Back to Journals » Journal of Hepatocellular Carcinoma » Volume 9
RNA Binding Motif Protein 3 Promotes Cell Metastasis and Epithelial–Mesenchymal Transition Through STAT3 Signaling Pathway in Hepatocellular Carcinoma
Authors Zhang L, Zhang Y, Shen D, Chen Y, Feng J, Wang X, Ma L, Liao Y , Tang L
Received 7 December 2021
Accepted for publication 21 April 2022
Published 9 May 2022 Volume 2022:9 Pages 405—422
DOI https://doi.org/10.2147/JHC.S351886
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Xin Wei Wang
Lu Zhang,1,* Yi Zhang,1,* Dongliang Shen,1 Ying Chen,1 Jianguo Feng,2 Xing Wang,1 Lunkun Ma,1 Yi Liao,3,4 Liling Tang1
1Key Laboratory of Biorheological Science and Technology, Ministry of Education, College of Bioengineering, Chongqing University, Chongqing, 400044, People’s Republic of China; 2Southwest Medical University, Department Anesthesiology, Affiliated Hospital, Luzhou, 646000, People’s Republic of China; 3The Central Laboratory, Shenzhen Second People’s Hospital/First Affiliated Hospital of Shenzhen University Health Science Center, Shenzhen, Guangdong, 518035, People’s Republic of China; 4Department of Thoracic Surgery, Southwest Hospital, Army Medical University, Chongqing, 400038, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Liling Tang; Yi Liao, Tel +86 139 9605 1730 ; +86 139 9656 6993, Fax +86-23-65111901 ; +86-23-68763333, Email [email protected]; [email protected]
Purpose: RNA binding motif protein 3 (RBM3) has been reported to be dysregulated in various cancers and associated with tumor aggressiveness. Epithelial–mesenchymal transition (EMT) is an important biological process by which tumor cells acquire metastatic abilities. This study aimed to explore the regulatory and molecular mechanisms of RBM3 in EMT process.
Methods: Western blotting, IHC, and qRT-PCR were performed to evaluate the expression of target genes. Transwell assay was used to investigate the migration and invasion. RNA immunoprecipitation and luciferase reporter assay were performed to explore the correlation of RBM3 with STAT3 or microRNA-383. Animal HCC models were used to explore the role of RBM3 in metastasis in vivo.
Results: RBM3 was highly expressed in HCC tissues compared to healthy tissues, and its level was negatively correlated with the prognosis of HCC patients. RBM3 overexpression accelerated migration and invasion, promoted EMT process, and activated STAT3 signaling. EMT induced by RBM3 was not only attenuated by inhibiting pSTAT3 via S3I-201 but also abolished by suppressing STAT3 expression via siRNAs. Mechanistically, RBM3 increased STAT3 expression by stabilizing STAT3 mRNA via binding to its mRNA. As an upstream target of RBM3, microRNA-383 inhibited RBM3 expression by binding to its 3ʹUTR and resulted in the inhibition of the EMT process. Inhibition of RBM3 in HCC animal models prolonged survival and ameliorated malignant phenotypes in mice.
Conclusion: Our findings support that RBM3 promotes HCC metastasis by activating STAT3 signaling.
Keywords: RBM3, metastasis, EMT, STAT3, microRNA-383
Introduction
Hepatocellular carcinoma (HCC), one of the four most common types of cancer, always holds a high morbidity and mortality.1 Although increasing number of studies have explored the relevant molecular mechanisms, the prognosis of HCC remains unsatisfactory, mainly due to metastasis and postoperative recurrence.2–4 Nowadays, the cancer therapy is still a very intractable problem because of its highly metastatic nature. Metastasis of tumor cells is life-threatening, and it could lead to the spread of tumor cells from situ tissues to other healthy tissues. Therefore, it is of great clinical value to explore the underlying mechanisms of tumor metastasis. Epithelial–mesenchymal transition (EMT) is an important biological process that cannot be ignored in the acquisition of aggressive capabilities by tumor cells. Epithelial cells acquire motility through EMT, and the migration speed of mesenchymal cells could also be improved by adopting amoeboid features.5 CD138, a positive marker of cell migration, was increased in human mammary epithelial cells undergoing EMT induced by TGFβ1 or 4-hydroxytamoxifen.6 Furthermore, the mesenchymal state allows cells to migrate to distant organs and maintain stemness.7 EMT is characterized by losing cell morphology, polarity, adhesion and down-regulation of E-cadherin, as well as up-regulation of N-cadherin and Vimentin.8 Primary tumors could acquire migratory and invasive abilities through EMT and form metastases.9,10 A large number of studies have shown that EMT is associated with metastasis of various cancers, such as breast cancer,11 nasopharyngeal carcinoma,12 head and neck squamous carcinoma,13 colorectal cancer14 and so on. EMT has been shown to play a vital role in the development and progression of HCC, providing the possibility for HCC to adapt to the tumor microenvironment and acquire the ability to migrate and invade.15–17 The importance of EMT in HCC is obvious, and thus it is vitally significant to explore the potential mechanisms of EMT in the regulation of HCC progression.
Signal transducer and activator of transcription 3 (STAT3) is known to be involved in cell proliferation,18,19 survival,20 metastasis21 and immune response.22,23 Numerous articles have demonstrated the great significance of STAT3 in the EMT process. For example, Guo et al24 found that miR-125b-5p inhibited the expression of STAT3 by binding to its 3’ UTR, thereby suppressing EMT in HCC cells. In breast cancer, SIX4 could induce the activation of EMT by directly interacting with STAT3 and increasing the phosphorylation expression of STAT3.25
RNA binding motif protein 3 (RBM3) is a member of the highly conserved RNA binding proteins that participate in cellular processes by binding to mRNA of target genes.26 Moreover, it acts as a cold shock response protein to protect cells from hypothermia-induced death.27 Under normal physiological conditions, RBM3 is essential for cell survival and proliferation. However, many preclinical studies have shown that RBM3 expression is dysregulated in various cancers, such as reduced expression in urothelial bladder cancer,28 colorectal cancer,29 non-small cell lung cancer,30 and increased expression in epithelial ovarian cancer,31 prostate cancer,32 and astrocytoma.33 The dysregulation of RBM3 suggests that RBM3 may be involved in tumor progression. Furthermore, RBM3 has been demonstrated to be associated with the migration and invasion of tumor cells. In urothelial carcinoma, RBM3 expression was extremely higher in metastases than in primary tumors.34 Similarly, RBM3 promoted the proliferation and metastasis of human breast cancer cells through binding to 3ʹUTR of actin-related protein 2/3 complex subunit 2 (ARPC2).35 Such a large number of studies have shown the positive regulation of RBM3 on the migration and invasion of various tumor cells. Interestingly, the relationship between RBM3 and EMT, the classical process by which cells acquire migration and invasion abilities, has not been reported. Moreover, the correlation between RBM3, EMT and STAT3 signaling has not been demonstrated.
