Back to Journals » Infection and Drug Resistance » Volume 14
Resistance Profiles and Biological Characteristics of Rifampicin-Resistant Staphylococcus aureus Small-Colony Variants
Authors Zheng X, Fang R, Wang C, Tian X, Lin J, Zeng W, Zhou T , Xu C
Received 18 January 2021
Accepted for publication 12 March 2021
Published 21 April 2021 Volume 2021:14 Pages 1527—1536
DOI https://doi.org/10.2147/IDR.S301863
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Xiangkuo Zheng,1,* Renchi Fang,2,* Chong Wang,3 Xuebin Tian,4 Jie Lin,1 Weiliang Zeng,4 Tieli Zhou,1 Chunquan Xu1
1Department of Clinical Laboratory, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, 325035, People’s Republic of China; 2Department of Laboratory Medicine, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, 310003, People’s Republic of China; 3Department of Laboratory Medicine, Qingdao Municipal Hospital, Qingdao, 266000, People’s Republic of China; 4School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, 325035, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chunquan Xu; Tieli Zhou
Department of Clinical Laboratory, The First Affiliated Hospital of Wenzhou Medical University, Wenzhou, 325000, People’s Republic of China
Tel +86-577-5557-9725
; +86-577-8668-9885
Email [email protected]; [email protected]
Background: Staphylococcus aureus (S. aureus) is a major contributor to nosocomial and community-acquired infections. S. aureus small colony variants (SCVs) which changed in relevant phenotype have made more limited and difficult for therapeutic options against S. aureus infections increasingly. Rifampicin is considered as the “last-resort” antibiotic against S. aureus. Our study investigated resistance profiles and biological characteristics of rifampicin-resistant S. aureus SCVs.
Methods: We collected S. aureus SCVs that were selected from 41 rifampicin-resistant clinical isolates. Then, biological characteristics, resistance spectrum, and rifampicin resistance mechanisms of tested S. aureus SCVs and corresponding parental strains were investigated by classic microbiological methods, agar dilution method, polymerase chain reaction (PCR). Moreover, the fitness cost of S. aureus SCVs, including growth, biofilm formation ability, and virulence profile, was also determined by bacterial growth curve assay, biofilm formation assay, and Galleria mellonella infection model.
Results: There were three S. aureus SCVs (JP310 SCVs, JP1450 SCVs, JP1486 SCVs) that were selected from 41 rifampicin-resistant S. aureus. S. aureus SCVs colonies were tiny, with decreased pigmentation, and the hemolysis circle was not obvious compared with corresponding parental strains. And SCVs could not be restored to normal-colony phenotype after hemin, menaquinone, or thymidine supplementation. Different rpoB mutations occurred in JP1486 SCVs. Antimicrobial susceptibility testing revealed MICs of SCVs were higher than corresponding parental strains. Besides, the growth ability and virulence of SCVs were lower, and biofilm formation ability of which increased compared with parental strains.
Conclusion: S. aureus SCVs share the rifampicin resistance mechanisms with parental strains, although there were some differences in the position of rpoB mutations. Moreover, we found that the biological characteristics of SCVs were significantly different from corresponding parental strains. In contrast, decreased susceptibility to other antibiotics of SCVs was observed during phenotype switch. Furthermore, SCVs incur the fitness cost.
