Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 18
RNA-Sequencing and Bioinformatics Analysis of Exosomal Long Noncoding RNAs Revealed a Novel ceRNA Network in Stable COPD
Authors Lin S, Liu C, Sun J, Guan Y
Received 24 April 2023
Accepted for publication 24 July 2023
Published 11 September 2023 Volume 2023:18 Pages 1995—2007
DOI https://doi.org/10.2147/COPD.S414901
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jill Ohar
Shan Lin,1 Caihong Liu,2 Jingting Sun,1 Yinghui Guan1
1Department of Respiratory Medicine, The First Hospital of Jilin University, Changchun, Jilin, People’s Republic of China; 2Department of Clinical Laboratory, The First Hospital of Jilin University, Changchun, Jilin, People’s Republic of China
Correspondence: Yinghui Guan, Email [email protected]
Purpose: Exosomes are able to exchange their bioactive RNA cargo to recipient cells. In COPD, exosomes can be controlled and engineered for its use as targeted diagnostic and therapeutic tool. Our study explored novel lncRNAs and mRNAs in plasma exosomes that could be involved in the pathogenesis of COPD.
Methods: High-throughput sequencing was conducted to detect the alterations in the expression of exosomal lncRNAs and mRNAs. Gene ontology (GO) functional analyses and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were used to determine the significant functions and pathways associated with differentially expressed (DE) lncRNAs. The mRNA expression profile dataset, GSE76925, and microRNA expression profile dataset, GSE70080, were obtained from the GEO database. Venn diagrams were used to find common DE mRNAs between my mRNAs dataset and GSE76925. These common DEGs were subjected to PPI analyses to identify Hub genes. Subsequently, Venn diagrams were used to identify common genes between the target genes of DE-miRNAs and Hub genes as well as DE-miRNAs and my lncRNAs dataset. Finally, a lncRNA–miRNA–mRNA co-expression network was constructed by prediction using proprietary software. The lncRNA and mRNA expressions were then validated by quantitative reverse-transcription polymerase chain reaction (qRT-PCR).
Results: We identified 1578 differentially regulated lncRNAs and 3071 differentially regulated mRNAs. GO and KEGG pathway analyses suggested that the DE lncRNAs are involved in the pathogenesis of COPD. A lncRNA–miRNA–mRNA meshwork was established to predict the potential interactions among these RNAs. RP3-329A5.8 and MRPS11 expression was then subjected to qRT-PCR for validation. Correlations between MRPS11 and clinic-pathological features were explored.
Conclusion: Our study provided a set of lncRNAs and mRNAs that may be involved in the pathogenesis of COPD, thereby highlighting the need for further research on both diagnostic biomarkers and molecular mechanisms.
Keywords: stable COPD, plasma, exosomes, lncRNA, mRNA
Introduction
COPD, a common preventable and treatable disease characterized by persistent airflow limitation and respiratory symptoms, is associated with exposure to harmful environments. COPD is currently the third leading cause of death globally. The high incidence and mortality of COPD, which seriously threaten human health, represent a public health problem to be solved urgently.1 Furthermore, the precise mechanisms of the occurrence and progression of COPD are still not clearly elucidated. It is pivotal to develop novel prognostic biomarkers and effective therapeutic targets and also to explore the underlying molecular mechanisms of COPD.
Exosomes are extracellular vesicles originating from endosomes with diameters of 30–100 nm. Exosomes, which can be synthesized and secreted by almost all kinds of cells, are widely distributed in physiological and pathological body fluids.2 They contain different kinds of nucleic acids, proteins, lipids, and glycoconjugates, and play a variety of biological roles by delivering bioactive substances to mediate cell communication.3 Recent studies have demonstrated that exosomes play a key role in the pathogenesis of various diseases such as malignant tumors, neural degenerative diseases, infectious diseases, and autoimmune diseases. In addition, exosomes have been shown to have great potential in the diagnosis, treatment, and prognosis of the aforementioned diseases.4
Long noncoding RNAs (lncRNAs) contain more than 200 nucleotides but have little or no protein-coding ability. LncRNAs have a wide range of biological effects, which can regulate gene expression at various levels, such as epigenetic regulation, transcriptional regulation, and post-transcriptional regulation.5 Currently, one of the important research directions is endogenous competitive RNA (ceRNA). Just like ceRNA, IncRNA can bind to miRNA through adsorption, similar to a “molecular sponge”, and indirectly regulate the degradation or translation process of the miRNA target protein. Increasing evidence has indicated that lncRNAs were selectively distributed to exosomes and were associated with the pathogenesis of lung diseases, such as lung cancer, COPD, asthma, ILD, and TB.6 For example, MALAT-1 in serum-derived exosomes promotes the proliferation and invasion of lung cancer cells by reducing apoptosis and facilitating cell cycle progression in these cells.7 Exosomal lncRNA TCONS_00064356 secreted by injured alveolar epithelial type II cells could affect the proliferation and migration of mesenchymal stem cells.8 Although lncRNAs have been an emerging focus in recent years, there is a paucity of published studies on the roles of exosomal lncRNAs in COPD.