In the present study, we identified the functional role of RBM3 on EMT process in HCC for the first time. Our data showed that RBM3 was overexpressed in human HCC tissues and HCC cell lines, and RBM3 overexpression was correlated with poor prognosis in HCC patients. Mechanistically, RBM3 bound directly to STAT3 mRNA to enhance its stability, and the resulting accumulation of STAT3 protein induced the EMT process and promoted the progression of HCC. The upstream target of RBM3 was predicted at TargetScan database and further validated.
Materials and Methods
Cell Culture
HEK-293T cell line, normal human liver cell HL-7702 and human hepatoma carcinoma cells MHCC97H were cultured in Dulbecco’s Modified Eagle Medium (HyClone, USA) containing 10% fetal bovine serum (FBS), 100U/mL penicillin, and 100 μg/mL streptomycin. HEK-293T and HL-7702 were obtained from The Third Military Medical University. MHCC97H was purchased from ShangHai MEIXUAN Biological Science and Technology (China). HepG2 cell line was kindly provided by Dr Tang from Chongqing Medical University and cultured in Minimum Eagle’s Medium (HyClone, USA) containing 10% FBS, 100U/mL penicillin, and 100 μg/mL streptomycin. For cells stably expressing RBM3, RBM3 shRNA, or scrambled shRNA, the medium was supplemented with 1 μg/mL puromycin. All cells were incubated in a humidified cell culture incubator with 5% CO2 at 37℃. All cell lines were assessed to be free of mycoplasma infection.
Plasmids
The cDNA of human RBM3 was cloned into the polyclonal site of pLenti-CMV-EGFP (Addgene) between the BamH I and Sal I restriction sites to construct plasmid pLenti-CMV-RBM3. In order to stably knock down RBM3 expression, shRNAs targeting two different positions of RBM3 mRNA were designed and cloned between the Age I and EcoR I restriction sites of the plasmid pLKO.1-TRC (Addgene) cloning vector, forming plasmid pLKO.1-RBM3-shRNA #1 and #2, respectively. A scrambled shRNA was designed as control (plasmid pLKO.1-scrambled-shRNA). The oligonucleotide primers used to generate RBM3 shRNAs are listed in Table 1.
 |
Table 1 Oligonucleotide Primer Sets Used to Generate Scrambled shRNA and RBM3 shRNA |
Establishment of Stable Cell Lines
HepG2 cell lines overexpressing EGFP or RBM3 (referred to as HepG2-EGFP or HepG2-RBM3) were established by lentiviral transfection of pLenti-CMV-EGFP or pLenti-CMV-RBM3 plasmids. In brief, HEK-293T packaging cells were co-transfected with pLenti-CMV-EGFP/RBM3, pCMV-VSV-G and pCMV-dR8.2 dvpr using Effectene Transfection Reagent (QIAGEN, Germany) according to the manufacturer’s protocol. After 48 hours of incubation, the viral supernatant was collected. HepG2 cells were infected with lentiviral particles for 24 hours and then screened with 2μg/mL puromycin for 1 week. MHCC97H cells were infected with lentiviral particles, which were established by lentiviral transfection of pLKO.1-RBM3-shRNA#s or pLKO.1-scrambled-shRNA in the same way as described above. Puromycin (2 μg/mL) was used to screen stable cell lines. MHCC97H cell lines expressing RBM3 shRNA or scrambled shRNA were referred to as MHCC97H-RBM3-shRNA#s or MHCC97H-scrambled-shRNA. HepG2-pLNCX2 and HepG2-pLNCX2-RBM3 cells were established by retroviral transfection with pLNCX2/pLNCX2-RBM3, pCMV-VSV-G and pUMVC as above. Geneticin (2 mg/mL) was used to screen the stable cell lines.
RNA Interference (RNAi)
MHCC97H cells were grown overnight to approximately 40% confluence and transfected with human RBM3 short interfering RNA (siRNA) for 6 hours. The transfection medium was then replaced with a fresh medium, and the cells were cultured for another 48 hours. In order to knock down STAT3 expression, HepG2-EGFP and HepG2-RBM3 cells were transfected with human STAT3 siRNAs. Transfection was performed with X-tremeGENE siRNA Transfection Reagent (Roche, Switzerland) according to the manufacturer’s instructions. The RBM3 siRNA sequences were as follows:
RBM3-siRNA #1 UUUAACACCGACGAGCAGGTT
RBM3-siRNA #2 GGACCUAUCUCUGAGGUGGTT
RBM3-siRNA #3 AUAUGGAUAUGGACGUUCCTT
RBM3-siRNA #4 CUUCCAAAUGGCUGTAUUUTT
The STAT3 siRNA sequences were as follows:
STAT3-siRNA #1 CCACUUUGGUGUUUCAUAATT
STAT3-siRNA #2 CCGUGGAACCAUACACAAATT
mRNA Decay Analyses
MHCC97H cells were grown overnight to approximately 40% confluence and transfected with RBM3-siRNA or control siRNA for 6 hours. Then, the transfection medium was replaced with a fresh medium, and cells were cultured for another 48 hours. Cells were lysed with lysis buffer (1% Triton-100, 0.1% sodium dodecyl sulfate) and added with 5 μM Actinomycin D (APE×BIO, USA) for 0, 20, 40 and 60 min. RNAs were extracted at specific time and reversed into cDNAs. qRT-PCR was performed to detect RBM3 and STAT3 expression.