Keywords: Staphylococcus aureus, small-colony variant, rifampicin resistance, resistance profile, biological characteristic
Introduction
Staphylococcus aureus (S. aureus), the Gram-positive opportunistic pathogen, is one of the most common causatives of nosocomial infections (often associated with biofilm formation), included skin and skin structure infection (SSSI), nosocomial pneumonia, and life-threatening endocarditis.1,2 Despite the continuous improvement of effective antimicrobial and patient care conditions, many cases of S. aureus bacteremia or sepsis with significant morbidity and mortality have been reported worldwide.3,4 With the widespread use or even misuse of antibacterial agents, S. aureus has shown the resistance to a broad range of antibiotics, such as vancomycin and daptomycin. In particular, the widespread emergence of multidrug-resistant S. aureus and methicillin-resistant S. aureus (MRSA) has resulted in a limited number of therapeutic options against S. aureus infections increasingly.2
Fortunately, rifampicin is considered as the “last-resort” antibiotic that retains the antibacterial activity against multidrug-resistant S. aureus and MRSA. Rifampicin was used primarily for the treatment of tuberculosis at first, however, the potent antistaphylococcal activity of which has been analyzed since the 1970s.5 Tshefu et al found that rifampicin works to prevent experimental staphylococcal implant-associated infections subsequently.6 Currently, the combination therapy of certain antimicrobial agents (fluoroquinolone, clindamycin, etc.) with rifampicin has played an important role in eradicating S. aureus colonization and treating invasive and persistent S. aureus infections.7–9 However, the emergence of rifampicin-resistant mutants when rifampicin was used in combination with other antibiotics has long been reported.10,11 According to those reports, resistance to rifampicin is caused by mutations within the β-subunit of the rifampin binding site of bacterial DNA-dependent RNA polymerase in an 81-bp region of the rpoB gene, which named rifampicin resistance-determining region (RRDR).12,13 In the process of bacteria acquiring the rifampicin resistance, rpoB mutations promote clinically relevant phenotype switching through the generation of a subpopulation of S. aureus small colony variants (SCVs).8,14 S. aureus SCVs, as per definition, are derived from classical S. aureus strains and have the characteristics of small colony morphology, such as slow growth rate, virulence factor changes and enhanced antibiotic resistance.15,16 SCVs have an intracellular survival pattern that evades the host’s immune attack so that protected from antimicrobial agents. Studies have shown that SCVs are correlated with persistent infections, such as cystic fibrosis, osteomyelitis, implant infections, or chronic wounds.17,18 However, though the S. aureus SCVs have been regarded as the most common reason for persistent infection literally, few studies focus on the alterations in biological characteristics of rifampicin-resistant S. aureus SCVs, which remain to be fully interpreted.
In the present study, three rifampicin-resistant S. aureus SCVs were detected from 41 rifampicin-resistant S. aureus from the First Affiliated Hospital of Wenzhou Medical University, aiming to explorer the resistance mechanisms and characteristic changes of S. aureus SCVs pattern. This study provides a better understanding of the S. aureus SCVs phenotype as an effective strategy for bacterial survival and rifampicin resistance.
Materials and Methods
Bacterial Strains
Total 563 non-duplicated S. aureus strains were isolated from the First Affiliated Hospital of Wenzhou Medical University (Wenzhou, Zhejiang Province, China) from 2013 to 2015. All strains were identified by the matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) system (BioMerieux, Lyons, France), and the rifampicin-resistant S. aureus strains were detected using the agar dilution method.19 Among 41 rifampicin-resistant normal-colony (NC) S. aureus isolates, three S. aureus SCVs were classified and identified based on their phenotypic traits: hemolysis, growth phenotype, and colony morphology on 5% sheep blood agar plates. And we focused on three rifampicin-resistant S. aureus SCVs (JP310 SCVs, JP1450 SCVs, JP1486 SCVs) and corresponding parental strains (JP310, JP1450, JP1486). 41 rifampicin-resistant S. aureus isolates were mainly from sputum (58.5%, 24/41), followed by blood (9.8%, 4/41), cerebrospinal fluid (7.3%, 3/41), stool (4.8%, 2/41), and drainage (4.8%, 2/41). Besides, 14.6% (6/41) strains were isolated from other sample types. Moreover, these SCVs-producing clinical S. aureus isolates recovered from wound tissue (JP310), and sputum (JP1450, JP1486), respectively.
Catalase, Coagulase, and Hemolytic Activity
Biological characteristics of S. aureus SCVs (JP310 SCVs, JP1450 SCVs, JP1486 SCVs) and their parental strains (JP310, JP1450, JP1486) were performed by classic microbiological methods. The catalase activity was determined by following its ability to split hydrogen peroxide (H2O2), and bubble formation was considered positive for catalase production.20 The coagulase activity was measured by the classical tube coagulase test with rabbit plasma.21 Hemolytic activity was evaluated by streaking isolated colonies on sheep blood agar plates, and the hemolysis zone was read following incubation at 37°C for 24 h.22
Auxotrophy Determination
S. aureus SCVs (JP310 SCVs, JP1450 SCVs, JP1486 SCVs) and their parental strains (JP310, JP1450, JP1486) were cultured on Trypticase soy agar (TSA) plates supplemented with hemin (1 μg/mL), menadione (1 μg/mL), and thymidine (100 μg/mL), singly or in combination, and the colony sizes on the TSA plates were tested.23
Antimicrobial Susceptibility Testing
Minimum Inhibitory Concentrations (MICs) of 9 antimicrobial agents, including rifampicin (RIF), oxacillin (OXA), ciprofloxacin (CIP), gentamicin (GEN), erythromycin (ERY), tetracycline (TCY), clindamycin (CLI), vancomycin (VAN), and linezolid (LNZ), were performed by the agar dilution method according to the latest Clinical and Laboratory Standards Institute (CLSI).24 The breakpoint values of rifampicin for S. aureus were applied as a susceptible MIC of ≤ 1 μg/mL and a resistant MIC of ≥ 4 μg/mL by the recommendation of the European Committee on Antimicrobial Susceptibility Testing clinical breakpoints (http://www.eucast.org/). Staphylococcus aureus ATCC 29213 was used for quality control.
