Back to Journals » Infection and Drug Resistance » Volume 15
Risk Factors and Molecular Mechanism of Polymyxin B Resistance in Carbapenem-Resistant Klebsiella pneumoniae Isolates from a Tertiary Hospital in Fujian, China
Authors Xu X, Zhu R, Lian S, Zhang H, Chen X, Fan L, Chen P, Cao Y
Received 29 September 2022
Accepted for publication 10 December 2022
Published 15 December 2022 Volume 2022:15 Pages 7485—7494
DOI https://doi.org/10.2147/IDR.S391674
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Xiaohong Xu,1,* Rongping Zhu,1,* Siyan Lian,1 Hui Zhang,2 Xin Chen,2 Lingfang Fan,1 Peisong Chen,2 Yingping Cao1
1Department of Clinical Laboratory, Fujian Medical University Union Hospital, Fuzhou, Fujian, People’s Republic of China; 2Medical Technology and Engineering, Fujian Medical University, Fuzhou, Fujian, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yingping Cao, Department of Clinical Laboratory, Fujian Medical University Union Hospital, Fuzhou, Fujian, People’s Republic of China, Tel +86-133-6591-0806, Email [email protected]
Background: The emergence of polymyxin B resistance among carbapenem-resistant Klebsiella pneumoniae (CRKP) causes clinical treatment to be more difficult. We aimed to investigate the risk factors and resistance mechanisms in the polymyxin resistant CRKP (PR-CRKP) strains.
Methods: From January 2021 to January 2022, 239 CRKP strains were selected, all of which were analyzed using antimicrobial susceptibility testing and clinical data. Polymerase chain reaction (PCR) was performed for the detection of resistance genes. RT-qPCR was used to quantify transcriptional levels of polymyxin resistance genes. Risk factors for polymyxin B resistant isolates were identified by logistic regression analysis.
Results: The resistance rate of polymyxin B was 5.02%. In all CRKP strains, 41.84% came from the ICU. The percentage of carbapenemase producing strains was 93.72%. The main carbapenem resistance gene was blaKPC (90.79%). In the 12 strains of PR-CRKP screened, pmrB and pmrK were overexpressed in all samples which were linked with polymyxin B resistance. Multivariate analysis showed that coronary heart disease may be an independent risk factor predisposing patients to polymyxin B resistance.
Conclusion: We determine the multifaceted mechanism and risk factors of polymyxin B resistance in CRKP. Polymyxin resistance is a complex and changing problem, and more research is required.
Keywords: polymyxin B resistance, carbapenem-resistantKlebsiella pneumoniae, risk factors, mechanism
Introduction
With the widespread use of carbapenem antibiotics, there have been an increasing number of corresponding carbapenem-resistant Enterobacterales (CRE).1 The World Health Organization (WHO) and the Centers for Disease Control and Prevention (CDC) emphasize that CRE is an urgent threat to public health.2 Klebsiella pneumoniae, one of the most significant species of Enterobacterales, is second only to Escherichia coli in the clinical isolation rate among gram-negative bacilli.3 K. pneumoniae is a common pathogen of hospital infection, especially carbapenem-resistant K. pneumoniae (CRKP), leading to a high prevalence of nosocomial infection.4
The mechanism of CRKP resistance is well known to be complex, the most predominant mechanism of which is the production of carbapenemase.5 Common enzymes include class A (KPC), class B (VIM, IMP, NDM), and class D (OXA-48-like) types.6 Among these, blaKPC-2 and blaNDM-1 are the most common carbapenem enzyme genes that cause outbreaks in China.7 The blaKPC gene was first discovered in the United States in 1996,8 it had a long history and was highly contagious. According to the China Antimicrobial Surveillance Network (CHINET), the resistance rate of K. pneumoniae to carbapenems increased rapidly from 2.9% in 2005 to more than 25.3% in 2019,9 which means that the resistance mechanism of K. pneumoniae to carbapenems should be given serious attention.