Western Blotting
Cells were collected and lysed using RIPA buffer containing a mixture of PMSF and phosphatase inhibitors. Cell lysates were incubated on ice for 30 min with shaking every 5 min and then centrifuged at 13,000 revolutions per minute (rpm) for 10 min at 42℃. Protein lysates were used to measure the expression of target proteins. Equal amounts of proteins were resolved on SDS-PAGE and transferred to a 0.45 μM PVDF membrane. The membrane was then blocked by slowly shaking with 5% nonfat milk for 1 hour at room temperature, followed by washing with Tris-buffered saline containing 0.1% Tween-20 (TBST) for 10 min and repeated 3 times. The primary antibody was incubated overnight at 4℃, then washed with TBST for 10 min, repeated 3 times, followed by incubating with the corresponding secondary antibody for 1 hour at room temperature with slow shaking. The images were taken with a standard chemiluminescence. The anti-E-cadherin, anti-N-cadherin, anti-p-STAT3 and anti-STAT3 antibodies were purchased from Cell Signaling Technology. The anti-RBM3, anti-VEGF, anti-MMP2 and anti-MMP9 antibodies were purchased from Abcam. The anti-Vimentin antibody was purchased from Santa Cruz. The anti-GAPDH antibody was purchased from Proteintech.
Immunohistochemistry (IHC)
The expression of RBM3, STAT3, E-cadherin, N-cadherin and Vimentin in formalin-fixed, paraffin-embedded hepatocellular carcinoma tissue sections were examined by immunohistochemistry using a Rabbit Streptomycin-Biotin detection system (Zsbio, China). Briefly, tissue slides were deparaffinized in xylene and rehydrated in alcohol. Antigen retrieval was achieved using a hot water bath and sodium citrate buffer. Endogenous peroxidase blocker was added for 10 minutes. Sections were blocked with normal goat serum for 15 minutes. Sections were then incubated overnight at 4℃ with the indicated primary antibody. Then, biotin-labeled goat anti-rabbit IgG was added for 20 minutes. Next, horseradish enzyme labeled streptomycin buffer and DAB buffer (Zsbio, China) were added for 10–15 min, respectively. At last, sections were counterstained with hematoxylin for 2 minutes. The anti-E-cadherin, anti-STAT3 antibodies were purchased from Cell Signaling Technology. The anti-RBM3 antibody was purchased from Proteintech.
The hepatocellular carcinoma tissue microarray containing ninety tumor tissues and their paired noncancerous tissues were randomly collected from HCC patients who had undergone surgical resection. None of these patients received any preoperative antitumor therapy. RBM3 expression was evaluated using a graded semi-quantitative scoring system. Staining intensity was classified as none (0), weak (1), strong (2), or very strong (3), and staining patterns were classified as negative (0: ≤10%), sporadic (1: 11–25%), focal (2: 26–50%) or diffuse (3: ≥51%). The overall score was calculated by multiplying intensity and positivity scores as follows: 0 (negative), 1–4 (moderately positive), 5–9 (strongly positive). To analyze clinical significance and prognosis, malignant samples with strongly positive RBM3 staining were classified as RBM3/H, while RBM3/L indicated moderately positive and negative expression.
Reverse Transcription Polymerase Reaction (RT-PCR) and Quantitative Real-Time PCR (qRT-PCR)
RNA was extracted using RNAsimple Total RNA Kit (TIANGEN, China) according to the manufacturer’s instructions. cDNA was then obtained by RT-PCR using the PrimerScriptTM RT reagent Kit with Gdna Eraser (Takara, Japan). The concentration of cDNA was measured using Nanodrop 2000 (Thermo Fisher Scientific, USA). qRT-PCR was performed on the first strand of cDNA with primers using NovoStart SYBR qPCR SuperMix Plus (Novoprotein, China). The primers were as follows:
STAT3 #1: Forward 5’-ATCACGCCTTCTACAGACTGC-3’
Reverse 5’-CATCCTGGAGATTCTCTACCACT-3’
STAT3 #2: Forward 5’-CTCCAGGATGACTTTGATT-3’
Reverse 5’-TTGCCGCCTCTTCCAGT-3’
GAPDH: Forward 5’-CTGGGCTACACTGAGCACC-3’
Reverse 5’-AAGTGGTCGTTGAGGGCAATG-3’
RBM3: Forward 5’- TGAGAGCCATGAACGGAGAGT-3’
Reverse 5’- GTAGCGGTCATAACCACCCTG-3’
Transwell Migration and Invasion Assays
Cells were digested into a single cell suspension. 1×104 cells were seeded in the upper chambers with 200 μL serum-free medium. Transwell chamber (Corning, USA) was paved with matrigel mix (Corning, USA) for invasion assay. The bottom chamber was filled with a medium containing 10% FBS. After incubation at 37℃ for 24 hours, the upper chamber was washed with cold phosphate buffer saline (PBS) and fixed with cold methanol at 4℃, then stained with crystal violet (Kaigen, China) for 20 min. Finally, the cells in different fields were photographed and counted.
RNA Immunoprecipitation Assay
HepG2-RBM3 cells were cultured in 10 cm dishes for 48 hours to 80% confluence. Cells were washed with cold PBS and lysed on ice with a mixture of RIPA buffer, PMSF, RNase inhibitor and protease inhibitor. The cell extracts were then placed under ultraviolet light for 30 seconds and incubated on ice for 30 min, shaking every 10 min. Next, cell lysates were incubated with anti-RBM3 (Proteintech, USA) or anti-IgG (Santa Cruz, USA) overnight at 4℃. The cell lysate bound to the antibody was then incubated with Protein A+G Agarose (Beyotime, China). Finally, the isolated RNA was extracted with RNAsimple Total RNA Kit (TIANGEN, China), and the purified RNA was subjected to Reverse Transcription PCR to get cDNA, which was then used for semi-quantitative PCR and qRT-PCR analysis.
Luciferase Reporter Assay
The sequences of RBM3-3ʹUTR and corresponding mutation were designed, synthesized and inserted into pmirGLO Dual-luciferase miRNA Target Expression Vector (Promega, USA), named wt-RBM3 and mut-RBM3, respectively. These two plasmids were co-transfected with miR-NC or miR-383 mimics into HepG2 cells. Relative luciferase activity was examined with a Dual Luciferase Assay Kit (Promega, USA) according to the manufacturer’s protocol.
