;40(7):1347–1361. doi:10.1038/s41388-020-01614-3
32. Yang H, Beutler B, Zhang D. Emerging roles of spliceosome in cancer and immunity. Protein Cell. 2021;13(8):559–579.
33. Zhang Y, Qian J, Gu C, Yang Y. Alternative splicing and cancer: a systematic review. Signal Transduct Target Ther. 2021;6(1):78. doi:10.1038/s41392-021-00486-7
34. Aggarwal V, Das A, Bal A, et al. MYD88, CARD11, and CD79B oncogenic mutations are rare events in the Indian cohort of de novo nodal diffuse large B-cell lymphoma. AIMM. 2019;27(4):311–318. doi:10.1097/PAI.0000000000000585
35. Caeser R, Di Re M, Krupka JA, et al. Genetic modification of primary human B cells to model high-grade lymphoma. Natcommun. 2019;10(1):1–16.
36. Baliñas-Gavira C, Rodríguez MI, Andrades A, et al. Frequent mutations in the amino-terminal domain of BCL7A impair its tumor suppressor role in DLBCL. Leukemia. 2020;34(10):2722–2735. doi:10.1038/s41375-020-0919-5
37. Alizadeh AA, Eisen MB, Davis RE, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403(6769):503–511. doi:10.1038/35000501
38. Jallades L, Baseggio L, Sujobert P, et al. Exome sequencing identifies recurrent BCOR alterations and the absence of KLF2, TNFAIP3 and MYD88 mutations in splenic diffuse red pulp small B-cell lymphoma. Haematologica. 2017;102(10):1758–1766. doi:10.3324/haematol.2016.160192
39. Koens L, Zoutman WH, Ngarmlertsirichai P, et al. Nuclear factor-κB pathway-activating gene aberrancies in primary cutaneous large B-cell lymphoma, leg type. J Invest Dermatol. 2014;134(1):290–292. doi:10.1038/jid.2013.265
40. Cardona Gloria Y, Bernhart SH, Fillinger S, et al. Absence of non-canonical, inhibitory MYD88 splice variants in B cell lymphomas correlates with sustained NF-κB signaling. Front Immunol. 2021;12(8):616–651. doi:10.3389/fimmu.2021.616451
41. Kishimoto T, Ying B-W, Tsuru S, et al. Molecular clock of neutral mutations in a fitness-increasing evolutionary process. PLoS Genet. 2015;11(7):e1005392. doi:10.1371/journal.pgen.1005392
42. Pimentel H, Parra M, Gee S, et al. A dynamic alternative splicing program regulates gene expression during terminal erythropoiesis. Nucleic Acids Res. 2014;42(6):4031–4042. doi:10.1093/nar/gkt1388
43. Turton KB, Annis DS, Rui L, Esnault S, Mosher DF. Ratios of four STAT3 splice variants in human eosinophils and diffuse large B cell lymphoma cells. PLoS One. 2015;10(5):e0127243. doi:10.1371/journal.pone.0127243
44. Zhu F, Wang KB, Rui L. STAT3 activation and oncogenesis in lymphoma. Cancers. 2019;12(1):19. doi:10.3390/cancers12010019
45. Gao J, Sidiropoulou E, Walker I, et al. SGK1 mutations in DLBCL generate hyperstable protein neoisoforms that promote AKT Independence. Blood. 2021;138(11):959–964. doi:10.1182/blood.2020010432
46. Hartmann S, Schuhmacher B, Rausch T, et al. Highly recurrent mutations of SGK1, DUSP2 and JUNB in nodular lymphocyte predominant Hodgkin lymphoma. Leukemia. 2016;30(4):844–853. doi:10.1038/leu.2015.328
47. Pasqualucci L, Compagno M, Houldsworth J, et al. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. JEM. 2006;203(2):311–317. doi:10.1084/jem.20052204
48. Mandelbaum J, Bhagat G, Tang H, et al. BLIMP1 is a tumor suppressor gene frequently disrupted in activated B cell-like diffuse large B cell lymphoma. Cancer Cell. 2010;18(6):568–579. doi:10.1016/j.ccr.2010.10.030
49. Xu-Monette ZY, Medeiros LJ, Li Y, et al. Dysfunction of the TP53 tumor suppressor gene in lymphoid malignancies. Blood. 2012;119(16):3668–3683. doi:10.1182/blood-2011-11-366062
50. Zlamalikova L, Moulis M, Ravcukova B, et al. Complex analysis of the TP53 tumor suppressor in mantle cell and diffuse large B-cell lymphomas. Oncol Rep. 2017;38(4):2535–2542. doi:10.3892/or.2017.5891
51. De Miranda NF, Georgiou K, Chen L, et al. Exome sequencing reveals novel mutation targets in diffuse large B-cell lymphomas derived from Chinese patients. Blood. 2014;124(16):2544–2553. doi:10.1182/blood-2013-12-546309
52. Hegde VV. Zinc Finger Protein 36L1 (ZFP36L1): Gene Expression, Regulation and Interactions with Immune Receptors in Human Tumour Cells. India: University of Essex; 2020.