The overall resistance rate of CRKP continues to rise and, for patients with CRKP infection, there are few clinically available treatments. The cationic polypeptide antibiotic polymyxin is often used as a last-line drug.10 Although novel antibiotics such as ceftazidime-avibactam, ceftolozane/tazobactam, and imipenem/relebactam have now been introduced,11 new combinations with other beta-lactams/beta-lactamase inhibitors have not been approved or used for economic reasons; polymyxin remains the primary treatment option for CRE-induced infections.12 However, the emergence of polymyxin resistance further limits treatment options and increases the risk of death. Polymyxin resistance in CRKP strains is mediated by the cationic group phosphoethanolamine or 4-amino-4-deoxy-L-arabinose modification of the lipid A component of lipopolysaccharide mediated by overexpression of the pmrC or pmrHFIJKLM operon.13 Elevated expression of pmrC or pmrHFIJKLM is mainly due to chromosomal mutations in genes involved in lipopolysaccharide synthesis, mgrB mutation/inactivation, and/or dysfunction of the two-component system including phoPQ, pmrAB, and crab.14 In addition, the acquisition of plasmid-mediated transmissible colistin resistance (mcr) genes can confer resistance to colistin,15 although this is rare in polymyxin resistant CRKP (PR-CRKP) strains. PR-CRKP strains can arise from the development of endogenous polymyxin resistance or from external sources through transmission between patients or environmental contamination.16 We aimed to study strains of CRKP isolated at Fujian Medical University Union Hospital, reveal their clinical characteristics, determine their resistance, and identify risk factors and mechanisms for developing polymyxin resistance.
Methods
Research Design
This study was conducted at Fujian Medical University Union Hospital in Fuzhou, Fujian province, from January 2021 to January 2022. During the study period, cultured isolates were identified by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOFMS; Bruker Daltonics Inc., Billerica, MA), and carbapenem (meropenem or imipenem) antimicrobial susceptibility testing was performed by the microbroth dilution method to confirm CRKP. A total of 239 CRKP strains were collected, including 12 PR-CRKP strains.
A case was defined as a patient from whom a polymyxin resistant CRKP organism was isolated from clinical cultures from any source during the study period. Controls were defined as patients admitted to the ward in the same study period as when a polymyxin-susceptible CRKP was isolated from a clinical culture at least 48 h after admission. Controls were recruited in a 2:1 ratio to cases. Cases and controls were matched for age, clinical manifestation, pathogen, hospital ward, and date of admission. A case or control could only be included in the study once. Informed consent was obtained from all participants.
Clinical Data Collection
Demographic and clinical data were collected from the clinical medical record data system. We analyzed several variables as possible risk factors for the emergence of polymyxin resistance, including whether the patient was exposed to aminoglycosides, β-lactams, carbapenems, cephalosporins, quinolones, polymyxin B; and whether there were invasive operations. Additional variables were Eastern Cooperative Oncology Group (ECOG) scores and hypertension, diabetes, and coronary heart disease complications. The age distribution, ward distribution, and sample origin of all CRKP isolates were collected simultaneously.
Antibiotic Susceptibility Testing
According to the Clinical and Laboratory Standards Institute (CLSI) guidelines,17 we used the microbroth dilution method to determine the minimum inhibitory concentrations (MIC) of polymyxin B (resistant cutoff, MIC ≥ 4 ug/mL), ceftazidime-avibactam, amoxicillin/clavulanate, piperacillin/tazobactam, piperacillin, compound trimoxazole, minocycline, cefpiroxam, ceftazidime, cefoperazone/sulbactam, cefoxitin, ceftriaxone, cefazoline, cefuroxime ester, aztreonam, tobramycin, gentamicin, ciprofloxacin, levofloxacin, and nitrofurantoin. Escherichia coli ATCC25922 and Pseudomonas aeruginosa ATCC27853 were used as quality control standards.
Characterization of Polymyxin and Carbapenem Resistance Genes
Polymerase chain reaction (PCR) was used to detect resistance genes, including blaKPC,blaIMP,blaVIM,blaNDM,blaOXA-48, mcr-1, ompK35, ompK36, and ompK37. The positive products were sequenced, and the sequencing results were compared using the basic local alignment search tool (BLAST) available at http://www.ncbi. nlm.nih.gov/BLAST. Mutations in genes potentially involved in polymyxin resistance (mgrB, crrAB, pmrA/pmrB, and phoP/phoQ) were inspected by alignment with reference genome K. pneumoniae subsp. pneumoniae MGH78578 (# NC_009648.1). The PROVEAN tool v.1.1.5 (http://provean.jcvi.org/index.php) was used to predict the effect of amino acid substitutions on protein function.18 A PROVEAN score ≤ −2.5 was deleterious for protein function, and a score > −2.5 was considered to have a neutral effect on protein function.
