Back to Journals » International Journal of General Medicine » Volume 15
Role of MicroRNA-223 and MicroRNA-182 as Novel Biomarkers in Early Detection of Colorectal Cancer
Authors Mahmoud HA , El Amin HA , Ahmed ESM, Kenawy AG, El-Ebidi AM , ElNakeeb I, Kholef EFM, Elsewify WAE
Received 14 December 2021
Accepted for publication 10 March 2022
Published 25 March 2022 Volume 2022:15 Pages 3281—3291
DOI https://doi.org/10.2147/IJGM.S353244
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Hala A Mahmoud,1 Hussein Ahmed El Amin,2 Ehab Saleh Mahmoud Ahmed,1 Ahmed Gaber Kenawy,1 Abdallah M El-Ebidi,3 Islam ElNakeeb,4 Emad Farah Mohammed Kholef,4 Wael Abd Elgwad Elsewify1
1Department of Internal Medicine, Faculty of Medicine, Aswan University, Aswan, Egypt; 2Department of Internal Medicine, Faculty of Medicine, Assiut University, Assiut, Egypt; 3Department of Biochemistry, Faculty of Medicine, Aswan University, Aswan, Egypt; 4Department of Clinical Pathology, Faculty of Medicine, Aswan University, Aswan, Egypt
Correspondence: Wael Abd Elgwad Elsewify, Department of Internal Medicine, Faculty of Medicine, Aswan University, Aswan, 81528, Egypt, Tel +201001657295, Fax +20973480449, Email [email protected]
Background: Colorectal cancer is a common and lethal disease. It is estimated that approximately 145,600 new cases of large bowel cancer are diagnosed annually in the USA. MiRNA-223 and miRNA-182 have been reported in various cancers, such as lung, gastric, breast and colorectal cancer and proposed to be valid and reliable for diagnosis as well as prognosis.
Aim: This study aimed to determine the role of miR-223 and miR-182 as novel biomarkers for early detection and prognosis of CRC.
Patient and Methods: This case–control study was conducted at the department of Internal Medicine, Aswan University Hospital, in the period from the 1st of February 2020 to the 20th of April 2021. Thirty-five cases and thirty age- and sex-matched controls were included in the study. All patients were subject to complete clinical evaluation, routine investigations, occult blood in stool, serum levels of CEA and CA 19– 9, serum levels of miR-223 and miR-182 by quantitative PCR.
Results: Significant difference between the two studied groups regarding biomarker changes was found. ROC curve analysis showed that the new markers had excellent diagnostic as well as prognostic criteria. Micro-RNA-223 diagnostic accuracy, sensitivity, specificity, PPV, NPV, FDR and FOR were 97%, 97.1%, 96.7%, 97%, 97%, 3.3% and 2.9%, respectively. Also, micro-RNA-182 diagnostic accuracy, sensitivity, specificity, PPV, NPV, FDR and FOR were 97%, 98%, 96%, 96%, 98%, 3.9% and 2%, respectively.
Conclusion: MiR-223 and miR-182 have been discovered to be relevant and reliable biomarkers for the early identification and prognosis of CRC. Increased levels of miR-223 and miR-182 were associated with increased risk of disease progression, and the more accurate the value of miR-223 and miR-182, the earlier the diagnosis of colorectal cancer.
