Back to Journals » OncoTargets and Therapy » Volume 9
Role of p14ARF and p15INK4B promoter methylation in patients with lung cancer: a systematic meta-analysis
Authors Yang X, Yang L, Dai W, Ye B
Received 13 July 2016
Accepted for publication 31 August 2016
Published 11 November 2016 Volume 2016:9 Pages 6977—6985
DOI https://doi.org/10.2147/OTT.S117161
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr XuYu Yang
Xinmei Yang,1,* Lei Yang,2,* Wanrong Dai,3 Bo Ye4
1Department of Oncology, the First Affiliated Hospital of Jiaxing University, Jiaxing, 2Cancer Center, The First Hospital of Jilin University, Changchun, 3Department of Pharmacy, The First Affiliated Hospital, Zhejiang University, Zhejiang, 4Department of Thoracic Surgery, Hangzhou Red Cross Hospital, Hangzhou, People’s Republic of China
*These authors contributed equally to this work
Background: The cyclin-dependent kinase inhibitors p14ARF and p15INK4B are tumor suppressor genes that have been reported to be silenced through promoter methylation in many human cancers. However, the strength of association between p14ARF or p15INK4B promoter methylation and lung cancer remains unclear. Thus, we first determined whether p14ARF and p15INK4B promoter methylation played a key role in the carcinogenesis of lung cancer.
Methods: Eligible studies were selected from the online electronic databases. The pooled odds ratios or hazard ratios and 95% confidence intervals were calculated and summarized.
Results: Finally, 12 studies with 625 lung cancer samples and 488 nontumor samples were included under the fixed-effects model. The pooled odds ratio showed that p14ARF promoter methylation was observed to be significantly higher in non-small-cell lung cancer (NSCLC) than in nontumor samples (P<0.001). No significant correlation was found between p15INK4B promoter methylation and lung cancer (P=0.27). Subgroup analysis of ethnicity revealed that p14ARF promoter methylation was significantly related to the risk of NSCLC in Asian and Caucasian populations. Subgroup analysis of sample type demonstrated that p14ARF promoter methylation was correlated with the risk of NSCLC in tissue samples (P<0.001), but not in bronchoalveolar lavage fluid and blood samples. P14ARF promoter methylation from one study was not significantly correlated with overall survival of patients with NSCLC. Promoter methylation of p14ARF and p15INK4B was not correlated with clinicopathological characteristics, such as gender status, smoking status, tumor differentiation, and tumor stage (P>0.05).
Conclusion: Our findings suggested that p14ARF promoter methylation may play an important role in the carcinogenesis of lung cancer, but not p15INK4B promoter methylation. Promoter methylation of p14ARF and p15INK4B was not associated with clinicopathological parameters. However, more extensive large-scale studies are essential to further validate our study.
Keywords: p14ARF, p15INK4B, methylation, lung cancer, overall survival
Introduction
Lung cancer is one of the most common human malignant tumors and is the leading cause of cancer-related deaths in the world.1 Based on global cancer statistics, ~1,824,700 new cases of lung cancer were clinically diagnosed, with an estimated 1,589,900 deaths in 2012.1 Human lung cancer consists of two major types: small-cell lung cancer and non-small-cell lung cancer (NSCLC). NSCLC is the most common type and accounts for 85% of lung cancer cases, including adenocarcinoma, squamous cell carcinoma, large cell carcinoma, and adenosquamous carcinoma.2,3 Due to the lack of an early diagnostic method, >40% of NSCLC patients have developed metastases by the time of diagnosis,4 and the average 5-year survival rate is currently 18%.5 Therefore, early diagnosis is crucial for improving the survival rate.6
Epigenetic modifications are involved in humor cancers, DNA methylation, as a crucial mechanism of epigenetic changes, leads to target gene silencing, and plays a key role in the carcinogenesis and progression of cancer.7–9 A large number of studies have reported that genes with aberrant promoter methylation are significantly correlated with lung cancer.10–13 Located on the human chromosome 9p21, the INK4/ARF locus encodes three cell-cycle inhibitory proteins involving p15INK4B, p14ARF, and p16INK4A; p16INK4A is encoded by cyclin-dependent kinase inhibitor 2A (CDKN2A), p14ARF is encoded by an alternative reading frame of CDKN2A, and p15INK4B is encoded by cyclin-dependent kinase inhibitor 2B (CDKN2B), which plays roles in the regulation of p53 and retinoblastoma pathways.14–16 Inactivation of p15INK4B, p14ARF, and p16INK4A as tumor suppressor genes is one of the most common events in human cancers.17–19 The loss of p15INK4B expression via promoter methylation has been reported in lung cancer cell lines.20 Seike et al reported that aberrant methylation of p15INK4B was not detected in lung cancer tissues.21 Wang et al reported that aberrant DNA methylation may be the most common mechanism of inactivating cancer-related genes in lung cancer, including p15INK4B.22 Yanagawa et al reported that aberrant promoter methylation of p14ARF was correlated with lung cancer.23 Zhang et al reported that p14ARF promoter methylation was not correlated with lung cancer.24