Transcriptional Analysis Real-Time Quantitative PCR
RNA extraction and transcription were carried out as previously described.19 Real-time quantitative PCR (RT-qPCR) was used to measure the expression of the phoP, phoQ, pmrK, pmrA, pmrB, and pmrC genes using the primers as previously described.19 Normalization was performed against the 16SrRNA gene using the ΔΔCT method (relative) with the 16SrRNA gene as an internal control. The obtained values were then normalized against those obtained with polymyxin-susceptible strains.
DNA Fingerprint Technology
ERIC-1 and ERIC-2 primers were used to conduct ERIC-PCR in PR-CRKP. The ERIC-PCR conditions were adjusted according to the report published by Smith et al.20 The DNA fingerprint was analyzed using GelCompar II, version 6.5 (Applied Mathematics, NV, Keistraat, Belgium). A cutoff value of 80% similarity was applied to define the cluster. The similarity among species was evaluated by band-matching Dice coefficient, and the tree map of each species was drawn by the unweighted pair grouping method with arithmetic mean (UPGMA). According to the cluster diagram of the UPGMA system, same strains are defined as strains with > 97% similarity, and strains with < 95% similarity are defined as unrelated strains.
Statistical Analysis
Data were analyzed using IBM SPSS ver. 21.0 statistical software (IBM Co., Armonk, NY). Frequency tables (n, %) for categorical variables and descriptive statistics (mean, median, standard deviation) for numerical variables were used. Comparisons of categorical variables were analyzed by the Chi square test. Logistic regression (Backward LR) methods (univariate, multivariate) were used to determine the risk factors. Statistical significance was assigned to a P value < 0.05.
Results
Clinical Characteristics
Between January 2021 and January 2022, a total of 239 CRKP strains were collected from Fujian Medical University Union Hospital. The age range of patients ranged from 1 to 98 years old, mainly concentrated in 51–80 years old (159/239, 66.53%), the sources of specimens were mainly sputum (66/239, 27.62%), bronchoalveolar lavage fluid (47/239, 19.67%), and blood (38/239, 15.90%). The main ward source of the strains was ICU (100/239, 41.84%).
Among 239 CRKP infected patients, we identified nine PR-CRKP infected patients in whom PS-CRKP strains had been isolated 3–677 days earlier. Patients with three strains of PR-CRKP infection were already polymyxin resistant when CRKP was detected. The 12 PR-CRKP infected patients were matched to 24 PS-CRKP infected patients based on age, clinical presentation, pathogen, ward, and date of admission. In the case and control groups, the proportion of males (75.0% and 79.2%, respectively) was higher than females (25.0% and 20.8%, respectively), and the ECOG score in patients in the case group compared to the control group (5 vs 3; P = 0.0125) was significantly higher (Table 1).
 |
Table 1 Comparison of Demographic Factors Among of the Case and Control Groups |
Antimicrobial Carbapenem Resistance Characterization
All CRKP strains were found to be resistant to nearly all antibiotics tested in this study. High resistance was detected toward amoxicillin/clavulanate (97.10%), piperacillin/tazobactam (99%), piperacillin (100%), cefpiroxam (95.80%), cefoxitin (96.40%), ceftriaxone (98.70%), ceftazidime (100%), cefazoline (100%), cefoperazone/sulbactam (99%), cefuroxime ester (100%), aztreonam (97.50%), tobramycin (86.20%), gentamicin (86.30%), ciprofloxacin (97.90%), levofloxacin (96.70%), and nitrofurantoin (92.90%). Minocycline (50%) and compound neonolemin (35.10%) showed lower rates of resistance. Polymyxin B (5.02%) and ceftazidime-avibactam (5.44%) had the lowest rates of resistance. (Figure 1). The resistance rate of polymyxin B was 12/239 (5.02%) and the MIC distribution results of polymyxin B of the CRKP strains are shown in Figure 2. The resistance rate of ceftazidime-avibactam was 13/239 (5.44%). For ceftazidime-avibactam-resistant CRKP strains, the positive rate of blaNDM was 12/13 (92.31%); specifically, when carrying both blaKPC and blaNDM, the rate was 7/13 (53.85%), blaNDM alone was 5/13 (38.46%), and blaIMP alone was 1/13 (7.69%). In the CRKP strains, the detection rate of blaKPC was 217/239 (90.79%), and the detection rate of blaNDM was 16/239 (6.69%). In detail, the blaKPC gene was detected in combination with blaNDM and blaIMP in nine and one strains, respectively. Membrane pore protein genes testing showed the deletion of membrane pore protein as follows: only OmpK-35 deletion (6/239, 2.51%); only OmpK-36 deletion (2/239, 0.84%); OmpK-35, OmpK-36 simultaneous deletion (1/239, 0.42%); OmpK-35, OmpK-37 simultaneous deletion (1/239, 0.42%); and OmpK-35, OmpK36, and OmpK-37 simultaneous deletion (1/239, 0.42%) (Table 2). Among the strains that did not detect common carbapenem resistance genes such as blaKPC,blaNDM, and blaIMP, two strains found that membrane pore proteins were lost, and the remaining five strains did not.