Keywords: CRC, mirRNA-223, mirRNA-182, accuracy and prognosis
Introduction
Colorectal cancer (CRC) is a deadly disease that affects many people. In the United States, an estimated 145,600 new instances of large bowel cancer are identified each year. It is the third leading cause of cancer death in women and the second major cause of cancer death in men.1 CRC is caused mostly by lifestyle and aging-related factors. A tiny percentage of CRC cases, however, may be caused by underlying genetic disorders. Changes in bowel motions, blood in the stool, loss of appetite, weight loss, and feeling weary all the time are all signs and symptoms of CRC.2 Life-style risk factors included: obesity, diet, smoking and absence of physical activity.3 Other risk factors are inflammatory bowel disorders (IBD) such as ulcerative colitis and Crohn’s disease.3 It accounted for about 10–15% of deaths in patients with IBD. Familial adenomatous polyposis and hereditary non-poly colon cancer are two inherited genetic disorders that can lead to CRC. As a result, only about 5% of people are affected by these disorders.3
The currently available screening modalities for early detection of CRC, such as faecal occult blood test (FOBT), carcinoembryonic antigen (CEA) test and carbohydrate antigen 19–9 (CA19-9) are effective but limited by the low specificity and sensitivity.4 FOBT, which is non-specific but can be detected especially in larger polyps and CRC. Sigmoidoscopy and colonoscopy are invasive procedures that carry certain morbidity concerns and necessitate time-consuming preparation procedures, resulting in a low participation rate.4 The ideal CRC biomarker should be simple to assess quantitatively, highly specific and sensitive, as well as dependable and repeatable It should be able to distinguish between various risk-based groups, allowing it to identify patients who truly require a second-line test. In an ideal world, this might be accomplished via a non-invasive, low-cost technology based on readily available biological samples.5
The identification and creation of an optimal biomarker requires an understanding of the genetic and epigenetic modifications that characterize CRC carcinogenesis. DNA methylation, microRNA (miRNA) expression, histone modification, and chromatin remodelling are all examples of epigenetic changes.6 MiRNAs have the potential to be accurate biomarkers for the diagnosis and categorization of CRC, as well as for predicting treatment results, due to their high tissue specificity and crucial role in oncogenesis.7
MiRNAs are endogenous, single-stranded, noncoding RNAs that were first recognized as developmental regulators in 1993. Breast cancer, lung cancer, pancreatic cancer, ovarian cancer, and colorectal cancer have all been found to have abnormal expression patterns.8 In the plasma of CRC patients, many miRNAs are dysregulated. They are crucial in determining cancer risk, diagnosis, prognosis, and medication response. As a result, they could be used as biomarkers.9
MiRNA-223 is an onco-miRNA that has been demonstrated to target survival and death-related genes, as well as reducing cell proliferation, migration, and invasion in CRC cells when knocked down. It is important in CRC and could be used as a molecular biomarker and therapeutic gene for CRC treatment.10 MiR-182, a member of the miR-183 family located on 7q31-34, is one of the most frequently studied cancer-related oncogenic miRNAs that is dysregulated in various cancer tissues, miR-182 may function as an oncogenic miRNA to enhance cancer cell proliferation, survival, aggressiveness, tumorigenesis, and drug resistance.11 It has been found in a variety of malignancies, including lung, gastric, breast, and colorectal cancer. It was previously thought to be up regulated in CRC.12
This study aimed to determine the role of miR-223 and miR-182 as a novel biomarker for early detection of CRC and prognosis.
Patients and Methods
This case-control study was conducted at the department of Internal Medicine, Aswan University Hospital, in the period from the 1st of February 2020 to the 20th of April 2021. Sample size calculation was conducted using G-Power 3.1.9.7. A minimum of 60 participants (30 CRC cases and 30 age and sex matched controls) was sufficient to demonstrate relevant differences of 0.3 between the two groups, as regard comparing the mean level of mi-RNA biomarkers with α error of 0.05 and power of the study of 80%. The study included 35 CRC cases to compensate for missing data.
The following were inclusion criteria for recruitment: patients aged > 18 years of both sexes, recently diagnosed with CRC by histopathological examination. On the other hand, patients with malignancies other than CRC, those with metastatic/recurrent CRC and those refused to participate or unable to complete the study for any reason were excluded.
All patients were subject to complete clinical evaluation including history taking, physical examination with special emphasis on the data suggestive of metastases ie, enlarged tender liver, hemiplegia, bone pain, pathological fracture etc. Review of the results of routine investigations including complete blood count, liver function test, kidney function test, prothrombin time, concentration and international normalized ratio (INR), occult blood in stool, serum levels of CEA and CA 19–9, serum levels of miR-223 and miR-182 by quantitative PCR.