Thus, there were also contradictory results concerning the methylation frequency of p14ARF and p15INK4B promoter in lung cancer samples. The current study was first analyzed to better identify the association between p14ARF and p15INK4B promoter methylation and lung cancer.
Materials and methods
Search for eligible studies
A systematic search was performed through online literature databases (PubMed, EMBASE, EBSCO, Wangfang, and Cochrane Library). The following search strategy was used: “methylation,” “hypermethylation,” or “promoter methylation”; and “lung cancer,” “lung carcinoma,” or “lung tumor”; and “p14,” “p14ARF,” “p15,” “p15INK4B,” “CDKN2B,” “cyclin-dependent kinase inhibitor 2.” The search was updated till June 24, 2016. Additionally, we manually searched the references of the selected studies to obtain additional studies. The full texts of all of the qualified studies were published.
Inclusion criteria
Studies were included if they met all the following criteria: 1) Study was an original case-control study on the association between p14ARF or p15INK4B gene promoter methylation and lung cancer; 2) Patients were diagnosed as lung cancer based on histopathological examination; 3) Study had sufficient data to calculate the pooled odds ratios (ORs) or hazard ratios (HRs) and 95% confidence intervals (CIs) for the meta-analysis; and 4) The most recent study with more information was selected when several publications contained duplicated data.
Data extraction
The data obtained from the publications included first author’s name, year of publication, country, ethnicity, sample type, methylation detection method, histology, number of participants, overall survival (OS), and methylation frequency. The various sample types included tissue, bronchoalveolar lavage fluid (BALF), and blood samples, clinicopathological parameters, such as gender status, smoking status, tumor differentiation, and tumor stage. The selection of eligible studies and data extraction were independently performed by two reviewers (LY and WD).
Statistical analysis
The current meta-analysis was performed using the STATA software (version 12.0, Stata Corporation, College Station, TX, USA). The pooled ORs and 95% CIs were calculated to determine the correlation between p14ARF or p15INK4B gene promoter methylation and lung cancer. The pooled HR with 95% CI was used to evaluate the impact of p14ARF promoter methylation on OS of NSCLC patients. Between-study heterogeneity was estimated based on the Cochran’s Q-test and I2 statistic.25 A random-effects model was applied for the meta-analysis with significant heterogeneity (I2>50% and P<0.1); otherwise, the fixed-effects model was used.26,27 P-value <0.05 was considered to be significant.
Results
Study characteristics
As shown in Figure 1, 269 potentially relevant articles were obtained from the PubMed, EMBASE, EBSCO, Wangfang, and Cochrane Library databases. After a series of selection procedures, a total of 12 studies involving 625 lung cancer samples and 488 nontumor samples were included in the current study. Eight studies with 505 NSCLC samples and 419 nontumor samples analyzed the relationship between p14ARF promoter methylation and NSCLC.23,24,28–33 Of 8 studies, 7 studies used methylation-specific polymerase chain reaction and 1 study used quantitative fluorogenic real-time polymerase chain reaction. Four studies with 120 lung cancer samples and 69 nontumor samples evaluated the relationship between p15INK4B promoter methylation and lung cancer.21,22,34,35 Of 4 studies, 3 studies used methylation-specific polymerase chain reaction detection and 1 study used 3-dimensional, polyacrylamide gel-based DNA microarray coupled with linker-polymerase chain reaction. Four studies evaluated the correlation of p14ARF promoter methylation with clinicopathological features.23,30,32,33 Two studies assessed the correlation of p15INK4B promoter methylation with clinicopathological features.22,34 The general characteristics of the included studies are presented in Table 1.