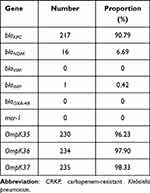 |
Table 2 Molecular Characterization of Resistance-Related Genes in 239 CRKP Isolates |
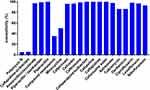 |
Figure 1 Susceptibility of CRKP isolates to different antimicrobial agents. |
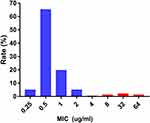 |
Figure 2 Distribution of MIC values of polymyxin B by broth micro-dilution method. Abbreviations: MIC, minimal inhibitory concentration; µg/mL, micrograms per milliliter. |
Risk Factors for Developing Polymyxin Resistance in CRKP Strains
Risk factors for the development of polymyxin B resistance in CRKP strains were determined by assessing the effects of all independent variables that showed statistically significant differences (p < 0.05) in comparative analyses of case and control patients. This final multivariable model also showed significant predictors for each group. Univariate analysis showed that hypertension, coronary heart disease, and tracheostomy were risk factors for the development of polymyxin B resistance in CRKP strains. In multivariate analysis, coronary heart disease (odds ratio [OR] 7.822; 95% confidence interval [CI]: 1.040–58.819; p = 0.046) was identified as an independent risk factor for polymyxin B resistance (Table 3).
 |
Table 3 Univariate and Multivariate Analyses of Risk Factors for the Emergence of Polymyxin Resistance in Carbapenem-Resistant Klebsiella pneumoniae (CRKP) |
Polymyxin Resistance Mechanism and Strain Affinity
Depending on the mechanism by which polymyxin resistance appeared in vivo, the expression of pmrB and pmrK mRNA of all 12 PR-CRKP strains was higher than the PS-CRKP counterparts; the expression of phoP and phoQ mRNA of most PR-CRKP strains was lower than the PS-CRKP counterparts. Mcr-1 was not detected in any of the strains. We detected a 51 G > A change in mgrB (1/12, 8.3%), a change in 310 A > G in phoP (3/12, 25.0%), a change in phoQ of 1185 C > T (3/12, 25.0%), and a change in 1194 C > T (1/12, 8.3%). We also detected a change in pmrB of 147 T > C (3/12, 25.0%) and a change in 149 T > C, leading to isoleucine mutation to alanine in one sample (1/12, 8.3%), and a change in 766 C > G in three samples, causing arginine mutations to changes in glycine (3/12, 25.0%). The replacement of amino acids present was a neutral alteration (Table 4). ERIC-PCR typing showed four fingerprint patterns for PR-CRKP strains; 66.7% were type A1, 8.3% were B1, 8.3% were C1, and 16.7% were type D1 (Figure 3).