Metastatic work up whenever indicated as abdominal ultrasound, CT chest, abdomen, mammography, bone scan etc. The biopsy was taken from colorectal mass by colonoscopy from patients after an informed verbal consent. The biopsy was preserved in formalin (10%) for histopathological examination.
Serum samples collection: from each patient and control subject, 5 mL venous blood were obtained, centrifuged at 3000 × g for 10 min at 4 °C and then the serum was separated. All serum samples were stored at −80°C until used. RNA extraction and qRT-PCR: total RNA including miRNAs was extracted from sera using the miRNeasy Mini Kit (Qiagen, Germany). According to the manufacturer’s instructions, c-DNA Synthesis was performed using miScript II RT kit (Qiagen). Quantitative real-time PCR (qRT-PCR) was performed using miScript syber green PCR kit (Qiagen).
Gene expression profiles were generated in arrays using the custom miScript miRNA PCR array for miR-223 and miR182 (Qiagen) according to manufacturers’ instruction as follows: 15 min at 95°C for 1 cycle, 15s at 94°C, 30s at 55°C, and 30s at 70°C for 40 cycles using AB 7500 Fast Real-Time PCR system. Threshold cycle data were analyzed using the RT2 Profiler software (version 3.4; SABiosciences). Relative gene expression levels were normalized to housekeeping gene (SNORD 68) and the fold changes of the target gene(s) expression relative to those of the control group were analyzed by the 2−ΔΔCT method.
Statistical Analysis
Data was verified, coded by the researcher, and analysed using IBM-SPSS 24.0 (IBM-SPSS Inc., Chicago, IL, USA).13 Descriptive statistics: Means, standard deviations, medians, ranges, and percentages were calculated. Test of significances: Chi-square/Fisher’s exact test was used for frequency difference comparison. For continuous variables two categories student’s t-test analysis was carried out to compare the means of dichotomous data that follow the normal distribution while Mann Whitney U-test was used to compare the medians of dichotomous data that did not follow the normal distribution. For continuous variables with more than two categories; ANOVA test was calculated to test the mean differences. ROC curve was used to assess the diagnostic accuracy of serum miRNAs, analysed as area under the curve (AUC), standard error (SE) and 95% CI. Validity statistics (sensitivity, specificity, positive and negative predictive value –PPV & NPV-) were calculated. A significant p value was considered when it is < 0.05.
Ethical Consideration
IRB approval was obtained from the Medical Ethic Committee, Faculty of Medicine, Aswan University (IRB no. 368/4/19). The study was carried out in accordance with the Helsinki Declaration guidelines. An official written administrative permission letter was obtained from dean of faculty of medicine, Aswan University Hospital, head of internal medicine department. The title and objectives of the study were explained to them to ensure their cooperation.
A written informed consent was obtained from the patient before the participation in the study. All collected data was confidential and was used for the purpose of scientific research only. Every research participant had the complete right and freedom to withdraw at any time from the study without any consequences on the medical service provided.
Results
After reviewing the inclusion and exclusion criteria patients (n=35) were recruited from the outpatient clinic of the Internal Medicine Department, Aswan University Hospital, in the period from February 1, 2020, to April 20, 2021. Age and sex matched controls (n=30) from the relatives of patients were added. Figure 1 showed the flow diagram of the current study.
 |
Figure 1 Flow chart diagram for the study participants. Abbreviation: CRC, colorectal cancer. |
The two groups were comparable with respect to the age, sex, smoking, BMI, and low/fiber diet (p>0.05). On the other hand, positive family history of CRC was significantly (p=0.006) higher in cases (23%) compared with control (0%) (Table 1). CRC cases showed higher significant (p<0.001) percentage of abdominal tenderness (71.4%) compared with control (0%) while all other clinical signs were comparable (p>0.05) (Table 2).