PCR Amplification and Sequencing of rpoB Gene
The genomic DNAs of tested SCVs and corresponding parental strains were extracted from fresh bacterial colonies using the Biospin Bacterial Genomic DNA Extraction kit (Bioflux, Tokyo, Japan). The rpoB gene was identified by polymerase chain reaction (PCR) using the rpoB forward primer −5ʹ-TTATGCTGCACCTTCGTG-3ʹ and rpoB reverse primer −5ʹ-CAAGTGCCCATACCTCCCATC-3ʹ. An annealing temperature of 50°C was used for PCR reactions, and the extension time was set (1 min/1 kb). Positive PCR products were sent to Shanghai BGI Technology Co. (Shanghai, China) for sequencing. The sequences obtained were analyzed using the BLAST program (www.ncbi.nlm.nih.gov/BLAST).
Multi-Locus Sequence Typing (MLST)
According to the protocols provided at http://saureus.mLst.net/, MLST analysis of the S. aureus isolates was carried out. Briefly, 7 housekeeping genes (arcC, aroE, glpF, gmk, pta, tpi, and yqiL) were analyzed. Allelic profiles and sequence types (STs) of tested S. aureus SCVs and corresponding parental isolates were further confirmed by comparing their nucleotide sequences with the MLST database (https://pubmlst.org).25
Bacterial Growth Curve Assay
The bacterial growth curve was carried out as previously described.26 Briefly, three tested S. aureus SCVs (JP310 SCVs, JP1450 SCVs, JP1486 SCVs) and their parental strains (JP310, JP1450, JP1486) were grown in Luria-Bertani (LB) broth at 37°C with shaking (180 rmp) overnight. These cultures were then transferred to 100mL of fresh LB broth (1:100 dilutions) and incubated with shaking (180 rmp) at 37°C. OD600 values for each isolate were measured at 0, 4, 8, 16, 20, and 24 h. All experiments were done in triplicate and repeated three times independently, and we used the averages values for estimating growth parameters.
Biofilm Formation Assay
Biofilm formation assay was performed by crystal violet staining as described previously.27 Briefly, three tested S. aureus SCVs and corresponding parental strains (1×106 CFU/mL) were inoculated in a 96-well polystyrene micro-test plate (Flat bottom with lid, Sterile; Corning, USA) containing fresh LB broth. Following all plates were incubated statically at 37°C for 24 h, planktonic bacteria were discarded carefully and the wells were washed twice with sterile phosphate-buffered saline (PBS) and then stained with 100 μL of 1% (w/v) crystal violet solution (lot number: NO.20190324, Beijing Solarbio Biotechnology Co., Ltd., China) for 15 min. The bound dye was solubilized for 30 min with 100 μL of eluent (95% absolute ethanol and 5% glacial acetic acid) and subsequently measured the OD595 values by the Multiskan FC microplate reader (Thermo Scientific, USA). Each isolate was assayed in three replicate wells and the experiments were repeated in triplicate independently.
Galleria mellonella Infection Model
To evaluate the virulence differences between three tested S. aureus SCVs and corresponding parental strains, G. mellonella larvae were used as an in vivo infection model.28 12 G. mellonella larvae of approximately 200 to 250 mg weight were selected for each isolate. Hamilton syringes and needles were used for all injections and were purchased (Hamilton, Nevada, U.S.). To ensure repeatability and accuracy, specialty Hamilton syringe (no. 80,401, 702LT, volume 25 μL) and needles (no. 90,534, KF, 22-gauge, 2 in.) were used to inject 10 μL of diluted bacterial suspension (5×108 CFU/mL) into the G. mellonella last left proleg. In contrast, Control larvae (n=12) were injected with 10 μL of sterile PBS. Afterward, all larvae were incubated for 5 days in the dark at 37°C. Insects were considered dead when they did not move upon touch or when they displayed a black body color. Survival data were plotted on a Kaplan-Meier curve.