 |
Table 4 Mechanisms of Polymyxin Resistance in 12 PR-CRKP Strains |
 |
Figure 3 ERIC-PCR dendrogram of all PR-CRKP clinical isolates. |
Discussion
Clinical detection rates of CRKP have been increasing worldwide, leading to more difficult antimicrobial therapy, and causing higher disease-related mortality.21 In this study, the department with the highest prevalence of CRKP was the ICU, with sputum as the main source. This is consistent with previous research in Shanghai.5
K. pneumoniae resistance to carbapenems was mediated by different resistance mechanisms, mainly including the production of carbapenemase, changes in pore proteins, and increased activity of the external pump.22,23 We studied the production of common carbapenemase (blaKPC,blaNDM,blaIMP,blaVIM, and blaOXA-48) and common membrane pore protein (OmpK-35, OmpK36, and OmpK-37) changes. When CRKP was detected in a global outbreak, the most common enzyme was CRKP producing KPC.24,25 In China, the detection rate of blaKPC-2 is approximately 73%.26 An epidemiology analysis showed that K. pneumoniae carbapenemase (KPC) was the predominant carbapenemase in CRKP strains (90.79%), which was consistent with current prevalence.27 The positive rate of OmpK35 was 96.23%, OmpK36 was 97.9%, and OmpK37 was 98.33%. Common carbapenemase genes were not detected in two strains of membrane pore protein loss, and five strains did not detect membrane pore protein loss; however, other mechanisms of resistance may be present.
In this study, CRKP had the high susceptibility to ceftazidime-avibactam (6.69%). For ceftazidime-avibactam-resistant strains, the carrying rate of NDM was 92.31% and the IMP was 7.69%. This, once again, verifies the fact that avibactam had no inhibitory effect on metallo-β-lactamase (MBL)-B type carbapenemase,28 meaning that the treatment of ceftazidime-avibactam was targeted and could not be fully covered. Coupled with the listing of ceftazidime-avibactam in 2019, and the short clinical use time, there is a lack of more effective clinical data.11 Therefore, polymyxin is still most commonly used in the clinical treatment of CRE.12
We observed that coronary heart disease may be the essential risk factor related to polymyxin B resistance. This is inconsistent with the results observed in previous epidemiological studies that polymyxin was the only risk factor for CRKP to develop polymyxin resistance.29 However, the development of polymyxin resistance in patients without polymyxin treatment was explored in a multicenter study.30 Our study is the first to suggest that coronary heart disease may be a risk factor for CRKP in the development of polymyxin resistance. There is a report31 that the mortality rate of patients infected with K. pneumoniae in heart disease is higher than other complications, and the clinical application of polymyxin was because K. pneumoniae had developed multiple resistance which required the use of the last line of defense, polymyxin B, in which case the patient’s condition had deteriorated and death was more likely. This conclusion was not completely unreasonable, and the practical significance still needs to be demonstrated by further studies.
In 2015, a plasmid-mediated polymyxin resistant gene, mcr-1, was first reported,15 with the highest prevalence of mcr-positive strains in China.32 However, mcr-1 was not detected in any isolates in the study. The reasons of this resistance need further research. While this study found a variety of genetic mutations in mgrB, pmrB, phoP, and phoQ, none of them affected changes in protein function, suggesting that the emergence of polymyxin resistance was more likely a random event, possibly a pre-existing polymyxin resistance mechanism in the CRKP strains, rather than being driven by a common mechanism. mgrB is a negative feedback regulator of the phoP/phoQ signaling system.33 Therefore, in strains with mgrB alterations, it is generally accompanied by upregulation of phoP and phoQ in PR-CRKP. However, no alterations in mgrB were found in this study; correspondingly, the expression of phoP/phoQ in most strains was downregulated. PmrB and pmrK overexpression was found in all PR-CRKP isolates, which implied that pmrB and pmrK overexpression might be the main reason for polymyxin resistance in CRKP. However, there was no way as yet to determine the possible exact mechanism of polymyxin resistance.
ERIC-PCR is a cost-effective, easy to perform, and fast method that can be used to compare generated clusters.34 ERIC-PCR was made against PR-CRKP strains, and we found that the 12 strains were divided into four types; 66.7% of the isolates showed the same ERIC profile, which indicated the possibility of clonal transmission between strains. This phenomenon reminded us that we must pay more attention to the transmission of PR-CRKP strains between patients.
This study has several limitations. First, our study was influenced by a retrospective analysis with a relatively small number of cases, thus reducing the statistical capacity of the study. Second, because the screening and monitoring of polymyxin resistance was not a routine examination in the Fujian Medical University Union Hospital, it was largely affected by the subjective choice of doctors to examine for it, and it was not possible to accurately assess the time when polymyxin resistance appeared. Third, mcr was the only plasmid gene we studied, and unknown mechanisms of infectious polymyxin resistance may be useful for observations of this study. Whole genome sequencing was required to determine other unknown mechanisms of polymyxin resistance.