Animal Experiments
Eight-week-old female BALB/c–nu mice were purchased from the Third Military Medical University. MHCC97H-RBM3-shRNA cells and MHCC97H-scrambled-shRNA cells (2 × 105) or HepG2-pLNCX2-RBM3 cells and HepG2-pLNCX2 cells (2 × 105) were injected into BALB/c–nu mice via tail vein. Mice were sacrificed after natural death, livers were isolated and fixed in 4% neutral-buffered formalin, and the survival days were recorded. All animal studies were performed in strict accordance with the Laboratory Animal Welfare and Ethics Committee of Chongqing University.
Statistical Analysis
The data were expressed as the mean ± SEM. Cumulative survival curves were calculated with the Kaplan–Meier method and analyzed with the Log rank test. The statistical significance of differences for the mean values of group was determined with Student’s t-test. Differences with a P-value of less than 0.05 were considered significant.
Results
Elevated Expression of RBM3 in HCC Samples Correlates with Poor Prognosis
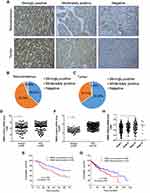
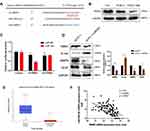
To investigate the clinical significance of RBM3 in HCC, we measured the expression of RBM3 in 90 HCC samples and their paired noncancerous tissues. Strong, moderate or weak expression of RBM3 in HCC and noncancerous samples are shown in the representative images (Figure 1A). In total, 33 noncancerous samples (36.67%) showed strong RBM3 expression, while the percentage of moderate and negative RBM3 expression was 57.78% (52 cases) and 5.56% (5 cases), respectively (Figure 1B). In contrast, RBM3 was relatively high in HCC tissues. Fifty-nine HCC samples (65.55%) showed strong RBM3 expression, while 28 HCC samples (31.33%) and 3 HCC samples (3.33%) showed moderate and negative RBM3 expression, respectively (Figure 1C). Samples with strongly positive RBM3 staining were identified as high RBM3 expression (RBM3/H), and moderately positive and negative expression were identified as low RBM3 expression (RBM3/L). Further expression analysis showed that RBM3 was highly expressed in HCC tumor tissues compared to the corresponding adjacent noncancerous tissues (Figure 1D). Kaplan–Meier analysis indicated that high expression of RBM3 was correlated with significantly shorter overall survival (Figure 1E). These results were consistent with the analysis of the Cancer Genome Atlas (TCGA) database (Figure 1F and G). TCGA database also showed that HCC patients with high RBM3 expression exhibited advanced tumor progression (Figure 1H). Additionally, RBM3 expression was associated with various clinical clinicopathological parameters, including age, death, tumor grade and recurrence in HCC patients (Table 2).
 |
Table 2 Correlation of Clinical Features and Clinicopathological Parameters of the Hepatocellular Carcinoma Patients and RBM3 Expression Analyzed by IHC |
RBM3 Promotes the EMT Process in HCC Cells
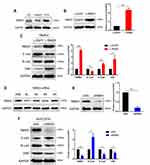
Protein level of RBM3 was evaluated in HCC cell lines and normal human liver cell HL-7702. Relatively high expression of RBM3 was observed in the HCC cell lines, HepG2 and MHCC97H (Figure 2A). We constructed a HepG2-RBM3 cell line to overexpress RBM3 (Figure 2B) and found that overexpression of RBM3 down-regulated the expression of E-cadherin while up-regulating the expression of N-cadherin and Vimentin (Figure 2C). In order to knock down RBM3 expression in MHCC97H cells, four RBM3-specific siRNAs were designed. Western blotting results showed that RBM3-siRNA#1 and #3 successfully reduced the expression of RBM3 (Figure 2D). To inhibit the expression of RBM3 in MHCC97H cells more effectively, we mixed RBM3-siRNA#1 and #3 (Figure 2E) and found that inhibition of RBM3 expression increased the expression of E-cadherin and decreased the expression of N-cadherin and Vimentin (Figure 2F). Moreover, we constructed MHCC97H-RBM3-shRNA#s cells to knock down RBM3 stably (Figure S2A). As with MHCC97H cells transfected with RBM3-siRNAs, inhibition of RBM3 expression by shRNAs also significantly suppressed N-cadherin and Vimentin (Figure S2B). Morphological evidence showed that RBM3 induced the transformation of cell morphology from irregular shape to long spindle-shaped form (Figure S1). These results indicate that RBM3 promotes the EMT process in HCC cells.
RBM3 Accelerates the Migration and Invasion in HCC Cells
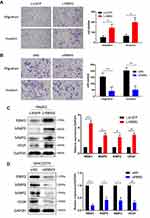
EMT is an essential process by which tumor cells acquire the ability to migrate and invade, and we further explored the effect of RBM3 on the metastasis of HepG2 and MHCC97H cells. Our data showed that overexpression of RBM3 significantly increased the migratory and invasive ability of HepG2 cells (Figure 3A), whereas down-regulation of RBM3 decreased the migratory and invasive ability of MHCC97H cells (Figures 3B and S2C). Furthermore, we evaluated the expression of two matrix metalloproteinases (MMPs), MMP2 and MMP9, as well as vascular endothelial growth factor (VEGF). Our data showed that overexpression of RBM3 upregulated the expression of MMP2, MMP9 and VEGF (Figure 3C), whereas inhibition of RBM3 down-regulated the expression of MMP2, MMP9 and VEGF (Figures 3D and S2B). These results suggest that RBM3 promotes the metastasis of HCC cells.
Involvement of STAT3 Signaling in RBM3-Induced EMT
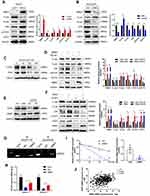
Numerous studies have proven that STAT3 signaling pathway is involved in the activation and regulation of EMT.24,36,37 In order to further explore the potential mechanism of RBM3 regulating the EMT process, we investigated the involvement of STAT3 signaling pathway in the EMT process regulated by RBM3 in HCC cells. Western blotting analysis showed that the ratio of pSTAT3/STAT3 and the total STAT3 expression were both up-regulated in EMT-activated HepG2-RBM3 cells (Figure 4A). In contrast, in MHCC97H cells transfected with RBM3-specific siRNAs, the ratio of pSTAT3/STAT3 and total STAT3 expression were both down-regulated (Figure 4B).