 |
Table 1 Differences in the Baseline Characteristics of the Studied Cohort |
 |
Table 2 Examination Data Differences Between Groups |
Table 3 showed the difference in laboratory findings between groups. The mean hemoglobin level in CRC cases group (10.16 ± 1.8 g/dl) was significantly higher than the level in the control group (12.15 ± 2.3 g/dl). Also, platelet Count was significantly different between the two study groups. Likewise, the mean serum albumin level was 3.44 ± 0.9 g/dl in CRC cases which was significantly lower than 4.28 ± 0.5 g/dl in the control groups. Moreover, the mean total bilirubin level was significantly higher in CRC cases (1.07 ± 0.6 mg/dl) than in control groups (0.72 ± 0.2 mg/dl), respectively. Additionally, there was statistically significant difference in the mean INR level between the two studied groups. The mean INR level in CRC group was higher (1.05 ± 0.3) than in the control group (0.82 ± 0.2). Furthermore, the means of microRNA-223 and microRNA-182 fold change were significantly (p<0.001) higher (2.28 and 3.03) in CRC cases group than in the control group (1.00 and 0.7) (Figure 2).
 |
Table 3 Laboratory Findings of the Studied Cohort |
 |
Figure 2 Biomarkers fold change differences between CRC cases and control. Abbreviation: CRC, colorectal cancer. |
Table 4 and Figure 3 showed the diagnostic criteria of the Biomarkers for CRC Prediction. It was found that AUC for CEA was 0.944 at cut off 2.5 with sensitivity and specificity (83% and 100%), respectively. Also, AUC of CA 19–9 was 0.837 at cut off 30 with sensitivity and specificity (89% and 77%), respectively. MiRNA-223 AUC was 0.963 at cut off 1.1 with sensitivity and specificity (97.1% and 96.7%), respectively. It had a diagnostic accuracy 97% with positive predicted value (PPV) 97%, negative predicted value (NPV) 97%, FDR (False Detection Rate) 3.3% and FOR (False Omission Rate) 2.9%. Likewise, AUC of MiRNA-182 was 0.949 at cut off 1.1 with sensitivity and specificity (98% and 96%), respectively. It had an accuracy of 97% with PPV of 96%, NPV of 98%, FDR of 3.9% and FOR of 2%.
 |
Table 4 Diagnostic Criteria of Biomarkers for CRC Prediction |
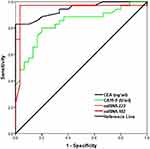 |
Figure 3 ROC curve for biomarkers for CRC prediction. |
The correlation between Micro-RNAs and other laboratory data was depicted in Figures 4 and 5. MiRNA-223 has a significant positive correlation with CEA level, CA 19–9 level and TNM Staging. Unlikely, it has a significant negative correlation with serum ALT level. For the MiRNA-182, it has a significant positive correlation with the Platelet count, CEA level, CA 19–9 level and TNM Staging. Contrarily, it has a significant negative correlation with serum ALT level and Serum Albumin level. Neither WBCs level/hemoglobin levels/AST level/INR nor total bilirubin showed significant correlation with micro-RNA-223 or micro-RNA-182 levels.
 |
Figure 4 Correlation between miRNA-223 and platelet count, ALT and serum albumin. Abbreviation: ALT, alanine aminotransferase. |
 |
Figure 5 Correlation between miRNA-182 and platelet count, ALT and serum albumin. Abbreviation: ALT, alanine aminotransferase. |
Moreover, the mean micro-RNA-223 level was higher (2.45 ± 0.5) in tumour stage > T3 than in tumour stage ≤ T3 (2.06 ± 0.5). Also, the mean Micro-RNA-182 level was higher in stage > T3 (3.25 ± 0.8) than in tumour stage ≤ T3 (2.64 ± 0.6). According to the present study results the mean micro-RNA-223 level was higher in tumour stage > N0 (2.66 ± 0.3) than in tumour stage ≤ N0 (1.98 ± 0.5). Likely, the mean micro-RNA-182 levels was higher in tumour stage > N0 (3.41 ± 0.3) than in tumour stage ≤ N0 (2.65 ± 0.5). Regarding the mean micro-RNA-223 in different TNM-Stages; it was 2.66 ± 0.3, 2.21 ± 0.4 and 1.46 ± 0.1 in stages III, II and I respectively. As well, the mean micro-RNA-223 levels were 2.63 ±0.3, 2.38 ± 0.5 and 1.92 ± 0.5 in adenocarcinoma GIII-IV, GII-III and GI-II, respectively. For the mean micro-RNA-182 in different TNM-Stages; it was 3.41 ± 0.3, 3.09 ± 0.6 and 1.62 ± 0.2 in stages III, II and I respectively. The mean Micro-RNA-182 levels were 3.42 ± 0.2, 3.15 ± 0.7 and 2.49 ± 0.8 in GIII-IV, GII-III and GI-II, respectively (Table 5).