53. Oliveira C, Faoro H, Alves LR, Goldenberg S. RNA-binding proteins and their role in the regulation of gene expression in Trypanosoma cruzi and Saccharomyces cerevisiae. Genet Mol Biol. 2017;40(12):22–30. doi:10.1590/1678-4685-gmb-2016-0258
54. Jespersen DS, Schönherz AA, Due H, Bøgsted M, Sondergaard TE, Dybkær K. Expression of NOTCH3 exon 16 differentiates Diffuse Large B-cell Lymphoma into molecular subtypes and is associated with prognosis. Sci Rep. 2019;9(1):335. doi:10.1038/s41598-018-36680-x
55. Nowell CS, Radtke F. Notch as a tumour suppressor. Nat Rev Cancer. 2017;17(3):145–159. doi:10.1038/nrc.2016.145
56. Zhang R, Lin P, Yang X, et al. Survival associated alternative splicing events in diffuse large B-cell lymphoma. Am J Transl Res. 2018;10(8):2636.
57. Zhang Y, Dong W, Wang J, Cai J, Wang Z. Human omental adipose-derived mesenchymal stem cell-conditioned medium alters the proteomic profile of epithelial ovarian cancer cell lines in vitro. OncoTargets Ther. 2017;10(18):1655. doi:10.2147/OTT.S129502
58. Fujimoto A, Okada Y, Boroevich KA, Tsunoda T, Taniguchi H, Nakagawa H. Systematic analysis of mutation distribution in three dimensional protein structures identifies cancer driver genes. Sci Rep. 2016;6(1):1–9. doi:10.1038/srep26483
59. Challa-Malladi M, Lieu YK, Califano O, et al. Combined genetic inactivation of β2-Microglobulin and CD58 reveals frequent escape from immune recognition in diffuse large B cell lymphoma. Cancer Cell. 2011;20(6):728–740. doi:10.1016/j.ccr.2011.11.006
60. Li Q, Ma L, Wu Z, et al. Zinc finger E‑box binding homeobox 2 functions as an oncogene in human laryngeal squamous cell carcinoma. Mol Med Rep. 2019;19(6):4545–4552. doi:10.3892/mmr.2019.10126
61. Schmidt A, Schmitz R, Giefing M, et al. Rare occurrence of biallelic CYLD gene mutations in classical Hodgkin lymphoma. Genes Chromosom Cancer. 2010;49(9):803–809. doi:10.1002/gcc.20789
62. Xu X, Wei T, Zhong W, et al. Down-regulation of cylindromatosis protein phosphorylation by BTK inhibitor promotes apoptosis of non-GCB-diffuse large B-cell lymphoma. Cancer Cell Int. 2021;21(1):195. doi:10.1186/s12935-021-01891-2
63. Zahn M, Kaluszniak B, Möller P, Marienfeld R. The PTP1B mutant PTP1B∆ 2–4 is a positive regulator of the JAK/STAT signalling pathway in Hodgkin lymphoma. J Carcinog. 2021;42(4):517–527. doi:10.1093/carcin/bgaa144
64. Zahn M, Marienfeld R, Melzner I, et al. A novel PTPN1 splice variant upregulates JAK/STAT activity in classical Hodgkin lymphoma cells. Blood. 2017;129(11):1480–1490. doi:10.1182/blood-2016-06-720516
65. Walker MP, Stopford CM, Cederlund M, et al. FOXP1 potentiates Wnt/β-catenin signaling in diffuse large B cell lymphoma. Sci.Signal. 2015;8(362):ra12. doi:10.1126/scisignal.2005654
66. Nutt SL, Kee BL. The transcriptional regulation of B cell lineage commitment. Immunity. 2007;26(6):715–725. doi:10.1016/j.immuni.2007.05.010
67. Møller MB, Kania P, Ino Y, et al. Frequent disruption of the RB1 pathway in diffuse large B cell lymphoma: prognostic significance of E2F-1 and p16INK4A. Leukemia. 2000;14(5):898–904. doi:10.1038/sj.leu.2401761
68. Alvarado Y, Giles FJ, Swords RT. The PIM kinases in hematological cancers. Expert Rev Hematol. 2012;5(1):81–96. doi:10.1586/ehm.11.69