To demonstrate whether RBM3 promotes the EMT process through STAT3 signaling pathway, we used a selective STAT3 inhibitor S3I-201, which inactivates STAT3 by blocking its phosphorylation and dimerization, and STAT3-specific siRNAs. S3I-201 inhibited STAT3 phosphorylation in a concentration-dependent manner at 72h. S3I-201 (50 μM) was used in further experiments due to its significant inhibitory effect on STAT3 phosphorylation without affecting the total STAT3 expression (Figure 4C). Our data showed that RBM3 overexpression decreased the expression of E-cadherin and increased the expression of N-cadherin, Vimentin, STAT3 and pSTAT3 in HepG2 cells. However, RBM3-induced EMT process was blocked by inhibiting the phosphorylation of STAT3 via S3I-201. Notably, STAT3 expression was not affected (Figure 4D). Since RBM3 significantly up-regulated STAT3 expression, we used two STAT3-specific siRNAs to inhibit STAT3 expression to explore the role of STAT3 in the RBM3-induced EMT process. STAT3-siRNA#1 was used for further studies because of its remarkable knockdown efficiency on STAT3 expression (Figure 4E). Our data showed that STAT3 depletion abrogated RBM3-induced E-cadherin down-regulation as well as N-cadherin and Vimentin up-regulation (Figure 4F). These results indicate that RBM3 promoted EMT process through STAT3 signaling pathway.
RBM3 is an RNA binding protein that interacts with coding and non-coding RNAs to involve in regulating RNA splicing, mRNA localization, translation and stability.38,39 In order to investigate the exact mechanism by which RBM3 regulates STAT3 expression, we conducted RNA immunoprecipitation assay to explore the combination of RBM3 and STAT3. Our results showed that RBM3 bound directly to STAT3 mRNA by reverse transcription PCR analysis (Figure 4G) and quantitative real-time PCR assay (Figure 4H). For exploring the role of RBM3 on the stability of STAT3 mRNA, we treated cells with actinomycin D to inhibit total RNA synthesis,40 and found that down-regulation of RBM3 accelerated STAT3 mRNA degradation (Figure 4I). Furthermore, RBM3 expression was positively correlated with STAT3 expression in 374 HCC patients in the TCGA database (Figure 4J). These results demonstrate that RBM3 upregulates STAT3 expression by stabilizing STAT3 mRNA via binding to its mRNA.
MicroRNA-383 Inhibits RBM3 Expression by Binding to the 3ʹUTR of RBM3 mRNA
MicroRNAs are a class of highly conserved single-stranded non-coding RNAs that can bind to the 3ʹUTR of target genes to inhibit their translation or induce their degradation.41 In order to uncover the regulatory mechanism of RBM3 in HCC cells, we used the TargetScan database to make predictions and found that microRNA-383 could be complementary to the bases at positions 41–47 of the RBM3 3ʹUTR (UCUGAUC) (Figure 5A). The inhibition of microRNA-383 on RBM3 expression was further confirmed by Western blotting (Figure 5B). In addition, dual-luciferase reporter assay showed that microRNA-383 markedly inhibited luciferase activity in HepG2 cells transfected with the predicted RBM3 3ʹUTR sequence, but this inhibition was attenuated in HepG2 cells transfected with mutant RBM3 3ʹUTR sequence (Figure 5C). Furthermore, the suppression of EMT progression by microRNA-383 was very similar to that of RBM3 inhibition (Figure 5D). The TCGA database showed that microRNA-383 expression was extremely low in primary HCC tumors compared to normal healthy participants (Figure 5E). Further correlation analysis from TCGA database indicated that microRNA-383 was strongly negatively correlated with RBM3 (Figure 5F). These findings suggest that microRNA-383 is an upstream target of RBM3.
Inhibition of RBM3 Prolongs Survival of Mice Burdened with HCC Xenograft Tumors
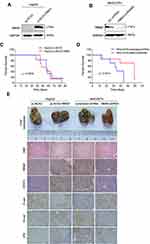
To investigate the role of RBM3 on tumor progression in vivo, we constructed RBM3 overexpression cells (HepG2-pLNCX2-RBM3) (Figure 6A) and RBM3 knockdown cells (MHCC97H-RBM3-shRNA) (Figure 6B). Mice were injected with the indicated cells, and survival days were recorded. Kaplan–Meier curves of cumulative survival plots showed that although overexpression of RBM3 showed no effect on the survival of xenografted mice (Figure 6C), inhibition of RBM3 prolonged the survival days of xenografted mice from 44 to 60 days (Figure 6D). We collected the livers from mice and found that RBM3 overexpression resulted in a higher incidence of tumor metastasis, increased STAT3, N-cadherin and Vimentin expression, as well as decreased E-cadherin expression. In contrast, RBM3 inhibition suppressed the formation of tumor metastases and the expression of STAT3, N-cadherin and Vimentin while increasing the expression of E-cadherin (Figure 6E). These results suggest that suppression of RBM3 inhibits the EMT process and metastasis of HCC cells in vivo.
Discussion
The unfavorable prognosis of cancer is largely attributed to the aggressiveness and metastasis of cancer cells. Increasing evidence indicated that RBM3 is dysregulated in various types of human cancers and plays a significant role in tumor cell proliferation and cancer patient outcome. Different tumor types exhibited opposing trends in RBM3 expression. Not only that, different cancer subtypes may hold discrepant RBM3 expression. In two subtypes of non-small cell lung cancer, the expression of RBM3 in adenocarcinoma was considerably higher than that in squamous cell carcinoma.42 The contradictory trend of RBM3 expression in different cancer types may be related to the complex tumor microenvironment. Using immunohistochemical assays, we found that RBM3 was significantly higher in HCC tissues than in adjacent non-tumor tissues. This finding was coincident with the analysis from TCGA database and the only two previous studies on RBM3 expression in HCC.43 In addition, RBM3 was also correlated with the prognosis of cancer patients. Some studies have reported that low expression of RBM3 was associated with poor prognosis in breast,44 bladder45 and colorectal cancers.29 Conversely, prostatic cancer patients with high RBM3 expression exhibited better survival,46 while survival in esophageal cancer was independent of RBM3 expression.47 Our data showed that HCC patients with low RBM3 expression exhibited higher overall survival compared to those with high RBM3 expression, which was also similar to the analysis of TCGA. These results indicate that high expression of RBM3 is an independent biomarker of poor prognosis in HCC.