Statistical Analysis
All data were analyzed using the GraphPad Prism v8.01 statistical software package (GraphPad Software, La Jolla, CA, USA). The unpaired Student’s t-test (two-tailed) was used for comparing the significance of the growth curves and biofilm formation between S. aureus SCVs and corresponding parental strains, while the G. mellonella survival data were analyzed by the log-rank (Mantel-Cox) test. Results with P-values <0.05 were considered to indicate statistical significance.
Results
Isolation and Identification of S. aureus SCVs
Three S. aureus SCVs (JP310 SCVs, JP1450 SCVs, JP1486 SCVs) were detected from 41 rifampicin-resistant S. aureus isolates. On the MHA with 5% sheep blood, the parental strains (JP310, JP1450, JP1486) grew normally while the three S. aureus SCVs grew slowly (Figure 1). After 32 hours of delayed culture, the colonies of SCVs were still tiny, with decreased pigmentation, and the hemolysis circle was not obvious; plasma coagulase assays also showed that all the parental strains were positive and SCVs strains were reduced. The auxotrophy tests for the agents listed were negative. The results showed that S. aureus SCVs could not be restored to NC phenotype after hemin, menaquinone, or thymidine supplementation (Table 1). MLST analysis revealed that JP310, JP1450, JP1486 belong to ST188, ST239, and ST238, respectively. Besides, STs of three S. aureus SCVs (JP310 SCVs, JP1450 SCVs, JP1486 SCVs) were consistent with corresponding parental strains, suggesting that they were isogenic (Table 2).
 |
Table 1 Characteristics of Three Rifampin-Resistant S. aureus Isolates and Corresponding SCVs |
 |
Table 2 ST Profile, rpoB Mutations, and Susceptibility of Three Rifampin-Resistant S. aureus Isolates and Corresponding SCVs to Other Antibiotics |
 |
Figure 1 Colony morphology of three rifampicin-resistant S. aureus strains and corresponding SCVs. |
Antimicrobial Susceptibility Patterns
Antimicrobial susceptibility testing revealed that the MIC values of oxacillin to JP1450 SCVs and JP1486 SCVs, ciprofloxacin to JP310 SCVs and JP1486 SCVs, gentamicin to JP1486 SCVs, tetracycline to JP310 SCVs and JP1486 SCVs, clindamycin to JP1486 SCVs were increased when compared with corresponding parental strains, as shown in Table 2. In contrast, there was no change in rifampicin susceptibility for all three S. aureus SCVs.
Distribution of Rifampicin-Resistance-Associated rpoB Mutations
As shown in Table 2, all isolates had at least one single amino acid substitution in RpoB. JP310 and JP310 SCVs had a mutation in codon 481 (from H to Y), JP1450 and JP1450 SCVs had two mutations in codon 466 (from L to S) and codon 481 (from H to N). There were differences between the rpoB gene mutations in JP1486 and JP1486 SCVs strains. When JP1486 had two amino acid substitutions of L466S and H481N, JP1486 SCVs only had H481N.
Fitness Cost of S. aureus SCVs
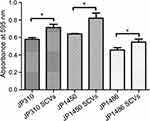
To assess whether the S. aureus SCVs were associated with the fitness cost, we investigated the growth rates and biofilm formation capacity for S. aureus SCVs and corresponding parental strains. Notably, the results of bacteria growth curves revealed that the growth rates of S. aureus SCVs were slowly than corresponding parental strains, respectively (Figure 2). Biofilm formation capacity of S. aureus SCVs was increased vs their corresponding parental strains (p <0.05) (Figure 3).
Galleria mellonella Infection Model
To evaluate the differences in virulence between the parental strains and S. aureus SCVs, G. mellonella larvae were inoculated with bacterial suspension as described above, and their survival rate was monitored daily (Figure 4). During the duration of the experiments, the mortality of larvae for S. aureus SCVs was lower than the corresponding parental strains, respectively. No mortality was observed in the control larvae. These results suggested that the phenotype switching in S. aureus SCVs was accompanied by the alterations in virulence.