 |
Table 5 Mean Micro-RNA Biomarkers in Different Tumour Staging |
Discussion
Despite recent and ongoing advances in diagnosis and therapy, more than half of colon and rectal tumors metastasis to the liver, lung, and lymph nodes, and the 5-year survival rate for patients with metastatic CRC remains low (less than 10%).14 Screening techniques for early diagnosis of CRC now available, such as gFOBT, CEA, and CA19-9, are effective but have low specificity and sensitivity. Sigmoidoscopy and colonoscopy are invasive procedures that carry certain morbidity concerns and necessitate time-consuming preparation procedures, resulting in a low participation rate.4 As a result, there is a pressing need for innovative biomarkers that can aid in diagnosis as well as enhance prognosis and therapy prediction.
The aim of this study was to determine role of miR-223 and miR-182 as a biomarker for early detection of colorectal cancer and prognosis. It is hypnotized that the more accurate the value of miR-223 and miR-182, the earlier the diagnosis CRC. This case-control study was conducted at internal medicine department, faculty of medicine, Aswan University after approval of hospital ethics committee. It included thirty five patients diagnosed as colorectal cancer by endoscopy and histopathology and thirty controls with the same age and sex as a control group.
The current study results showed that there were non-significant differences between the two groups regarding smoking, obesity, low fiber diet consumption and high fiber diet consumption. The results of the study by Papadimitriou et al,15 that aimed to investigate the associations of circulating tryptophan and its two major metabolites, serotonin and kynurenine, with colon cancer risk, come in agreement with the current study results. In contrast Park et al in a study stated that the Westernized dietary pattern (meats, fast foods rich in carbohydrates, oil and sugar) was associated with an elevated risk of CRC, whereas the traditional and prudent patterns were associated with a decreased risk of rectal cancer and all types of CRC, respectively.16
There was a highly significant difference between CRC cases and control group in this study regarding CRC family history. In certain circumstances, the reasons for the elevated risk are unclear. Inherited genes, shared environmental and nutritional factors, or a combination of these factors might cause cancer to “run in the family”.17 On clinical examination a highly significant difference was detected between the two groups regarding abdominal tenderness. Regarding laboratory findings; significant higher mean parameters (WBCs, platelet, s. albumin, t. bilirubin and INR) was found in CRC cases than controls. It can be explained by systemic inflammatory response to tumors, which is associated with abnormalities of several blood components, especially neutrophils and lymphocytes.18
According to the current study results there were significantly higher mean levels in CRC cases than controls. Zhang et al in a study reported that the overexpression of miR-223 in CRC was identified with a significant P-value.19 Also, Liu et al found that miR-223 level was upregulated in CRC patients.17 According to a previous study miR-182 expression was significantly higher in CRC patients than in control group, ranging from 0.03 to 217.24 RQU with a mean ± SE of 50.08 ± 4.53 in the former, while varying between 2.31 and 70.72 RQU with a mean ± SE of 18.78 ± 1.72 in the latter.20 In the current study ROC curve analysis was used to evaluate the diagnostic performance of biomarkers for prediction of CRC between CRC patients and normal subjects. Compared with normal subjects, the AUCs for CEA, CA 19–9, Micro-RNA-223 and Micro-RNA-182 were 0.944, 0.837, 0.963 and 0.949, respectively. The cut-off values were 2.5, 30 for CEA and CA 19–9 respectively. It was 1.1 for both Micro-RNA-223 and Micro-RNA-182. Bagaria et al conducted a study to determine the clinical serum levels of CEA and CA19-9, individually and in combination, for the diagnosis of 50 healthy subjects and 150 cases of esophageal, gastric, and colon cancer. They discovered that CEA had a sensitivity of 74%, a specificity of 100%, and NPV of 79.36% in CRC cases. CA19-9 has a sensitivity of 26% and NPV of 57.47%.21 Yang et al in a study reported that the AUC for CEA and CA19-9 were approximately 0.7654 and 0.6123.22