In this study, we used RNA-sequencing and screened differentially expressed (DE) lncRNA and DE messenger RNAs of plasma exosomes between sCOPD and healthy controls (HCs). In addition, we searched the hub mRNA and constructed interaction networks of the lncRNA–miRNA–mRNA via specific bioinformatics in the pathogenesis of COPD. To confirm sequencing results with the identified DE genes, we performed quantitative reverse-transcription polymerase chain reaction (qRT-PCR) between sCOPD and HC groups.
Materials and Methods
Sample Collection
We selected 26 patients diagnosed with COPD and 10 normal healthy individuals at the First Hospital of Jilin University from January 2022 to September 2022. Our inclusion criteria were as follows: (i) diagnosis of sCOPD per the GOLD guidelines; (ii) clinical stability and freedom from exacerbation for at least three months; (iii) voluntary participation in the study. A total of 10 healthy donors (sex- and age-matched with COPD patients) were recruited among healthy adults. Table 1 shows the clinical features of the study participants. Blood samples were collected using EDTA anti-coagulation tubes and centrifuged at 1900 × g at 4°C for 10 minutes. Then the plasma was collected and centrifuged again (3000 × g;15 min) at 4°C and stored at −80°C until use. The study was approved by The Ethics Committee of the First Hospital of Jilin University (protocol no. 2022–467), and all methods were performed per the principles of the Declaration of Helsinki. Written informed consent was obtained from each participant.
 |
Table 1 Demographic and Clinical Characteristics of the Two Groups |
Exosome Isolation and Identification
Exosomes were isolated using an exoEasy Maxi Kit (Qiagen) per the manufacturer’s instructions. Exosomes were identified via transmission electron microscopy (TEM) and Western blot analysis. The exosomal morphology was characterized via TEM (Hitachi, HT7700) per the manufacturer’s instructions. Plasma exosome proteins were used for SDS-PAGE. Specific exosomal markers (CD63 and GRP94) were used to detect exosomes via Western blot analysis.
RNA Isolation, Library Construction, and Sequencing
Total RNA from exosomes was isolated using the TRIzol reagent (Invitrogen Life Technologies) per the manufacturer’s instructions. The quantification and quality of the RNA were confirmed using a NanoDrop ND-1000 system. The purity of the RNA was measured by the O.D. A260/A280 ratio, which ranged from 1.8 to 2.1. RNA integrity was determined using denatured agarose gel electrophoresis. Ribosomal RNA (rRNA) was removed from the total RNA using the NEBNext rRNA Depletion Kit. RNA libraries were constructed by using the NEBNext Ultra II Directional RNA Library Prep Kit (New England Biolabs, Massachusetts, USA) per the manufacturer’s instructions. Library sequencing was constructed on an Illumina HiSeq instrument with 150-bp paired-end reads.
RNA-Sequencing Analysis
Connectors and low-quality reads were removed, and high-quality clean reads were obtained using Cutadapt software v1.9.3 (http://code.google.com/p/cutadapt/). The results were compared with the reference genome (UCSC hg19) and sequenced using HISAT2 software v2.04 (http://www.ccb.jhu.edu/software/hisat/). Cuffdiff software v2.2.1 (http://cufflinks.cbcb.umd.edu/) was used to obtain DE lncRNAs and mRNAs. DE lncRNAs and mRNAs were screened by calculating the fold-change (FC) and P-values based on fragments per kilobase million mapped reads. The Gene Ontology (GO) (http://www.geneontology.org) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses (https://www.genome.jp/kegg/) were employed to determine the functional roles and signaling pathways of DEGs and P-value <0.05 was considered significant.