Discussion
Nowadays, the emergence of rifampicin-resistant S. aureus can lead to persistent bacteremia or even severe infections in clinical, which has posed a threat globally.29–31 The limited options of treatment seriously affect the therapeutic effect of those resistant pathogens. To make things worse, under the selective pressure of antimicrobial drugs, the colony size tends to be smaller and bacteria grow slowly, followed by the emergence of SCVs, resulting in serious infection due to the survives of intracellularly and evades the immune system.15 In addition, some products of bacteria and the metabolic regulation mechanisms of the bacteria themselves may also lead to the emergence of SCVs.32 In the past few decades, many studies have shown that persistent infection caused by S. aureus is associated with SCVs, but the biological characteristics of those SCVs have not been fully understood yet.
In this study, three stable SCVs were screened from 41 rifampicin-resistant S. aureus. Compared with their parental strains, the three SCVs grew slowly. The colonies were tiny, along with the inconspicuous hemolysis-reduced coagulase activity, and decreased pigmentation. These characteristics were consistent with those reported literarily. In general, the slower growth of SCVs was related to changes in the metabolic pathways, and the types of auxotrophs found in clinical, mainly involved in electron transport chain defects and thymine synthesis defects.15,33 Three SCVs were incubated with TSA plates supplemented with hemin, menaquinone, or thymidine at 37°C for 48 h. According to our data, the growth rates did not accelerate, and the colonies did not revert to the parental phenotypes. In morphology, our results indicated that these three compounds could not stimulate the growth of the strains. It might be that some clinical SCVs were still unable to identify the specific types of auxotrophy.
Three point-mutations (H481Y, H481N, and L466S) were found in the rpoB gene of both SCVs and their parental strains, and they were all located in the rifampicin resistance determining region. Mutations H481Y and H481N were reported as the most frequent substitutions that are associated with rifampicin resistance.14 In another of our studies, H481Y also caused high-level resistance to rifampicin in S. aureus selected in vitro.34 H481N mutation was found to promote the emergence of a subpopulation of stable rifampicin-resistant SCVs with reduced susceptibility to vancomycin and daptomycin.14 However, although the H481N mutation was detected in two of the three SCVs, we did not observe the decrease in the susceptibility of SCVs to vancomycin. An interesting finding of this study was that compared to JP1486, the JP1486 SCVs lacked the L466S mutation. L466S mutation has been reported to induce low-level resistance to rifampicin but high-level resistance to rifampicin with other mutations, particularly amino acids at positions 455, 481, and 529. In another study, the L466S mutation did not cause S. aureus resistance to rifampicin.35,36 We suggested that L466S was not the main cause of high levels of rifampicin resistance, which was consistent with previous reports. And genetic changes in rpoB of S. aureus might emerge when switched from NC phenotype to SCVs.
Through the antimicrobial susceptibility test, we found that the MIC of oxacillin to JP1450 SCVs, the MICs of ciprofloxacin and tetracycline to JP310 SCVs, and the MIC of ciprofloxacin, tetracycline, gentamicin, oxacillin, and clindamycin to JP1486 SCVs were increased by 2 to 32 folds compared with their parental strains. In general, the reduction in the growth rate of SCVs usually affects the efficacy of antibacterial drugs, especially those are active against microorganisms with strong cleavage abilities. For aminoglycosides such as gentamicin, the reduced susceptibility of SCVs might be due to a decrease in transmembrane potential, which impaired the uptake of aminoglycosides by S. aureus.37,38 Fluoroquinolones usually have desirable antibacterial activity against SCVs, but some SCVs have higher MIC values. For example, SCVs had higher ciprofloxacin MIC values than isogenic wild-type strains.39,40 Tuchscherr et al have shown that clindamycin could also induce the production of SCVs.41 We speculated that S. aureus might increase resistance to certain antibiotics during the switching process from the NC phenotype to SCVs phenotype. However, the relationship between the SCVs and increased resistance to antibacterial drugs still needs to be further researched.
SCVs are an important morphological shift in bacteria-infected the human body, which greatly affects the interaction between the host and the pathogen, leading to chronic and recurrent infections. This change usually occurs with a bacterial metabolic shift. Subsequently, the suitability of SCVs has been studied to understand the specific metabolic changes of SCVs. Through the growth curve analysis, we found that the growth rate of SCVs was lower than their corresponding parental strains, which might be related to the decreased cell division ability of SCVs.42 The slower growth of this phenotype reduced the chances of detection, so the risk of clinically inappropriate antibiotic treatment was increased. Biofilm is another form of infection persistence. After detecting the biofilm formation abilities, SCVs had stronger biofilm formation abilities than their parental strains (Figure 3). Studies had shown that the sub-inhibitory concentration of gentamicin not only triggered the appearance of SCVs, but also the biofilm formation ability of S. aureus, which was enhanced by the activation of SCVs sigma factor B.43 Therefore, SCVs which survived in cells had a strong biofilm-forming ability and were more likely to cause persistent infection. Finally, we constructed the G. mellonella infection model to simulate the virulence of SCVs in vivo. According to the survival rate of the larvae, the virulence of SCVs was lower than their parental strains, which might be due to the virulence regulator agr not activated in SCVs and the α-toxin encoding gene hla was expressed at a low level.44 It has also been reported that the H481Y mutation could cause global transcriptional changes, leading to upregulation of capsule production, along with attenuated virulence in a murine bacteremia model.13 The declining virulence of SCVs has largely prevented activation of host immune responses or cytotoxicity during persistent infection, and this explains why chronic infections of SCVs often lack significant inflammatory symptoms.