69. de Miranda NF, Peng R, Georgiou K, et al. DNA repair genes are selectively mutated in diffuse large B cell lymphomas. J Exp Med J EXP MED. 2013;210(9):1729–1742. doi:10.1084/jem.20122842
70. Elfrink S, de Winde CM, van den Brand M, et al. High frequency of inactivating tetraspanin C D37 mutations in diffuse large B-cell lymphoma at immune-privileged sites. Blood. 2019;134(12):946–950. doi:10.1182/blood.2019001185
71. Fan Z, Pei R, Sha K, Chen L, Wang T, Lu Y. Comprehensive characterization of driver genes in diffuse large B cell lymphoma. Oncol Lett. 2020;20(1):382–390. doi:10.3892/ol.2020.11552
72. Krysiak K, Gomez F, White BS, et al. Recurrent somatic mutations affecting B-cell receptor signaling pathway genes in follicular lymphoma. Blood. 2017;129(4):473–483. doi:10.1182/blood-2016-07-729954
73. Guo B, Huang Y, Duan Y, Liao C, Cen H. SGK1 mutation status can further stratify patients with germinal center B‐cell‐like diffuse large B‐cell lymphoma into different prognostic subgroups. Cancer Med. 2022;11(5):1281–1291. doi:10.1002/cam4.4550
74. Gribben JG. The sequence of events in diffuse large B-cell lymphoma. Blood. 2013;122(7):1097–1098. doi:10.1182/blood-2013-06-506089
75. Zhang J, Gu Y, Chen B. Drug-resistance mechanism and new targeted drugs and treatments of relapse and refractory DLBCL. Cancer Manag Res. 2023;245–255. doi:10.2147/CMAR.S400013
76. Vaqué JP, Martínez N, Batlle-López A, et al. B-cell lymphoma mutations: improving diagnostics and enabling targeted therapies. Haematologica. 2014;99(2):222–231. doi:10.3324/haematol.2013.096248
77. Shen R, Fu D, Dong L, et al. Simplified algorithm for genetic subtyping in diffuse large B-cell lymphoma. Signal Transduct Target Ther. 2023;8(1):145. doi:10.1038/s41392-023-01358-y
78. Xu-Monette ZY, Wu L, Visco C, et al. Mutational profile and prognostic significance of TP53 in diffuse large B-cell lymphoma patients treated with R-CHOP: report from an International DLBCL Rituximab-CHOP Consortium Program Study. Blood. 2012;120(19):3986–3996. doi:10.1182/blood-2012-05-433334
79. Kusowska A, Kubacz M, Krawczyk M, Slusarczyk A, Winiarska M, Bobrowicz M. Molecular aspects of resistance to immunotherapies advances in understanding and management of diffuse large B-cell lymphoma. Int J Mol Sci. 2022;23(3):1501. doi:10.3390/ijms23031501
80. Wilson WH, Young RM, Schmitz R, et al. Targeting B cell receptor signaling with ibrutinib in diffuse large B cell lymphoma. Nat.Med. 2015;21(8):922–926. doi:10.1038/nm.3884
81. Yu H, Sotillo E, Harrington C, et al. Repeated loss of target surface antigen after immunotherapy in primary mediastinal large B cell lymphoma. Am J Hematol. 2017;92(1):E11. doi:10.1002/ajh.24594
82. Sciarrillo R, Wojtuszkiewicz A, Assaraf YG, et al. The role of alternative splicing in cancer: from oncogenesis to drug resistance. Drug Resist Updat. 2020;53:100728. doi:10.1016/j.drup.2020.100728
83. Danilov AV, Magagnoli M, Matasar MJ. Translating the biology of diffuse large B-cell lymphoma into treatment. Oncologist. 2022;27(1):57–66. doi:10.1093/oncolo/oyab004
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