Conclusion
The CRKP isolates carrying blaKPC are still widely present in Eastern Fujian in Southeast China. The development of polymyxin resistance appears to be a complex multifactorial process that is unlikely to be explained solely by existing polymyxin resistance mechanisms. Therefore, the mechanisms of polymyxin resistance need to be further explored, and hospitals should take effective measures to prevent the further expansion of PR-CRKP.
Ethics Approval and Informed Consent
All procedures of this study involving humans (individuals, medical records, human samples, and clinical isolates) were reviewed and approved by the Medical Ethics Committee of Fujian Medical University Union Hospital (2022KY156). All the patients participating in this study signed informed consent, while the guardians of children aged less than 18 years signed on behalf of them. We confirm that this study was conducted in accordance with the Declaration of Helsinki.
Acknowledgments
The authors would like to thank those who participated in this study.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Jin Z, Wang Z, Gong L, et al. Molecular epidemiological characteristics of carbapenem-resistant Klebsiella pneumoniae among children in China. AMB Express. 2022;12(1):89. doi:10.1186/s13568-022-01437-3
2. Raro OHF, Collar GS, Da Silva RMC, et al. Performance of polymyxin B agar-based tests among carbapenem-resistant Enterobacterales. Lett Appl Microbiol. 2021;72(6):767–773. doi:10.1111/lam.13467
3. Hu F, Guo Y, Yang Y, et al. Resistance reported from China antimicrobial surveillance network (CHINET) in 2018. Eur J Clin Microbiol Infect Dis. 2019;38(12):2275–2281. doi:10.1007/s10096-019-03673-1
4. Holt KE, Wertheim H, Zadoks RN, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112(27):E3574–E3581. doi:10.1073/pnas.1501049112
5. Zhang WX, Chen HY, Chen C, et al. Resistance phenotype and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae isolates in Shanghai. Microb Drug Resist. 2021;27(10):1312–1318. doi:10.1089/mdr.2020.0390
6. Zhao Y, Liao Y, Zhang N, et al. Four types of ST11 novel mutations from increasing carbapenem-resistant Klebsiella pneumoniae in Guangdong, 2016–2020. Front Microbiol. 2021;12:702941. doi:10.3389/fmicb.2021.702941
7. Zhang X, Li F, Cui S, et al. Prevalence and distribution characteristics of blaKPC-2 and blaNDM-1 genes in Klebsiella pneumoniae. Infect Drug Resist. 2020;13:2901–2910. doi:10.2147/IDR.S253631
8. Yigit H, Queenan AM, Anderson GJ, et al. Novel carbapenem-hydrolyzing beta-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45(4):1151–1161. doi:10.1128/AAC.45.4.1151-1161.2001
9. Liao W, Liu Y, Zhang W. Virulence evolution, molecular mechanisms of resistance and prevalence of ST11 carbapenem-resistant Klebsiella pneumoniae in China: a review over the last 10 years. J Glob Antimicrob Resist. 2020;23:174–180. doi:10.1016/j.jgar.2020.09.004
10. Cain AK, Boinett CJ, Barquist L, et al. Morphological, genomic and transcriptomic responses of Klebsiella pneumoniae to the last-line antibiotic colistin. Sci Rep. 2018;8(1):9868. doi:10.1038/s41598-018-28199-y
11. Van Duin D, Lok JJ, Earley M, et al. Colistin versus ceftazidime-avibactam in the treatment of infections due to carbapenem-resistant Enterobacteriaceae. Clin Infect Dis. 2018;66(2):163–171. doi:10.1093/cid/cix783
12. Perez F, El Chakhtoura NG, Yasmin M, et al. Polymyxins: to combine or not to combine? Antibiotics. 2019;8(2):38. doi:10.3390/antibiotics8020038
13. Poirel L, Jayol A, Nordmann P. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev. 2017;30(2):557–596. doi:10.1128/CMR.00064-16
14. Poirel L, Jayol A, Bontron S, et al. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother. 2015;70(1):75–80. doi:10.1093/jac/dku323
15. Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-7
16. Giamarellou H. Epidemiology of infections caused by polymyxin-resistant pathogens. Int J Antimicrob Agents. 2016;48(6):614–621. doi:10.1016/j.ijantimicag.2016.09.025