According to the current study, it was found that Micro-RNA-223 diagnostic accuracy, Sensitivity, Specificity, PPV, NPV, FDR and FOR was 97%, 97.1%, 96.7%, 97%, 97%, 3.3% and 2.9%, respectively. Nearly similar to this study results, Zheng et al found that Micro-RNA-223 had 95% sensitivity and 94% specificity for CRC detection.23 Wang et al in a previously conducted study reported that Sensitivity and Specificity of mRNA 223 in CRC patients were 76.5% and 96.4%, respectively.24 This was in contrast to the results found in study conducted by Yau et al was lower than the current study results Micro-RNA-223 Sensitivity was 67.9% and the Specificity was 87.4%.25 Pesta et al study results were different than that found in the current study. Sensitivity, Specificity, Positive Predictive Value and Negative Predictive Value were 33.3%, 54.6%, 23.1% and 66.7%, respectively.26
The current study results showed that Micro-RNA-182 diagnostic accuracy, Sensitivity, Specificity, PPV, NPV, and FDR and FOR were 97%, 98%, 96%, 96%, 98%, 3.9% and 2%, respectively. In agreement with the current study results, Carter et al conducted a meta-analysis, it was found that in CRC patients Micro-RNA-182 sensitivity ranged from 46% to 92.79% and specificity from 41% to 93.2%.7
Possible correlation between mi-RNA and other laboratory data was done. Micro-RNA-223 has a significant positive correlation with TNM Staging, CEA level and CA 19–9 level. Micro-RNA-182 has significant positive correlation with TNM Staging, the Platelet count, CEA level and CA 19–9 level.
In the current study the mean micro-RNA-223 and 182 level was higher in tumor stage > T3 than stage ≤ T3.
Also, the mean micro-RNA-223 levels were 1.98 ± 0.5 and 2.66 ± 0.3 in ≤ N0 and > N0 stages, respectively. The mean micro-RNA-182 levels were 2.65 ± 0.5 and 3.41 ± 0.3 in ≤ N0 and > N0 stages, respectively. Perilli et al conducted a study to evaluate miR-182 dysregulation in CRC, miR-182 expression was investigated by qPCR in a series of 20 stage IV CRCs. A significant overexpression of miR-182 was observed in primary CRCs (5.3-fold change) compared to normal subjects.27
Regarding the mean micro-RNA-223 in different TNM-Stages, there was steady increase 1.46, 2.21 and 2.66 in stages I, II and III, respectively. Li et al conducted a study to investigate microRNA-223 (miR-223) expression in CRC and its relationship with tumorigenesis and disease prognosis. In agreement with this study the results showed that miR-223 level in poorly differentiated tumor was 6.024 (0.558–10.284), the level in moderately differentiated tumor was 3.203 (1.324–4.934) and in well differentiated tumor it was 1.965 (1.163–2.772). No significant correlation was detected.28 Also, it was reported in previous study that the mean miR-223 level was 1.513 (0.410–2.160), 2.430 (1.350–3.152) and 7.698 ((2.035–13.146) in tumor stages T1 + T2, T3 and T4, respectively, which was significantly different.29
The mean micro-RNA-182 levels among this study population were 1.62 ± 0.2, 3.09 ± 0.6 and 3.41 ± 0.3 in stages I, II and III respectively. It was reported that the mean micro-RNA-223 levels were 1.92 ± 0.5, 2.38 ± 0.5 and 2.63 ±0.3 in adenocarcinoma GI-II, GII-III and GIII-IV, respectively. The mean Micro-RNA-182 levels were 2.49 ± 0.8, 3.15 ± 0.7 and 3.42 ±0.2 in adenocarcinoma GI-II, GII-III and GIII-IV, respectively. Liu et al evaluated the correlation between miR-182 expression and clinicopathological characteristics, and the data showed that the up-regulation of miR-182 was significantly correlated with large tumor size (p = 0.016), positive regional lymph node metastasis (p = 0.008), and advanced TNM stage (p = 0.020). In contrast, no correlation was detected in the expression level of miR-182 with age or other clinicopathological parameters (all at p > 0.05, respectively). Taken together, these results suggested that a higher level of miR-182 expression may be related with colorectal cancer progression.30–32
Conclusion
MiR-223 and miR-182 were found to be accurate and reliable biomarkers for early identification and prognosis of CRC in this study. It was also discovered that the more accurate the value of miR-223 and miR-182 was, the earlier colorectal cancer was diagnosed. Increased levels of miR-223 and miR-182 were also found to be linked to an increased risk of disease progression in colorectal cancer patients.