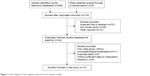  | Figure 1 Flow diagram of the stepwise selection from relevant studies. |
Promoter methylation of p14ARF and p15INK4B in lung cancer
Substantial heterogeneity among studies was not detected (p14ARF: I2=0.0%, P=0.900; p15INK4B: I2=0.0%, P=0.992), as shown in Figures 2 and 3, indicating that our results were stable and reliable, with no evidence of heterogeneity. The pooled OR from 8 studies with 505 NSCLC samples and 419 nontumor samples showed that p14ARF promoter methylation status was significantly higher in NSCLC than in nontumor samples (OR =4.94, 95% CI: 2.79–8.76, P<0.001) (Figure 2), which suggested that p14ARF promoter methylation may play a key role in the initiation of NSCLC. The pooled OR from 4 studies involving 120 lung cancer samples and 69 nontumor samples demonstrated that p15INK4B promoter methylation status had a similar OR among lung cancer and nontumor samples (OR =2.76, 95% CI: 0.46–16.70, P=0.27) (Figure 3), indicating that p15INK4B promoter methylation was not significantly associated with a risk of lung cancer.
Subgroup analyses of p14ARF promoter methylation
Subgroup analyses based on the ethnic population (Asian population and Caucasian population) and sample type (tissue, BALF, and blood) were analyzed to find the different strength of association. Subgroup analysis of ethnicity revealed that p14ARF promoter methylation was significantly correlated with an increased risk of NSCLC in Asians and Caucasians (OR =4.54, 95% CI: 2.47–8.32, P<0.001; OR =8.05, 95% CI: 1.39–46.50, P=0.02; respectively) (Figure 4). Subgroup analysis of sample type demonstrated that p14ARF promoter methylation had a significantly increased risk of NSCLC in tissue samples (OR =4.55, 95% CI: 2.54–8.18, P<0.001), but not in BALF and blood samples (Figure 5). The results of BALF and blood samples should be carefully considered using smaller sample sizes.
Prognostic value of p14ARF promoter methylation
The pooled HR from 1 study with 92 NSCLC patients demonstrated that p14ARF promoter methylation was not significantly correlated with OS of patients with NSCLC (HR =0.57, 95% CI: 0.19–1.73).29 Additional studies with larger subjects are needed to assess the correlation between p14ARF promoter methylation and OS in NSCLC in the future.
Association of p14ARF and p15INK4B promoter methylation with clinicopathological features
We determined whether p14ARF and p15INK4B promoter methylation was associated with clinicopathological characteristics, including four studies with 259 NSCLC patients and 2 studies with 73 patients with NSCLC, respectively. Our results showed that aberrant promoter methylation of p14ARF was not correlated with clinicopathological characteristics (P>0.05), including tumor stage, tumor histology, gender status, tumor differentiation, smoking status, and lymph node status (Figure 6). Aberrant promoter methylation of p15INK4B was also not associated with clinicopathological features (P>0.05), including gender status, tumor histology, tumor differentiation, and tumor stage (Figure 7).