Microarray Data Resources and Preprocessing
We searched for miRNA expression profiles of COPD serum and mRNA expression profiles of COPD lung tissues in the GEO database and selected GSE70080 and GSE76925. We used the limma R package to identify DEGs between COPD patients and HCs. An adjusted p-value of < 0.05 was considered the threshold for statistical significance.
Protein–Protein Interaction (PPI) Network Analysis
The STRING database (https://cn.string-db.org/) was used to construct the PPI network of mRNAs, while cytohubba-MCC in Cytoscape v3.6.1 was used to identify hub genes.
Prediction of lncRNA-miRNA-mRNA Interactions
We used miRDB (http://mirdb.org) and DIANA microT-CDS (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index) to predict the potential downstream mRNA of pan-miRNA. LncACTdb3.0 (http://bio-bigdata.hrbmu.edu.cn/LncACTdb/index.html) was used to predict miRNA-lncRNA interactions with high stringency. The interaction networks of mRNAs, miRNAs, and lncRNAs were constructed and visualized.
Reverse-Transcription-Quantitative RT-q(PCR)
Total RNA was extracted from exosomes using the TRIzol reagent. cDNA was synthesized using the Hifair® II 1st Strand cDNA Synthesis kit (Shanghai Yeasen Biotechnology Co., Ltd.) per the manufacturer’s specifications. RT-qPCR was performed using SYBR® Green Master Mix (Shanghai Yeasen Biotechnology Co., Ltd.) with the QuantStudio 5 Real-Time PCR System (Thermo Fisher Scientific, Inc.) per the manufacturer’s instructions. The lncRNA and mRNA expression levels were verified using GAPDH as the internal control via the 2−ΔΔCT method.
Statistical Analysis
Data are expressed as mean ± standard deviation. The Student t-test was performed to analyze normal distribution between the two groups. Correlations were analyzed by using Spearman’s rank correlation coefficient analysis. All statistical analyses were performed using SPSS software, ver. 21.0. A two-sided p-value ≤ 0.05 was considered statistically significant.
Results
Identification of Isolated Exosomes
TEM was used to determine the morphology and size of each exosome. Exosomes had irregular spherical appearances with clearly defined and intact membranes and diameters of 30–100 nm (Figure 1A). The exosome component (CD63) was detected via Western blot analysis to confirm the presence of exosomes; however, the marker, GRP94, was not detected (Figure 1B). These analyses verified the purity of the exosomes.
DE lncRNAs and mRNAs
Plasma lncRNAs and mRNAs from five patients with COPD and five control participants were sequenced. Volcano and scatter plots depicted the distinguished and clustered expression levels of lncRNA and mRNA in COPD patients and HCs, respectively (Figure 2A–D). In total, 24,719 exosomal lncRNAs and 20,308 exosomal mRNAs were identified. A total of 1578 lncRNAs and 3071 mRNAs were differentially regulated by fold change ≥2.0 and p < 0.05. Among these, 1351 lncRNAs were downregulated and 227 were upregulated, while 664 mRNAs were downregulated and 2407 were upregulated.
GO and KEGG Pathway Analyses
To understand the biological functions of the DE lncRNAs and mRNAs, GO, and KEGG enrichment analyses were used to map all of them. GO analyses indicated that biological processes of upregulated DE lncRNAs were mainly involved in neurogenesis, anatomical structure development, and multicellular organism development. In downregulated groups, DE lncRNAs were found to be related to hemostasis, megakaryocyte differentiation, and thrombin-activated receptor signaling pathways. GO molecular function analysis indicated that upregulated DE lncRNAs were enriched in ion binding, cation binding, and cytoskeletal protein binding. Calcium ion binding, the structural constituent of muscle tissues, and protease binding have been determined to share a significant amount of downregulated DE lncRNAs with the “molecular function” GO term. For cellular components, upregulated DE lncRNAs were correlated with the synaptic membrane, cell junction, and postsynaptic membrane. In downregulated groups, the contractile fiber part, myosin complex, and intracellular part were primary enriched categories (Figure 3A–F). Furthermore, the top 10 KEGG-enriched pathways of upregulated DE lncRNAs were: morphine addiction, malaria, the glycosphingolipid biosynthesis-ganglio series, the glutamatergic synapse, the GABA-ergic synapse, the dopaminergic synapse, the calcium signaling pathway, axon guidance, arrhythmogenic right ventricular cardiomyopathy, and African trypanosomiasis (Figure 4A). The downregulated DE lncRNAs were mainly concentrated in vascular smooth muscle contraction, tight junctions, and small-cell lung cancer (Figure 4B).