Conclusion
In summary, we had found the difference in biological characteristics between S. aureus SCVs and corresponding parental strains. S. aureus SCVs shared the rifampicin resistance mechanisms with the parental strains, although there were some different mutations in rifampicin resistance-related gene rpoB. In addition, SCVs could acquire new antibiotic resistance during the phenotype switching process.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethical Statement
The whole investigation protocols in this study were approved by The Ethics Committee of the First Affiliated Hospital of Wenzhou Medical University. There are no studies with humans or animals performed by any of the authors in this article. Informed consent was waived because this study with observational nature mainly focused on bacteria and did no interventions to patients.
Acknowledgments
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work. Xiangkuo Zheng and Renchi Fang contributed equally to this work and are the co-first authors of this article. Tieli Zhou and Chunquan Xu are joint corresponding authors.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sharafutdinov IS, Trizna EY, Baidamshina DR, et al. Antimicrobial effects of sulfonyl derivative of 2(5H)-furanone against planktonic and biofilm associated methicillin-resistant and -susceptible Staphylococcus aureus. Front Microbiol. 2017;8:2246. doi:10.3389/fmicb.2017.02246
2. Lee AS, de Lencastre H, Garau J, et al. Methicillin-resistant Staphylococcus aureus. Nat Rev Dis Primers. 2018;4(1):18033. doi:10.1038/nrdp.2018.33
3. Bassetti M, Carnelutti A, Righi E. The role of methicillin-resistant Staphylococcus aureus in skin and soft tissue infections. Curr Opin Infect Dis. 2017;30(2):150–157. doi:10.1097/QCO.0000000000000353
4. Roch M, Varela MC, Taglialegna A, Rose WE, Rosato AE. Activity of telavancin against Staphylococcus aureus isolates, including those with decreased susceptibility to ceftaroline, from cystic fibrosis patients. Antimicrob Agents Chemother. 2018;62(9). doi:10.1128/AAC.00956-18
5. Zimmerli W, Widmer AF, Blatter M, Frei R, Ochsner PE. Role of rifampin for treatment of orthopedic implant-related staphylococcal infections: a randomized controlled trial. Foreign-Body Infection (FBI) Study Group. JAMA. 1998;279(19):1537–1541. doi:10.1001/jama.279.19.1537
6. Tshefu K, Zimmerli W, Waldvogel FA. Short-term administration of rifampin in the prevention or eradication of infection due to foreign bodies. Rev Infect Dis. 1983;5(Suppl 3):S474–80. doi:10.1093/clinids/5.supplement_3.s474
7. Wouthuyzen-Bakker M, Tornero E, Morata L, et al. Moxifloxacin plus rifampin as an alternative for levofloxacin plus rifampin in the treatment of a prosthetic joint infection with Staphylococcus aureus. Int J Antimicrob Agents. 2018;51(1):38–42. doi:10.1016/j.ijantimicag.2017.04.011
8. Melendez-Carmona MA, Munoz-Gallego I, Viedma E, Lora-Tamayo J, Chaves F. Intraosteoblastic activity of levofloxacin and rifampin alone and in combination against clinical isolates of meticillin-susceptible Staphylococcus aureus causing prosthetic joint infection. Int J Antimicrob Agents. 2019;54(3):356–360. doi:10.1016/j.ijantimicag.2019.06.018