17. Institute CaLS. Performance Standards for Antimicrobial Susceptibility Testing. In: CLSI Supplement M100.
18. Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31(16):2745–2747. doi:10.1093/bioinformatics/btv195
19. Haeili M, Javani A, Moradi J, et al. MgrB alterations mediate colistin resistance in Klebsiella pneumoniae isolates from Iran. Front Microbiol. 2017;8:2470. doi:10.3389/fmicb.2017.02470
20. Smith JL, Drum DJ, Dai Y, et al. Impact of antimicrobial usage on antimicrobial resistance in commensal Escherichia coli strains colonizing broiler chickens. Appl Environ Microbiol. 2007;73(5):1404–1414. doi:10.1128/AEM.01193-06
21. Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9(4):228–236. doi:10.1016/S1473-3099(09)70054-4
22. Lan P, Jiang Y, Zhou J, et al. A global perspective on the convergence of hypervirulence and carbapenem resistance in Klebsiella pneumoniae. J Glob Antimicrob Resist. 2021;25:26–34. doi:10.1016/j.jgar.2021.02.020
23. Hansen GT. Continuous evolution: perspective on the epidemiology of carbapenemase resistance among enterobacterales and other gram-negative bacteria. Infect Dis Ther. 2021;10(1):75–92. doi:10.1007/s40121-020-00395-2
24. Yu X, Zhang W, Zhao Z, et al. Molecular characterization of carbapenem-resistant Klebsiella pneumoniae isolates with focus on antimicrobial resistance. BMC Genomics. 2019;20(1):822. doi:10.1186/s12864-019-6225-9
25. Rodrigues ACS, Santos ICO, Campos CC, et al. Non-clonal occurrence of pmrB mutations associated with polymyxin resistance in carbapenem-resistant Klebsiella pneumoniae in Brazil. Mem Inst Oswaldo Cruz. 2019;114:e180555. doi:10.1590/0074-02760180555
26. Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical Carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi:10.1016/j.ebiom.2017.04.032
27. Hu N, Wang D, Lin Y, et al. Molecular analysis and antimicrobial resistance pattern of tigecycline-non-susceptible K. pneumoniae isolated from a tertiary care hospital of East Asia. Infect Drug Resist. 2021;14:4147–4155. doi:10.2147/IDR.S334098
28. Cortazzo V, Posteraro B, Menchinelli G, et al. Susceptibility of meropenem-resistant and/or carbapenemase-producing clinical isolates of enterobacterales (Enterobacteriaceae) and pseudomonas aeruginosa to ceftazidime-avibactam and ceftolozane-tazobactam as assessed by in vitro testing methods. Antibiotics. 2022;11(8):1023. doi:10.3390/antibiotics11081023
29. Huang PH, Cheng YH, Chen WY, et al. Risk factors and mechanisms of in vivo emergence of colistin resistance in carbapenem-resistant Klebsiella pneumoniae. Int J Antimicrob Agents. 2021;57(6):106342. doi:10.1016/j.ijantimicag.2021.106342
30. zhang X, qu F, jia W, et al. Polymyxin resistance in carbapenem-resistant Enterobacteriaceae isolates from patients without polymyxin exposure: a multicentre study in China. Int J Antimicrob Agents. 2021;57(2):106262. doi:10.1016/j.ijantimicag.2020.106262
31. Goncalves Barbosa LC, Silva ESJA, Bordoni GP, et al. Elevated mortality risk from CRKp associated with comorbidities: systematic review and meta-analysis. Antibiotics. 2022;11(7):874. doi:10.3390/antibiotics11070874
32. Nang SC, Li J, Velkov T. The rise and spread of mcr plasmid-mediated polymyxin resistance. Crit Rev Microbiol. 2019;45(2):131–161. doi:10.1080/1040841X.2018.1492902
33. Cannatelli A, Giani T, Dapos Andrea MM, et al. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother. 2014;58(10):5696–5703. doi:10.1128/AAC.03110-14
34. Adzitey F, Huda N, Ali GR. Molecular techniques for detecting and typing of bacteria, advantages and application to foodborne pathogens isolated from ducks. Biotech. 2013;3(2):97–107.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