Identification of DE mRNAs Both in COPD Plasma Exosomes and Tissues
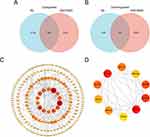
In this study, the microarray dataset, GSE76925, was included to identify DE mRNAs in COPD tissues. We examined the intersections of DE mRNAs in COPD plasma exosome and GSE76925. Venn diagram software was used to identify the common DEGs between DE mRNAs and GSE76925. Results showed that 343 common DEGs were detected, including 52 downregulated genes and 291 upregulated genes (Figure 5A and B).
PPI Network Analysis and Hub Gene Recognition
STRING was used to construct DEGs PPI networks with confidence scores of ≥0.4 and visualized using Cytoscape 3.7.1 software (Figure 5C). Cytohubba, a plug-in Cytoscape, was performed to identify the top 10 hub genes among the DEGs, including RCL1, NOP14, RPS19, MRPS12, MRPS11, TFB1M, MRPS6, MRPL24, MRPL21, and MRPS22 (Figure 5D). Furthermore, we analyzed the relationship between the expression of the 10 hub genes and lung function in COPD. We obtained the expression data of the 10 hub genes, FEV1/FVC, and FEV1 predictive data from the GSE76925 dataset. Except for TFB1M, we found that the nine other genes had significant correlations with FEV1/FVC and the predicted FEV1 in COPD per the Spearman relationship analysis (Table 2 and Table 3).
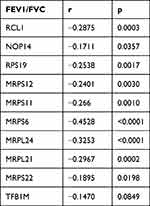 |
Table 2 The SPEARMAN Relationship Between FEV1/FVC and Top 10 Hub Genes |
 |
Table 3 The SPEARMAN Relationship Between FEV1 Predict and Top 10 Hub Genes |
Construction lncRNA-miRNA-mRNA ceRNA Network
We obtained GSE70080 from serum samples of patients with COPD. This dataset came from the GPL20591 platform and contained 16 control samples and 16 COPD samples. Then we used the limma R package to obtain the DE-miRNAs (Figure 6A). There were nine upregulated DE-miRNAs and 11 downregulated DE-miRNAs in GSE70080 (adj.p<0.05 and |logFC|>1). Since the abovementioned hub genes are upregulated DEGs, we used DIANA and miRDB to predict the target genes of downregulated DE-miRNAs in GSE70080 and obtained target genes. The overlap between hub genes and predicted target genes was only MRPS11 (Figure 6B). We chose the MRPS11 gene and structured an MRPS11-related ceRNA network. We obtained hsa-miR-532 and hsa-miR-122 as predicted miRNAs of MRPS11 via the DIANA and miRDB. LncACTdb3.0 was used to predict the lncRNA of hsa-miR-532 and hsa-miR-122. Venn software was used to obtain the overlapping five lncRNAs between our seq and predicted lncRNAs (Figure 6C). Eventually, the MRPS11-related ceRNA network was constructed and consisted of five lncRNAs, as well as two miRNAs (Figure 6D).
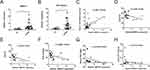
LncRNAs and mRNAs Validation by qRT-PCR
To validate the reliability of our dataset, the expression levels of MPRS11 and RP3-329A5.8 were further identified via qRT-PCR in clinical samples. Similar to the results of our dataset, the expression levels of MPRS11 and RP3-329A5.8 were significantly higher in sCOPD serum exosomes than in normal blood samples (Figure 7A and B). MPRS11 was positively correlated with RP3-329A5.8 (P < 0.001) (Figure 7C). MPRS11 negatively correlated with FEV1/FVC, the predicted FEV1%, and PaO2 in sCOPD blood samples (P < 0.05) (Figure 7D–F). However, the association between the expression level of MPRS11 and the serum levels of C-reactive protein (CRP) and procalcitonin (PCT) in sCOPD patients was not statistically significant (Figure 7G and H).