9. Albrecht J, Baine PA, Ladizinski B, Jemec GB, Bigby M. Long-term clinical safety of clindamycin and rifampicin combination for the treatment of hidradenitis suppurativa. A critically appraised topic. Br J Dermatol. 2019;180(4):749–755. doi:10.1111/bjd.17265
10. Ju O, Woolley M, Gordon D. Emergence and spread of rifampicin-resistant, methicillin-resistant Staphylococcus aureus during vancomycin-rifampicin combination therapy in an intensive care unit. Eur J Clin Microbiol Infect Dis. 2006;25(1):61–62. doi:10.1007/s10096-005-0063-1
11. Zhou YF, Xiong YQ, Tao MT, et al. Increased activity of linezolid in combination with rifampicin in a murine pneumonia model due to MRSA. J Antimicrob Chemother. 2018;73(7):1899–1907. doi:10.1093/jac/dky129
12. Campbell EA, Korzheva N, Mustaev A, et al. Structural mechanism for rifampicin inhibition of bacterial rna polymerase. Cell. 2001;104(6):901–912. doi:10.1016/s0092-8674(01)00286-0
13. Gao W, Cameron DR, Davies JK, et al. The RpoB H(4)(8)(1)Y rifampicin resistance mutation and an active stringent response reduce virulence and increase resistance to innate immune responses in Staphylococcus aureus. J Infect Dis. 2013;207(6):929–939. doi:10.1093/infdis/jis772
14. Guerillot R, Goncalves da Silva A, Monk I, et al. Convergent evolution driven by rifampin exacerbates the global burden of drug-resistant Staphylococcus aureus. mSphere. 2018;3(1). doi:10.1128/mSphere.00550-17
15. Proctor RA, von Eiff C, Kahl BC, et al. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol. 2006;4(4):295–305. doi:10.1038/nrmicro1384
16. Kahl BC, Becker K, Loffler B. Clinical significance and pathogenesis of staphylococcal small colony variants in persistent infections. Clin Microbiol Rev. 2016;29(2):401–427. doi:10.1128/CMR.00069-15
17. Wong Fok Lung T, Monk IR, Acker KP, et al. Staphylococcus aureus small colony variants impair host immunity by activating host cell glycolysis and inducing necroptosis. Nat Microbiol. 2020;5(1):141–153. doi:10.1038/s41564-019-0597-0
18. Proctor R. Respiration and small colony variants of Staphylococcus aureus. Microbiol Spectr. 2019;7(3). doi:10.1128/microbiolspec.GPP3-0069-2019
19. Lim KT, Teh CS, Yusof MY, Thong KL. Mutations in rpoB and fusA cause resistance to rifampicin and fusidic acid in methicillin-resistant Staphylococcus aureus strains from a tertiary hospital in Malaysia. Trans R Soc Trop Med Hyg. 2014;108(2):112–118. doi:10.1093/trstmh/trt111
20. Zhi S, Banting G, Li Q, et al. Evidence of naturalized stress-tolerant strains of Escherichia coli in municipal wastewater treatment plants. Appl Environ Microbiol. 2016;82(18):5505–5518. doi:10.1128/AEM.00143-16
21. Compernolle V, Verschraegen G, Claeys G. Combined use of pastorex staph-plus and either of two new chromogenic agars, MRSA ID and CHROMagar MRSA, for detection of methicillin-resistant Staphylococcus aureus. J Clin Microbiol. 2007;45(1):154–158. doi:10.1128/JCM.01115-06
22. Pino A, Russo N, Van Hoorde K, et al. Piacentinu ennese PDO cheese as reservoir of promising probiotic bacteria. Microorganisms. 2019;7(8):254. doi:10.3390/microorganisms7080254
23. Wellinghausen N, Chatterjee I, Berger A, et al. Characterization of clinical Enterococcus faecalis small-colony variants. J Clin Microbiol. 2009;47(9):2802–2811. doi:10.1128/JCM.00485-09