Tumor cells could acquire aggressiveness through the EMT process, thus entering the circulation of the blood and lymphatic system, and metastasizing from the primary site to new tissues and organs, leading to incurable or even death of tumor patients.48 RBM3 has been reported to be positively associated with the metastatic process of various tumor cells.34,35,49 However, the biological effects of RBM3 on EMT, a developmental process by which tumor cells acquire pro-metastatic properties, have not been reported. Only one study has found that overexpression of RBM3 could upregulate the expression of Vimentin in neuroblastoma cells.49 We demonstrate for the first time that RBM3 could promote the EMT process in HCC cells. The transwell assay, a classic method for measuring tumor cell metastasis,50 showed that RBM3 accelerated the migration and invasion of HCC. Matrix metalloproteinases (MMPs) are well known for their role in degrading histological barrier during tumor metastasis.51 MMP2 and MMP9 could specifically degrade type IV collagen, a main component of the basement membrane, providing the possibility for tumor cells to metastasize into the vascular system.52 Vascular endothelial growth factor (VEGF) plays a crucial role in angiogenesis, which induces endothelial cell proliferation and creates suitable conditions for tumor cell metastasis.53 MMP2, MMP9 and VEGF were often used as metastatic markers of tumor progression.54,55 We found that RBM3 increased the expression of MMP2, MMP9 and VEGF.
STAT3 signaling pathway plays a key role in tumor-associated EMT process.56 As a kind of RNA-binding protein, RBM3 could modulate the expression of target genes by binding to their mRNAs.38 By blocking total mRNA synthesis using actinomycin D, our data suggested that RBM3 indeed bonds to STAT3 mRNA, thereby attenuating STAT3 mRNA degradation. Previous studies have demonstrated that microRNA-383 regulated proliferation and metastasis by binding to the 3ʹUTR of target genes in cholangiocarcinoma57 and colorectal cancer.58 The regulatory role of microRNA-383 on RBM3 is not yet understood. We found that the inhibitory effect of RBM3 on EMT in HCC cells may be modulated by microRNA-383.
Conclusions
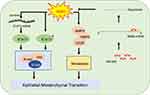
Our results showed that RBM3 accelerated the migration and invasion of HCC cells and promoted the EMT process by upregulating pSTAT3 and total STAT3 expression (Figure 7). As the first study to explore the RBM3/STAT3/EMT axis, our results may provide new understanding and strategies for the metastasis and therapy of HCC.
Abbreviations
ARPC2, actin related protein 2/3 complex subunit 2; EMT, epithelial–mesenchymal transition; FBS, fetal bovine serum; HCC, hepatocellular carcinoma; MMP, matrix metalloproteinases; mRNA, message RNA; qRT-PCR, quantitative real-time PCR; RBM3, RNA binding motif protein 3; RT-PCR, reverse transcription polymerase reaction; shRNA, short hairpin RNA; siRNA, short interfering RNA; STAT3, signal transducer and activator of transcription 3; TCGA, the Cancer Genome Atlas; TAM, tumor-associated macrophages; UTR, untranslated region; VEGF, vascular endothelial growth factor.
Ethics Approval and Informed Consent
The study protocol was approved by the Ethics Committee of the First Affiliated Hospital of Third Military Medical University (Army Medical University), PLA (2015; Chongqing, China). Procedures for the collection of human samples, and their use for tissue arrays and gene expression studies were approved by the Ethical Committee of the Army Medical University (Chongqing, China). All the animal studies were performed strictly in accordance with the Animal Care Guidelines approved by the Laboratory Animal Welfare and Ethics Committee of Chongqing University (IACUC Issue No.: CQU-IACUC-RE-202111-002). The cell lines used in this study have been proven to meet the ethical requirements issued by the affiliated Chongqing University Cancer Hospital Ethics Committee (ethical code: CZLS2022038-A).
Consent for Publication
The content of this manuscript has not been published or submitted for publication elsewhere. All the authors have contributed significantly, and that all authors are in agreement with the content of the manuscript.
Acknowledgments
The authors wish to acknowledge Dr Ni Tang from Chongqing Medical University for providing HepG2 cell line.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Natural Science Foundation of China [grant number 82172831]; the General Project of Chongqing Natural Science Foundation [grant number cstc2021jcyj-msxmX0642]; the Graduate Research and Innovation Foundation of Chongqing, China [grant number CYB21069]; the natural science foundation of Chongqing [grant number 2021ycjh-bgzxm0169; cstc2021jcyj-cxttX0002cstc]; and the China Postdoctoral Science Foundation [grant number 2019M663106].
Disclosure
The authors report no conflicts of interest in this work.
References
1. Zeng H, Zheng R, Guo Y, et al. Cancer survival in China, 2003–2005: a population-based study. Int J Cancer. 2015;136(8):1921–1930. doi:10.1002/ijc.29227
2. Fu J, Wang H. Precision diagnosis and treatment of liver cancer in China. Cancer Lett. 2018;412:283–288. doi:10.1016/j.canlet.2017.10.008
3. Siegel RL, Miller KD, Jemal A. Jemal A Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
4. Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
5. Graziani V, Rodriguez-Hernandez I, Maiques O, Sanz-Moreno V. The amoeboid state as part of the epithelial-to-mesenchymal transition programme. Trends Cell Biol. 2021;32(3):228–242. doi:10.1016/j.tcb.2021.10.004
6. Wagner J, Masek M, Jacobs A, et al. Mass cytometric and transcriptomic profiling of epithelial-mesenchymal transitions in human mammary cell lines. Sci Data. 2022;9(1):44. doi:10.1038/s41597-022-01137-4
7. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi:10.1016/j.cell.2009.11.007
8. Vu T, Datta PK. Regulation of EMT in colorectal cancer: a culprit in metastasis. Cancers. 2017;9(12):12. doi:10.3390/cancers9120171
9. Yang J, Antin P, Berx G, et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2020;21(6):341–352. doi:10.1038/s41580-020-0237-9
10. Marcucci F, Stassi G, De MR. Epithelial-mesenchymal transition: a new target in anticancer drug discovery. Nat Rev Drug Discov. 2016;15(5):311–325. doi:10.1038/nrd.2015.13
11. Carroll CP, Bolland H, Vancauwenberghe E, et al. Targeting hypoxia regulated sodium driven bicarbonate transporters reduces triple negative breast cancer metastasis. Neoplasia. 2022;25:41–52. doi:10.1016/j.neo.2022.01.003
12. Wang J, Zhong Q, Zhang H, et al. Nogo-B promotes invasion and metastasis of nasopharyngeal carcinoma via RhoA-SRF-MRTFA pathway. Cell Death Dis. 2022;13(1):76. doi:10.1038/s41419-022-04518-0
13. Yu D, Pan M, Li Y, et al. RNA N6-methyladenosine reader IGF2BP2 promotes lymphatic metastasis and epithelial-mesenchymal transition of head and neck squamous carcinoma cells via stabilizing slug mRNA in an m6A-dependent manner. J Exp Clin Cancer Res. 2022;41(1):6. doi:10.1186/s13046-021-02212-1
14. Stehr AM, Wang G, Demmler R, et al. Neutrophil extracellular traps drive epithelial-mesenchymal transition of human colon cancer. J Pathol. 2021;256(4):455–467.