Discussion
COPD, a condition associated with a person’s smoking habits, is characterized by chronic airway inflammation. It reduces the potential for physical activity by interfering with normal breathing. The symptoms of patients with early COPD are relatively mild, and they seldom seek medical treatment before they reach the advanced stage, which significantly reduces the efficacy of any treatment administered. Furthermore, in the most severe cases, it is challenging to relieve the acute exacerbations and airflow limitation, resulting in a decrease in the patient’s quality of life. Therefore, finding new biomarkers which can diagnose COPD in the early stage and identify patients with a high risk of frequent exacerbations is helpful to develop specific targeted therapies and provide guidance for the prevention of COPD.
Exosomes are a subset of EVs with an average diameter of ~100 nm. Their kinds of constituents, including proteins, lipids, amino acids, nucleic acids, and metabolites, reflect their cell of origin. Exosomes have diverse activities such as transmitting molecules and signals to other cells and remodeling the extracellular matrix. Exosomes are associated with development, tissue homeostasis, immunity, viral pathogenicity, cardiovascular diseases, cancer, and neurodegenerative diseases.4 Exosomes have been reported to exist in all biological fluids, and their composition is can be determined via biological fluid analyses. Recent studies have shown that exosomes have the potential to aid in the diagnosis and prognosis determination of patients with cancer and other diseases. In addition to their diagnostic potential, exosomes also have potential utility in medical treatment. Their natural compositions may play an important role in minimizing adverse reactions and enhancing bioavailability. Exosomes can deliver diverse therapeutic payloads, such as short-interfering RNAs, chemotherapeutic agents, antisense oligonucleotides, and immune modulators.9 LncRNAs lack important open reading frames and are transcripts longer than 200 nucleotides. LncRNAs play an important role in multiple life activities, including epigenetic regulation, cell cycle regulation, and cell differentiation regulation. It is an emerging type of regulatory RNA that can be selectively packaged into exosomes to act as a messenger in intercellular communication. For example, a recent study revealed that lnc-matrix metalloproteinase 2–2 (lncMMP2-2) in A549 cell-derived exosomes promotes the invasion and migration of lung cancer cells by increasing MMP2 expression.10 Although lncRNAs have been an emerging focus in recent years, there is a paucity of published works on the functions of exosomal lncRNAs in COPD. Nevertheless, some researchers have revealed that certain lncRNAs also play significant roles in COPD, which may offer some clues for the further discovery of exosomal lncRNAs that exert effects on the occurrence and development of COPD in the future. Hence, we sequenced the lncRNA and mRNA of plasma exosomes in patients with COPD and found 1578 DE lncRNAs and 3071 DE mRNA among patients with COPD and HCs, respectively.
We predicted the potential functions of these DE lncRNAs using GO and KEGG enrichment analyses, with respect to COPD occurrence and development. According to the GO analysis, the DE, upregulated lncRNAs were located in synaptic membranes and cell junctions, and they were associated with biological processes involved in airway remodeling, such as anatomical structure development and multicellular organism development. Hogg et al found that FEV1 was negatively associated with the accumulation of mucous exudates in small airway lumens and an increase in the wall thickness of these small airways.11 Epithelial abnormalities, including squamous metaplasia, epithelial proliferation, and goblet cell hyperplasia, are likely responsible for an increased volume of tissue in the walls of small airways. In addition, peribronchiolar fibrosis is also a vital feature leading to the narrowing of the small airways in patients with COPD. It is suggested that these anatomical structural changes in small airways are associated with repair mechanisms to protect the small airways from the destruction caused by cigarette smoke.12 Furthermore, downregulated DE lncRNAs, which were located in the contractile fiber part and myosin complex, were associated with biological processes involved in systemic disease with pulmonary and extrapulmonary features, such as hemostasis, thrombin-activated receptor signaling pathways, and structural constituents of muscle tissues. COPD pro-inflammatory mediators increase persistent systemic inflammation by activating vascular endothelial inflammation, blood coagulation, and plaque formation (with plaque instability).13 Damage to the endothelium and inflammation resulted in a pro-thrombotic state via the induction of a disorder between pro- and anti-coagulant factors.14 In particular, cigarette smoking increased platelet activation, triggered the coagulation cascade, and subsequently led to thrombotic effects.15 Skeletal muscle dysfunction is one of the systemic manifestations of COPD which encompasses a decrease in strength and endurance but an increase in fatigability. Skeletal muscle dysfunction is characterized by atrophy, a fiber-type shift from type I to type IIx fibers, and impairments in mitochondrial function and capillarization.16