Discussion
Tumor suppressor genes p16INK4A, p14ARF, and p15INK4B encoding cell-cycle regulatory proteins play a crucial role in the negative regulation of the cell cycle and the inhibition of the cell proliferation.36 The silencing of p16INK4A, p14ARF, and p15INK4B genes by DNA methylation of the C-phosphate-G (CpG) islands of the promoter regions has been reported to be involved in the carcinogenesis and be an early biologic event in many cancers.37–40 Based on a meta-analysis, Gu et al reported that p16INK4A promoter methylation may play a key role in the tumorigenesis of lung cancer.41 However, whether p14ARF or p15INK4B promoter methylation plays a crucial role in the carcinogenesis of lung cancer, which remains to be certified. The results were inconsistent with regard to p14ARF and p15INK4B promoter methylation rate in lung cancer. For example, Topaloglu et al reported that p14ARF promoter was absent in methylation in NSCLC.30 Fischer et al reported that p14ARF promoter had a methylation frequency of 30.4% in NSCLC.29 In addition, Seike et al showed that p15INK4B promoter had no methylation,21 while Chaussade et al showed that p15INK4B promoter had a methylation rate of 100% in lung cancer.35 Therefore, we performed a meta-analysis to evaluate the strength of p14ARF and p15INK4B promoter methylation on lung cancer risk.
There was no evidence of the heterogeneity in the current meta-analysis, presenting no obvious publication bias. Our findings demonstrated that p14ARF promoter methylation status had a significantly higher OR in NSCLC than in nontumor samples, while significant correlation was not observed between p15INK4B promoter methylation and lung cancer, suggesting that p14ARF promoter methylation may play an important role in the initiation of NSCLC. However, the result of p15INK4B promoter methylation should be prudent as only 120 lung cancer samples were included in our analysis.
Next, subgroup analyses of the ethnic population (Asians and Caucasians) and sample type (tissue, BALF, and blood) were conducted to find the different association between p14ARF promoter methylation and different subgroups in NSCLC versus nontumor samples. Significant correlation between Asian population and Caucasian population in p14ARF promoter methylation was found in subgroup analysis of ethnicity, which suggested that Asian and Caucasian populations were susceptible to p14ARF promoter methylation. According to subgroup analysis of sample type, a significant association between p14ARF promoter methylation and NSCLC was observed in tissue subgroup, but not in BALF and blood subgroups. The results should be carefully considered as only one or two studies with smaller subjects were analyzed in BALF and blood subgroups.
Previous several meta-analyses had evaluated the correlation of p16INK4A methylation with clinicopathological parameters in lung cancer,11,42,43 which suggested that p16INK4A methylation was associated with smoking status and tumor histology. However, the correlation of p14ARF and p15INK4B promoter methylation with clinicopathological features was not determined. Our findings suggested that p14ARF and p15INK4B promoter methylation was not correlated with clinicopathological characteristics, such as gender status, tumor histology, tumor differentiation, and tumor stage. Based on small sample sizes (p14ARF: 259 NSCLC patients, p15INK4B: 73 patients with NSCLC), more studies with larger sample size should be done in the future.
Limitations
There were several limitations in the present meta-analysis. First, there might be selection bias because eligible studies were restricted to articles published in English and Chinese, studies with other language and other styles, such as conference abstracts were missed. Second, the main ethnic population consisted of Asian and Caucasian populations, and other ethnicities, such as Africans, were insufficient. Third, only smaller subjects were included in BALF and blood subgroups; more studies with larger sample sizes are essential to further determine whether p14ARF promoter methylation can become a promising biomarker based on BALF or blood detection. Fourth, only 1 study reported that p14ARF promoter methylation was not associated with the prognosis of NSCLC patients in OS; further large-scale studies with larger subjects are very necessary in the future. Finally, sample sizes on clinicopathological features were smaller in this study.
Conclusion
The results suggested that p14ARF promoter methylation may play a pivotal role in the carcinogenesis of lung cancer, but not p15INK4B promoter methylation. In addition, p14ARF promoter methylation was a susceptible gene for Asians and Caucasians. Aberrant promoter methylation of p14ARF and p15INK4B was not associated with clinicopathological features. Additional studies with larger subjects are needed to further validate our results.
Acknowledgments
This study was supported by grants from Development Center for Medical Science and Technology National Health Commission of the People’s Republic of China (W2012FZ120), Key Disciplines of Jiaxing (Oncology) (04-F-14), Science and Technology Projects of Jiaxing (2014AY21030-4), and Innovation team of Lung Cancer in Early Diagnosis and Comprehensive Treatment (2014).