Disclosure
There are no potential conflicts of interest for the authors to disclose.
References
1. Siegel R, Miller K, Jemal A. Cancer statistics. CA Cancer J Clin. 2019;69:7. doi:10.3322/caac.21551
2. Thompson M, O’Leary D, Flashman K, et al. Clinical assessment to determine the risk of bowel cancer using symptoms, age, mass and iron deficiency anaemia (SAMI). Br J Surg. 2017;104:1393. doi:10.1002/bjs.10573
3. Barral M, Dohan A, Allez M, et al. Gastrointestinal cancers in inflammatory bowel disease: an update with emphasis on imaging findings. Crit Rev Oncol Hematol. 2016;97:30–46. doi:10.1016/j.critrevonc.2015.08.005
4. Solomon C, Inadomi J, Solomon CG. Screening for colorectal neoplasia. N Engl J Med. 2017;376:149–156. doi:10.1056/NEJMcp1512286
5. Symonds E, Pedersen S, Baker R, et al. A blood test for methylated BCAT1 and IKZF1 vs a fecal immunochemical test for detection of colorectal neoplasia. Clin Transl Gastroenterol. 2016;7:e137. doi:10.1038/ctg.2015.67
6. Inamura K. Colorectal cancers: an update on their molecular pathology. Cancers. 2018;10(1):26. doi:10.3390/cancers10010026
7. Carter J, Galbraith N, Yang D, et al. Blood-based microRNAs as biomarkers for the diagnosis of colorectal cancer: a systematic review and meta-analysis. Br J Cancer. 2017;116(6):762–774. doi:10.1038/bjc.2017.12
8. Bracken C, Scott H, Goodall G. A network-biology perspective of microRNA function and dysfunction in cancer. Nat Rev Genet. 2016;17:719–732. doi:10.1038/nrg.2016.134
9. Vychytilova-Faltejskova P, Radova L, Sachlova M, et al. Serum-based microRNA signatures in early diagnosis and prognosis prediction of colon cancer. Carcinogenesis. 2016;37:941–950. doi:10.1093/carcin/bgw078
10. Deng L, Luo X, Li H, et al. MicroRNA-223 functions as an oncogene in human colorectal cancer cells. Oncol Rep. 2018;32:115120.
11. Si W, Shen J, Zheng H, et al. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics. 2019;11(1):25. doi:10.1186/s13148-018-0587-8
12. Liu X, Xu T, Hu X, et al. Elevated circulating miR-182 acts as a diagnostic biomarker for early colorectal cancer. Cancer Manag Res. 2018;10:857. doi:10.2147/CMAR.S158016
13. IBM_SPSS. Statistical package for social science. Ver.21. Standard version. Copyright © SPSS Inc., 2012–2016. NY, USA; 2016.