These signaling pathways that are enriched in the KEGG analysis in this current study were also associated with the pathogenesis and prognosis of COPD, such as the arrhythmogenic right ventricular (RV) cardiomyopathy and small-cell lung cancer signaling pathway. COPD is often associated with cardiovascular comorbidities. Cigarette smoke (CS) directly affects the pulmonary vasculature and leads to pulmonary hypertension (PH). Furthermore, PH can result in hypertrophy, dilatation, and dysfunction of the RV by increasing its workload.17 A large proportion of patients with lung cancer have a history of COPD. It has been acknowledged that cigarette-associated COPD should be regarded as a vital risk factor for lung cancer. CS causes lung barrier inflammation and dysfunction, and endothelial injury is an important mediator in this process. Protracted inflammation is associated with EMT, which gives rise to aberrant tissue repair, which might help interpret the close relationship between COPD and lung cancer.18
A combination of GSE76925 and our RNA-seq analysis identified 343 common DE mRNAs. To systemically analyze the relationships of common DEGs in COPD, we mapped them into the STRING database, obtained PPI networks, and identified the top 10 hub genes via cytoHubba. MRPS12, MRPS11, MRPS6, MRPL24, MRPL21, and MRPS22 are all essential components of mitochondrial ribosomal proteins (MRPs), which are involved in the structural and functional integrity of the mitoribosome complex.19 The remaining four hub genes (NOP14, TFB1M, RPS19, and RCL1) are also involved in regulating mitochondrial ribosome biogenesis.20–23 The mitoribosome is responsible for protein synthesis of mitochondrial DNA-encoded genes. These proteins play a central role in cellular activities, such as oxidative phosphorylation, ATP production, and respiration. Mitochondrial dysfunction can be caused by the inhibition of mitochondrial protein synthesis or mitoribosome assembly impairments, resulting in a decrease in mitochondrial translation. This decline can trigger mitochondrial ribosomal stress and lead to pulmonary cell injury, death, and diseases.24 In the quadriceps muscles of COPD patients, miR-542-3p/5p expression was elevated and negatively associated with mitochondrial gene expression. Impaired mitoribosome synthesis contributed to quadriceps muscle dysfunction.25
Recent studies have demonstrated that lncRNAs may function as miRNA sponges. GSE70080 was used to analyze the serum miRNA-seq of COPD patients. miRDB and DIANA were used to predict downstream mRNAs of DE-miRNAs. MRPS11 was the only overlap gene between hub genes and predicted target genes. MRPS11, as the related gene of lncRNA ZFHX4-AS1, was associated with immune checkpoints and could regulate the proliferation and invasion of ovarian cancer cells.26 Recent studies have reported that MRPS11 may contribute to the pathogenesis of Parkinson’s disease and ankylosing spondylitis.27,28 However, it was the first time it was pointed out that MRPS11 may be associated with COPD. hsa-miR-122 and hsa-miR-532-5p were predicted upstream miRNAs of MRPS11 separately via the DIANA and miRDB. After combining our RNA-seq and LncACTdb3.0, we finally constructed a lncRNA–miRNA–mRNA network. According to a previous study,29 hsa-miR-122-5p was significantly downregulated in COPD patients compared to HCs; moreover, downregulated miR-122 expression level associated with microbiota imbalance may play an important role on COPD progression by enhancing IL-17a production.30 Thus, we pay more attention to hsa-miR-122. lncRNA X–inactive specific transcript (XIST) expression was elevated in lung samples from patients with COPD. XIST promotes inflammatory responses and apoptosis in CS extract-stimulated 16HBE cells via the miR-200c-3p/EGR3 axis.31 So, we chose RP3-329A5.8 for subsequent validation.
To conclude, in this study, we demonstrated the upregulation of MRPS11 and RP3-329A5.8 in serum exosomes obtained from sCOPD patients. Additionally, MPRS11 was negatively correlated with FEV1/FVC and the predicted FEV1% in sCOPD patients. MRPS11 and RP3-329A5.8 could be potential diagnostic biomarkers for COPD, and they may help to improve the accuracy of diagnosis and assess the severity of COPD. The expression level of exosome MRPS11 showed no correlation with CRP and PCT, suggesting that the influence of MRPS11 on sCOPD is unrelated to the inflammatory response. MPRS11 encodes a mitochondrial ribosomal protein that binds to 12S rRNA and is involved in mitochondrial protein synthesis. Several studies have revealed that mitochondrial dysfunction has an influence on COPD by regulating oxidative stress, proliferation, apoptosis, fibrosis, and, certainly, metabolism.32 Hence, we predicted that the RP3-329A5.8/miR-122/MRPS11 ceRNA might contribute to clarifying the role of MRPS11 on the mitochondrial dysfunction in the pathogenesis of COPD. The specific mechanism and effect of ceRNA on COPD need to be further investigated.