Author contributions
All authors contributed toward data analysis, drafting and revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. | ||
Zhao Y, Zhou H, Ma K, et al. Abnormal methylation of seven genes and their associations with clinical characteristics in early stage non-small cell lung cancer. Oncol Lett. 2013;5(4):1211–1218. | ||
Pao W, Chmielecki J. Rational, biologically based treatment of EGFR-mutant non-small-cell lung cancer. Nat Rev Cancer. 2010;10(11):760–774. | ||
Schiller JH, Harrington D, Belani CP, et al; Eastern Cooperative Oncology Group. Comparison of four chemotherapy regimens for advanced non-small-cell lung cancer. N Engl J Med. 2002;346(2):92–98. | ||
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. | ||
Byers LA, Rudin CM. Small cell lung cancer: where do we go from here? Cancer. 2015;121(5):664–672. | ||
Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30(7):733–750. | ||
Khan SA, Reddy D, Gupta S. Global histone post-translational modifications and cancer: Biomarkers for diagnosis, prognosis and treatment? World J Biol Chem. 2015;6(4):333–345. | ||
Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. | ||
Diaz-Lagares A, Mendez-Gonzalez J, Hervas D, et al. A novel epigenetic signature for early diagnosis in lung cancer. Clin Cancer Res. 2016;22(13):3361–3371. | ||
Huang T, Chen X, Hong Q, et al. Meta-analyses of gene methylation and smoking behavior in non-small cell lung cancer patients. Sci Rep. 2015;5:8897. | ||
Brock MV, Hooker CM, Ota-Machida E, et al. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358(11):1118–1128. | ||
Kim DH, Nelson HH, Wiencke JK, et al. Promoter methylation of DAP-kinase: association with advanced stage in non-small cell lung cancer. Oncogene. 2001;20(14):1765–1770. | ||
Hirosue A, Ishihara K, Tokunaga K, et al. Quantitative assessment of higher-order chromatin structure of the INK4/ARF locus in human senescent cells. Aging Cell. 2012;11(3):553–556. | ||
Furonaka O, Takeshima Y, Awaya H, Ishida H, Kohno N, Inai K. Aberrant methylation of p14(ARF), p15(INK4b) and p16(INK4a) genes and location of the primary site in pulmonary squamous cell carcinoma. Pathol Int. 2004;54(8):549–555. | ||
Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83(6):993–1000. | ||
Mai H, Liu X, Chen Y, et al. Hypermethylation of p15 gene associated with an inferior poor long-term outcome in childhood acute lymphoblastic leukemia. J Cancer Res Clin Oncol. 2016;142(2):497–504. | ||
Venza M, Visalli M, Biondo C, et al. Epigenetic regulation of p14ARF and p16INK4A expression in cutaneous and uveal melanoma. Biochim Biophys Acta. 2015;1849(3):247–256. | ||
Venza M, Visalli M, Biondo C, et al. Epigenetic marks responsible for cadmium-induced melanoma cell overgrowth. Toxicol In Vitro. 2015;29(1):242–250. | ||
Herman JG, Jen J, Merlo A, Baylin SB. Hypermethylation-associated inactivation indicates a tumor suppressor role for p15INK4B. Cancer Res. 1996;56(4):722–727. | ||
Seike M, Gemma A, Hosoya Y, et al. Increase in the frequency of p16INK4 gene inactivation by hypermethylation in lung cancer during the process of metastasis and its relation to the status of p53. Clin Cancer Res. 2000;6(11):4307–4313. | ||
Wang Y, Zhang D, Zheng W, Luo J, Bai Y, Lu Z. Multiple gene methylation of nonsmall cell lung cancers evaluated with 3-dimensional microarray. Cancer. 2008;112(6):1325–1336. | ||
Yanagawa N, Tamura G, Oizumi H, et al. Promoter hypermethylation of RASSF1A and RUNX3 genes as an independent prognostic prediction marker in surgically resected non-small cell lung cancers. Lung Cancer. 2007;58(1):131–138. | ||