14. Sponholz S, Schirren M, Baldes N, et al. Repeat resection for recurrent pulmonary metastasis of colorectal cancer. Langenbecks Arch Surg. 2017;402:77–85. doi:10.1007/s00423-016-1547-4
15. Papadimitriou N, Gunter M, Murphy N, et al. Circulating tryptophan metabolites and risk of colon cancer: results from case‐control and prospective cohort studies. Intl j Cancer. 2021;149(9):1659–1669. doi:10.1016/j.cgh.2021.4.28
16. Park Y, Lee J, Oh JH, et al. Dietary patterns and colorectal cancer risk in a Korean population: a case-control study. Medicine. 2016;95(25). e3759.
17. Lu K, Wood M, Daniels M, et al. American society of clinical oncology expert statement: collection and use of a cancer family history for oncology providers. J Clin Oncol. 2014;32(8):833. doi:10.1200/JCO.2013.50.9257
18. Pedrazzani C, Mantovani G, Fernandes E, et al. Assessment of neutrophil-to-lymphocyte ratio, platelet-to-lymphocyte ratio and platelet count as predictors of long-term outcome after R0 resection for colorectal cancer. Sci Rep. 2017;7(1):1. doi:10.1038/s41598-017-01652-0
19. Zhang J, Luo X, Li H, et al. MicroRNA-223 functions as an oncogene in human colorectal cancer cells. Oncol Rep. 2014;32(1):115–120. doi:10.3892/or.2014.3173
20. Rapti S, Kontos C, Papadopoulos I, et al. Enhanced miR-182 transcription is a predictor of poor overall survival in colorectal adenocarcinoma patients. Clin Chem Lab Med. 2014;52(8):1217–1227. doi:10.1515/cclm-2013-0950
21. Bagaria B, Sood S, Sharma R, et al. Comparative study of CEA and CA19-9 in esophageal, gastric and colon cancers individually and in combination (ROC curve analysis). Cancer Biol Med. 2013;10(3):148.
22. Yang L, Jiang Q, Li D, et al. TIMP1 mRNA in tumor-educated platelets is diagnostic biomarker for colorectal cancer. Aging. 2019;11(20):8998. doi:10.18632/aging.102366
23. Zheng G, Du L, Yang X, et al. Serum microRNA panel as biomarkers for early diagnosis of colorectal adenocarcinoma. Br J Cancer. 2014;111:1985–1992. doi:10.1038/bjc.2014.489
24. Wang J, Song Y, Ma B, et al. Regulatory roles of non-coding RNAs in colorectal cancer. Int J Mol Sci. 2015;16(8):19886–19919. doi:10.3390/ijms160819886
25. Yau T, Tang C, Harriss E, et al. Fecal microRNAs as a non-invasive tool in the diagnosis of colonic adenomas and colorectal cancer: a meta-analysis. Sci Rep. 2019;9(1):1–3. doi:10.1038/s41598-019-45570-9
26. Pesta M, Kucera R, Topolcan O, et al. Plasma microRNA levels combined with CEA and CA19-9 in the follow-up of colorectal cancer patients. Cancers. 2019;11(6):864. doi:10.3390/cancers11060864
27. Perilli L, Vicentini C, Agostini M, et al. Circulating miR-182 is a biomarker of colorectal adenocarcinoma progression. Oncotarget. 2014;5(16):6611. doi:10.18632/oncotarget.2245
28. Li Z, Yang Y, Du L, et al. Overexpression of miR-223 correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Med Oncol. 2014;31(11):256. doi:10.1007/s12032-014-0256-5
29. Li Z, Xu R, Li N. MicroRNAs from plants to animals, do they define a new messenger for communication? Nutr Metab. 2018;15(1):68. doi:10.1186/s12986-018-0305-8
30. Liu H, Du L, Wen Z, et al. Up-regulation of miR-182 expression in colorectal cancer tissues and its prognostic value. Int J Colorectal Dis. 2013;28(5):697–703. doi:10.1007/s00384-013-1674-0
31. Liu H, Liu T, Wu H, et al. Serum micro RNA signatures and metabolomics have high diagnostic value in colorectal cancer using two novel methods. Cancer Sci. 2018;109(4):1185–1194. doi:10.1111/cas.13514
32. Liu M, Wang Q, Shen J, et al. Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol. 2019;16(7):899–905. doi:10.1080/15476286.2019.1600395
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