15. Dong T, Zhang Y, Chen Y, et al. FOXO1 inhibits the invasion and metastasis of hepatocellular carcinoma by reversing ZEB2-induced epithelial-mesenchymal transition. Oncotarget. 2017;8(1):1703–1713. doi:10.18632/oncotarget.13786
16. Han LL, Yin XR, Zhang SQ. miR-103 promotes the metastasis and EMT of hepatocellular carcinoma by directly inhibiting LATS2. Int J Oncol. 2018;53(6):2433–2444. doi:10.3892/ijo.2018.4580
17. Liu GM, Li Q, Zhang PF, et al. Restoration of FBP1 suppressed snail-induced epithelial to mesenchymal transition in hepatocellular carcinoma. Cell Death Dis. 2018;9(11):1132. doi:10.1038/s41419-018-1165-x
18. Zheng S, Liu J, Zhao Z, Song R. Role of STAT3/mTOR pathway in chronic kidney injury. Am J Transl Res. 2020;12(7):3302–3310.
19. Poholek CH, Raphael I, Wu D, et al. Noncanonical STAT3 activity sustains pathogenic Th17 proliferation and cytokine response to antigen. J Exp Med. 2020;217(10). doi:10.1084/jem.20191761
20. Wang C, Dou C, Wang Y, et al. TLX3 repressed SNAI1-induced epithelial-mesenchymal transition by directly constraining STAT3 phosphorylation and functionally sensitized 5-FU chemotherapy in hepatocellular carcinoma. Int J Biol Sci. 2019;15(8):1696–1711. doi:10.7150/ijbs.33844
21. Chen RY, Yen CJ, Liu YW, et al. CPAP promotes angiogenesis and metastasis by enhancing STAT3 activity. Cell Death Differ. 2020;27(4):1259–1273. doi:10.1038/s41418-019-0413-7
22. Balic JJ, Albargy H, Luu K, et al. STAT3 serine phosphorylation is required for TLR4 metabolic reprogramming and IL-1beta expression. Nat Commun. 2020;11(1):3816. doi:10.1038/s41467-020-17669-5
23. Goel RR, Nakabo S, Dizon BLP, et al. Lupus-like autoimmunity and increased interferon response in patients with STAT3-deficient hyper-IgE syndrome. J Allergy Clin Immunol. 2020;147(2):746–749.e9. doi:10.1016/j.jaci.2020.07.024
24. Guo R, Wu Z, Wang J, et al. Development of a non‐coding‐RNA‐based EMT/CSC inhibitory nanomedicine for in vivo treatment and monitoring of HCC. Adv Sci. 2019;6(9):1801885. doi:10.1002/advs.201801885
25. Sun XL, Ma J, Chen QZ, et al. SIX4 promotes metastasis through STAT3 activation in breast cancer. Am J Cancer Res. 2020;10(1):224.
26. Danno S, Nishiyama H, Higashitsuji H, et al. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem Biophys Res Commun. 1997;236(3):804–807. doi:10.1006/bbrc.1997.7059
27. Zargar R, Urwat U, Malik F, et al. Molecular characterization of RNA binding motif protein 3 (RBM3) gene from Pashmina goat. Res Vet Sci. 2015;98:51–58. doi:10.1016/j.rvsc.2014.11.016
28. Boman K, Segersten U, Ahlgren G, et al. Decreased expression of RNA-binding motif protein 3 correlates with tumour progression and poor prognosis in urothelial bladder cancer. BMC Urol. 2013;13(1). doi:10.1186/1471-2490-13-17
29. Melling N, Simon R, Mirlacher M, et al. Loss of RNA-binding motif protein 3 expression is associated with right-sided localization and poor prognosis in colorectal cancer. Histopathology. 2016;68(2):191–198. doi:10.1111/his.12726
30. Melling N, Bachmann K, Hofmann B, et al. Prevalence and clinical significance of RBM3 immunostaining in non-small cell lung cancers. J Cancer Res Clin Oncol. 2019;145(4):873–879. doi:10.1007/s00432-019-02850-1
31. Ehlen A, Brennan DJ, Nodin B, et al. Expression of the RNA-binding protein RBM3 is associated with a favourable prognosis and cisplatin sensitivity in epithelial ovarian cancer. J Transl Med. 2010;8(1). doi:10.1186/1479-5876-8-78
32. Jonsson L, Gaber A, Ulmert D, et al. High RBM3 expression in prostate cancer independently predicts a reduced risk of biochemical recurrence and disease progression. Diagn Pathol. 2011;6(1). doi:10.1186/1746-1596-6-91
33. Zhang HT, Zhang ZW, Xue JH, et al. Differential expression of the RNA-binding motif protein 3 in human astrocytoma. Chin Med J. 2013;126(10):1948–1952.