Zhang Y, Wang R, Song H, et al. Methylation of multiple genes as a candidate biomarker in non-small cell lung cancer. Cancer Lett. 2011;303(1):21–28. | ||
Coory MD. Comment on: Heterogeneity in meta-analysis should be expected and appropriately quantified. Int J Epidemiol. 2010;39(3):932. | ||
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ. 2003;327(7414):557–560. | ||
DerSimonian R. Meta-analysis in the design and monitoring of clinical trials. Stat Med. 1996;15(12):1237–1248; discussion 1249–1252. | ||
De Jong WK, Verpooten GF, Kramer H, Louwagie J, Groen HJ. Promoter methylation primarily occurs in tumor cells of patients with non-small cell lung cancer. Anticancer Res. 2009;29:363–369. | ||
Fischer JR, Ohnmacht U, Rieger N, et al. Prognostic significance of RASSF1A promoter methylation on survival of non-small cell lung cancer patients treated with gemcitabine. Lung Cancer. 2007;56(1):115–123. | ||
Topaloglu O, Hoque MO, Tokumaru Y, et al. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10(7):2284–2288. | ||
Jarmalaite S, Kannio A, Anttila S, Lazutka JR, Husgafvel-Pursiainen K. Aberrant p16 promoter methylation in smokers and former smokers with nonsmall cell lung cancer. Int J Cancer. 2003;106(6):913–918. | ||
Tian KH, Lin LS, Jia ZT, Guo XJ, Zhang L. Promoter methylation status and protein expression of p14ARF gene in squamous cell carcinoma and adenocarcinoma of the lung. Zhongguo Fei Ai Za Zhi. 2006;9(1):40–44. | ||
Hu ZL, Sheng Y, Tian KH, Li L, Li W. The methylation status detected and analysis in clinic about p14ARF from non-small lung cancer tissues and its surrounding normal part. J Jining Med Univ. 2014;37(1):27–32. | ||
Shimamoto T, Ohyashiki JH, Hirano T, Kato H, Ohyashiki K. Hypermethylation of E-cadherin gene is frequent and independent of p16INK4A methylation in non-small cell lung cancer: potential prognostic implication. Oncol Rep. 2004;12(2):389–395. | ||
Chaussade L, Eymin B, Brambilla E, Gazzeri S. Expression of p15 and p15.5 products in neuroendocrine lung tumours: relationship with p15(INK4b) methylation status. Oncogene. 2001;20(45):6587–6596. | ||
Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in cancer progression. Exp Cell Res. 2001;264(1):42–55. | ||
Serra RW, Fang M, Park SM, Hutchinson L, Green MR. A KRAS-directed transcriptional silencing pathway that mediates the CpG island methylator phenotype. Elife. 2014;3:e02313. | ||
Li J, Knobloch TJ, Poi MJ, et al. Genetic alterations of RD(INK4/ARF) enhancer in human cancer cells. Mol Carcinog. 2014;53(3):211–218. | ||
Endo M, Kobayashi C, Setsu N, et al. Prognostic significance of p14ARF, p15INK4b, and p16INK4a inactivation in malignant peripheral nerve sheath tumors. Clin Cancer Res. 2011;17(11):3771–3782. | ||
Boultwood J, Wainscoat JS. Gene silencing by DNA methylation in haematological malignancies. Br J Haematol. 2007;138(1):3–11. | ||
Gu J, Wen Y, Zhu S, et al. Association between P(16INK4a) promoter methylation and non-small cell lung cancer: a meta-analysis. PLoS One. 2013;8(4):e60107. | ||
Han JC, Xu F, Chen N, et al. Promoter methylations of RASSF1A and p16 is associated with clinicopathological features in lung cancers. J Cancer Res Ther. 2016;12(1):340–349. | ||
Huang T, Li J, Zhang C, et al. Distinguishing lung adenocarcinoma from lung squamous cell carcinoma by two hypomethylated and three hypermethylated genes: a meta-analysis. PLoS One. 2016;11(2):e0149088. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.