Our study has some limitations. We did not verify key results using biochemical experiments and clinical trials. Due to incomplete follow-up data of the samples that we analyzed and collected, the potential relationships between hub genes and comorbidities or prognosis were not conducted. Therefore, subsequent studies with larger samples and related follow-up data should validate and extend the clinical utility of the hub genes that were identified in this study. Further in vivo and in vitro studies are required to explore how hub genes in COPD are regulated.
Conclusion
We identified DE lncRNAs and mRNAs in plasma exosomes from patients with COPD through high-throughput sequencing. Moreover, RP3-329A5.8/miR-122/MRPS11 ceRNA might affect the physiopathology of COPD by regulating mitochondrial function. These results might be useful in exploring diagnosis biomarkers, predicting pathological progress and treating of COPD.
Funding
This research was supported by grant from the Science and Technology Department of Jilin Province (YDZJ202301ZYTS014).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Agusti A, Vogelmeier C, Faner R. COPD 2020: changes and challenges. Am J Physiol Lung Cell Mol Physiol. 2020;319(5):L879–L883. doi:10.1152/ajplung.00429.2020
2. Wang C, Li Z, Liu Y, et al. Exosomes in atherosclerosis: performers, bystanders, biomarkers, and therapeutic targets. Theranostics. 2021;11(8):3996–4010. doi:10.7150/thno.56035
3. Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73(10):1907–1920. doi:10.1016/j.jprot.2010.06.006
4. Pegtel DM, Gould SJ. Exosomes. Annu Rev Biochem. 2019;88:487–514. doi:10.1146/annurev-biochem-013118-111902
5. Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi:10.1016/j.cell.2018.01.011
6. Poulet C, Njock M-S, Moermans C, et al. Exosomal long non-coding RNAs in lung diseases. Int J Mol Sci. 2020;21(10):3580. doi:10.3390/ijms21103580
7. Zhang R, Xia Y, Wang Z, et al. Serum long non coding RNA MALAT-1 protected by exosomes is up-regulated and promotes cell proliferation and migration in non-small cell lung cancer. Biochem Biophys Res Commun. 2017;490(2):406–414. doi:10.1016/j.bbrc.2017.06.055
8. Song L, Peng J, Guo X. Exosomal lncRNA TCONS_00064356 derived from injured alveolar epithelial type II cells affects the biological characteristics of mesenchymal stem cells. Life Sci. 2021;278:119568. doi:10.1016/j.lfs.2021.119568
9. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478):6977. doi:10.1126/science.aau6977
10. Wu DM, Deng SH, Liu T, et al. TGF-beta-mediated exosomal lnc-MMP2-2 regulates migration and invasion of lung cancer cells to the vasculature by promoting MMP2 expression. Cancer Med. 2018;7(10):5118–5129.