34. Florianova L, Xu B, Traboulsi S, et al. Evaluation of RNA-binding motif protein 3 expression in urothelial carcinoma of the bladder: an immunohistochemical study. World J Surg Oncol. 2015;13(1). doi:10.1186/s12957-015-0730-3
35. Chen P, Yue X, Xiong H, Lu X, Ji Z. RBM3 upregulates ARPC2 by binding the 3ʹUTR and contributes to breast cancer progression. Int J Oncol. 2019. doi:10.3892/ijo.2019.4698
36. Kim MS, Lee HS, Kim YJ, et al. MEST induces twist-1-mediated EMT through STAT3 activation in breast cancers. Cell Death Differ. 2019;26(12):2594–2606. doi:10.1038/s41418-019-0322-9
37. Zhang X, Sai B, Wang F, et al. Hypoxic BMSC-derived exosomal miRNAs promote metastasis of lung cancer cells via STAT3-induced EMT. Mol Cancer. 2019;18(1):40. doi:10.1186/s12943-019-0959-5
38. Zhou RB, Lu XL, Zhang CY, Yin DC. RNA binding motif protein 3: a potential biomarker in cancer and therapeutic target in neuroprotection. Oncotarget. 2017;8(13):22235–22250. doi:10.18632/oncotarget.14755
39. Qin H, Ni H, Liu Y, et al. RNA-binding proteins in tumor progression. J Hematol Oncol. 2020;13(1):90. doi:10.1186/s13045-020-00927-w
40. Francart ME, Vanwynsberghe AM, Lambert J, et al. Vimentin prevents a miR-dependent negative regulation of tissue factor mRNA during epithelial-mesenchymal transitions and facilitates early metastasis. Oncogene. 2020;39(18):3680–3692. doi:10.1038/s41388-020-1244-1
41. Long H, Wang X, Chen Y, et al. Dysregulation of microRNAs in autoimmune diseases: pathogenesis, biomarkers and potential therapeutic targets. Cancer Lett. 2018;428:90–103. doi:10.1016/j.canlet.2018.04.016
42. Salomonsson A, Micke P, Mattsson JSM, et al. Comprehensive analysis of RNA binding motif protein 3 (RBM3) in non-small cell lung cancer. Cancer Med. 2020;9(15):5609–5619. doi:10.1002/cam4.3149
43. Dong W, Dai ZH, Liu FC, et al. The RNA-binding protein RBM3 promotes cell proliferation in hepatocellular carcinoma by regulating circular RNA SCD-circRNA 2 production. EBioMedicine. 2019;45:155–167. doi:10.1016/j.ebiom.2019.06.030
44. Kang SH, Cho J, Jeong H, Kwon SY. High RNA-binding motif protein 3 expression is associated with improved clinical outcomes in invasive breast cancer. J Breast Cancer. 2018;21(3):288–296. doi:10.4048/jbc.2018.21.e34
45. Boman K, Andersson G, Wennersten C, et al. Podocalyxin-like and RNA-binding motif protein 3 are prognostic biomarkers in urothelial bladder cancer: a validatory study. Biomark Res. 2017;5(1). doi:10.1186/s40364-017-0090-y
46. Grupp K, Wilking J, Prien K, et al. High RNA-binding motif protein 3 expression is an independent prognostic marker in operated prostate cancer and tightly linked to ERG activation and PTEN deletions. Eur J Cancer. 2014;50(4):852–861. doi:10.1016/j.ejca.2013.12.003
47. Grupp K, Hofmann B, Kutup A, et al. Reduced RBM3 expression is associated with aggressive tumor features in esophageal cancer but not significantly linked to patient outcome. BMC Cancer. 2018;18(1):1106. doi:10.1186/s12885-018-5032-z
48. Jingyu Z, Xiao-Jun T, Hang Z, et al. TGF-β-induced epithelial-to-mesenchymal transition proceeds through stepwise activation of multiple feedback loops. Sci Signal. 2014;7(345):ra91. doi:10.1126/scisignal.2005304
49. Pilotte J, Kiosses W, Chan SW, et al. Morphoregulatory functions of the RNA-binding motif protein 3 in cell spreading, polarity and migration. Sci Rep. 2018;8(1):7367. doi:10.1038/s41598-018-25668-2
50. Chai F, Li Y, Liu K, Li Q, Sun H. Caveolin enhances hepatocellular carcinoma cell metabolism, migration, and invasion in vitro via a hexokinase 2-dependent mechanism. J Cell Physiol. 2019;234(2):1937–1946. doi:10.1002/jcp.27074
51. Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi:10.1016/j.cell.2010.03.015
52. Hannocks MJ, Zhang X, Gerwien H, et al. The gelatinases, MMP-2 and MMP-9, as fine tuners of neuroinflammatory processes. Matrix Biol. 2019;75–76:102–113. doi:10.1016/j.matbio.2017.11.007
53. Mahdi A, Darvishi B, Majidzadeh AK, Salehi M, Farahmand L. Challenges facing antiangiogenesis therapy: the significant role of hypoxia-inducible factor and MET in development of resistance to anti-vascular endothelial growth factor-targeted therapies. J Cell Physiol. 2019;234(5):5655–5663. doi:10.1002/jcp.27414
54. Zhu Y, Yan L, Zhu W, et al. MMP2/3 promote the growth and migration of laryngeal squamous cell carcinoma via PI3K/Akt-NF-kappaB-mediated epithelial-mesenchymal transformation. J Cell Physiol. 2019;234(9):15847–15855.
55. Zhang Q, Agoston AT, Pham TH, et al. Acidic bile salts induce epithelial to mesenchymal transition via VEGF signaling in non-neoplastic Barrett’s cells. Gastroenterology. 2019;156(1):130–144. doi:10.1053/j.gastro.2018.09.046
56. Bao Q, Zhang B, Suo Y, et al. Intermittent hypoxia mediated by TSP1 dependent on STAT3 induces cardiac fibroblast activation and cardiac fibrosis. eLife. 2020:9:e49923.
57. Wan P, Chi X, Du Q, et al. miR-383 promotes cholangiocarcinoma cell proliferation, migration, and invasion through targeting IRF1. J Cell Biochem. 2018;119(12):9720–9729. doi:10.1002/jcb.27286
58. Li J, Smith AR, Marquez RT, et al. MicroRNA-383 acts as a tumor suppressor in colorectal cancer by modulating CREPT/RPRD1B expression. Mol Carcinog. 2018;57(10):1408–1420. doi:10.1002/mc.22866
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.