11. Hogg JC, Chu F, Utokaparch S, et al. The nature of small-airway obstruction in chronic obstructive pulmonary disease. N Engl J Med. 2004;350(26):2645–2653. doi:10.1056/NEJMoa032158
12. Saetta M, Turato G, Baraldo S, et al. Goblet cell hyperplasia and epithelial inflammation in peripheral airways of smokers with both symptoms of chronic bronchitis and chronic airflow limitation. Am J Respir Crit Care Med. 2000;161(3 Pt 1):1016–1021. doi:10.1164/ajrccm.161.3.9907080
13. Barnes PJ. Cellular and molecular mechanisms of chronic obstructive pulmonary disease. Clin Chest Med. 2014;35(1):71–86. doi:10.1016/j.ccm.2013.10.004
14. Lemichez E, Lecuit M, Nassif X, et al. Breaking the wall: targeting of the endothelium by pathogenic bacteria. Nat Rev Microbiol. 2010;8(2):93–104. doi:10.1038/nrmicro2269
15. Hunter KA, Garlick PJ, Broom I, et al. Effects of smoking and abstention from smoking on fibrinogen synthesis in humans. Clin Sci (Lond). 2001;100(4):459–465. doi:10.1042/cs1000459
16. Marklund S, Bui KL, Nyberg A. Measuring and monitoring skeletal muscle function in COPD: current perspectives. Int J Chron Obstruct Pulmon Dis. 2019;14:1825–1838. doi:10.2147/COPD.S178948
17. Andre S, Conde B, Fragoso E, et al. COPD and cardiovascular disease. Pulmonology. 2019;25(3):168–176. doi:10.1016/j.pulmoe.2018.09.006
18. Hou W, Hu S, Li C, et al. Cigarette smoke induced lung barrier dysfunction, EMT, and tissue remodeling: a possible link between COPD and lung cancer. Biomed Res Int. 2019;2019:2025636. doi:10.1155/2019/2025636
19. Cheong A, Lingutla R, Mager J. Expression analysis of mammalian mitochondrial ribosomal protein genes. Gene Expr Patterns. 2020;38:119147. doi:10.1016/j.gep.2020.119147
20. Kirsch VC, Orgler C, Braig S, et al. The cytotoxic natural product vioprolide a targets nucleolar protein 14, which is essential for ribosome biogenesis. Angew Chem Int Ed Engl. 2020;59(4):1595–1600. doi:10.1002/anie.201911158
21. Verma G, Bowen A, Gheibi S, et al. Ribosomal biogenesis regulator DIMT1 controls beta-cell protein synthesis, mitochondrial function, and insulin secretion. J Biol Chem. 2022;298(3):101692. doi:10.1016/j.jbc.2022.101692
22. Hiregange DG, Rivalta A, Yonath A, et al. Mutations in RPS19 may affect ribosome function and biogenesis in Diamond Blackfan anemia. FEBS Open Bio. 2022;12(7):1419–1434. doi:10.1002/2211-5463.13444
23. Zhu Q, Tao B, Chen H, et al. Rcl1 depletion impairs 18S pre-rRNA processing at the A1-site and up-regulates a cohort of ribosome biogenesis genes in zebrafish. Nucleic Acids Res. 2021;49(10):5743–5759. doi:10.1093/nar/gkab381
24. Karim L, Kosmider B, Bahmed K. Mitochondrial ribosomal stress in lung diseases. Am J Physiol Lung Cell Mol Physiol. 2022;322(4):L507–L517. doi:10.1152/ajplung.00078.2021
25. Farre-Garros R, Lee JY, Natanek SA, et al. Quadriceps miR-542-3p and −5p are elevated in COPD and reduce function by inhibiting ribosomal and protein synthesis. J Appl Physiol. 2019;126(6):1514–1524. doi:10.1152/japplphysiol.00882.2018
26. Wang X, Wang Y, Sun F, et al. Novel LncRNA ZFHX4-AS1 as a potential prognostic biomarker that affects the immune microenvironment in ovarian cancer. Front Oncol. 2022;12:945518. doi:10.3389/fonc.2022.945518
27. Zheng H, Qian X, Tian W, Cao L. Exploration of the common gene characteristics and molecular mechanism of Parkinson’s disease and crohn’s disease from. Transcript Data Brain Sci. 2022;12:6.
28. Najafzadeh L, Mahmoudi M, Ebadi M, et al. Co-expression network analysis reveals key genes related to ankylosing spondylitis arthritis disease: computational and experimental validation. Iran J Biotechnol. 2021;19(1):e2630. doi:10.30498/IJB.2021.2630
29. O’Farrell HE, Bowman RV, Fong KM, et al. Plasma extracellular vesicle miRNAs can identify lung cancer, current smoking status, and stable COPD. Int J Mol Sci. 2021;22(11):5803. doi:10.3390/ijms22115803
30. Zhu K, Zhou S, Xu A, et al. Microbiota imbalance contributes to COPD deterioration by enhancing IL-17a production via miR-122 and miR-30a. Mol Ther Nucleic Acids. 2020;22:520–529. doi:10.1016/j.omtn.2020.09.017
31. Chen P, Jiang P, Chen J, et al. XIST promotes apoptosis and the inflammatory response in CSE-stimulated cells via the miR-200c-3p/EGR3 axis. BMC Pulm Med. 2021;21(1):215. doi:10.1186/s12890-021-01582-8
32. Prakash YS, Pabelick CM, Sieck GC. Mitochondrial dysfunction in airway disease. Chest. 2017;152(3):618–626. doi:10.1016/j.chest.2017.03.020
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.