24. CLSI. Performance Standard for Antimicrobial Susceptibility Testing.
25. Tang YT, Cao R, Xiao N, et al. Molecular epidemiology and antimicrobial susceptibility of methicillin-resistant Staphylococcus aureus isolates in Xiangyang, China. J Glob Antimicrob Resist. 2018;12:31–36. doi:10.1016/j.jgar.2017.08.016
26. Zhou Y, Wang T, Guo Y, et al. In vitro/vivo activity of potential MCR-1 inhibitor in combination with colistin againsts mcr-1-positive Klebsiella pneumonia. Front Microbiol. 2018;9:1615. doi:10.3389/fmicb.2018.01615
27. Rossi Goncalves I, Dantas RCC, Ferreira ML, et al. Carbapenem-resistant Pseudomonas aeruginosa: association with virulence genes and biofilm formation. Braz J Microbiol. 2017;48(2):211–217. doi:10.1016/j.bjm.2016.11.004
28. Tsai CJ, Loh JM, Proft T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence. 2016;7(3):214–229. doi:10.1080/21505594.2015.1135289
29. Gao W, Chua K, Davies JK, et al. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 2010;6(6):e1000944. doi:10.1371/journal.ppat.1000944
30. Howden BP, McEvoy CR, Allen DL, et al. Evolution of multidrug resistance during Staphylococcus aureus infection involves mutation of the essential two component regulator WalKR. PLoS Pathog. 2011;7(11):e1002359. doi:10.1371/journal.ppat.1002359
31. Tenover FC, Sinner SW, Segal RE, et al. Characterisation of a Staphylococcus aureus strain with progressive loss of susceptibility to vancomycin and daptomycin during therapy. Int J Antimicrob Agents. 2009;33(6):564–568. doi:10.1016/j.ijantimicag.2008.12.010
32. Tuchscherr L, Medina E, Hussain M, et al. Staphylococcus aureus phenotype switching: an effective bacterial strategy to escape host immune response and establish a chronic infection. EMBO Mol Med. 2011;3(3):129–141. doi:10.1002/emmm.201000115
33. Garcia LG, Lemaire S, Kahl BC, et al. Pharmacodynamic evaluation of the activity of antibiotics against hemin- and menadione-dependent small-colony variants of Staphylococcus aureus in models of extracellular (broth) and intracellular (THP-1 monocytes) infections. Antimicrob Agents Chemother. 2012;56(7):3700–3711. doi:10.1128/AAC.00285-12
34. Wang C, Fang R, Zhou B, et al. Evolution of resistance mechanisms and biological characteristics of rifampicin-resistant Staphylococcus aureus strains selected in vitro. BMC Microbiol. 2019;19(1):220. doi:10.1186/s12866-019-1573-9
35. Yilmaz ES, Aslantas O. Antimicrobial resistance and underlying mechanisms in Staphylococcus aureus isolates. Asian Pac J Trop Med. 2017;10(11):1059–1064. doi:10.1016/j.apjtm.2017.10.003
36. Tang HJ, Lai CC, Hsueh PR, et al. RNA polymerase B subunit gene mutations in biofilm-embedded methicillin-resistant Staphylococcus aureus following rifampin treatment. J Microbiol Immunol Infect. 2016;49(3):394–401. doi:10.1016/j.jmii.2015.06.006
37. von Eiff C. Staphylococcus aureus small colony variants: a challenge to microbiologists and clinicians. Int J Antimicrob Agents. 2008;31(6):507–510. doi:10.1016/j.ijantimicag.2007.10.026
38. Garcia LG, Lemaire S, Kahl BC, et al. Antibiotic activity against small-colony variants of Staphylococcus aureus: review of in vitro, animal and clinical data. J Antimicrob Chemother. 2013;68(7):1455–1464. doi:10.1093/jac/dkt072
39. Idelevich EA, Kriegeskorte A, Stubbings W, et al. Comparative in vitro activity of finafloxacin against staphylococci displaying normal and small colony variant phenotypes. J Antimicrob Chemother. 2011;66(12):2809–2813. doi:10.1093/jac/dkr393
40. von Eiff C, Friedrich AW, Becker K, Peters G. Comparative in vitro activity of ceftobiprole against staphylococci displaying normal and small-colony variant phenotypes. Antimicrob Agents Chemother. 2005;49(10):4372–4374. doi:10.1128/AAC.49.10.4372-4374.2005
41. Tuchscherr L, Kreis CA, Hoerr V, et al. Staphylococcus aureus develops increased resistance to antibiotics by forming dynamic small colony variants during chronic osteomyelitis. J Antimicrob Chemother. 2016;71(2):438–448. doi:10.1093/jac/dkv371
42. Kriegeskorte A, Grubmuller S, Huber C, et al. Staphylococcus aureus small colony variants show common metabolic features in central metabolism irrespective of the underlying auxotrophism. Front Cell Infect Microbiol. 2014;4:141. doi:10.3389/fcimb.2014.00141
43. Mitchell G, Brouillette E, Seguin DL, et al. A role for sigma factor B in the emergence of Staphylococcus aureus small-colony variants and elevated biofilm production resulting from an exposure to aminoglycosides. Microb Pathog. 2010;48(1):18–27. doi:10.1016/j.micpath.2009.10.003
44. Kriegeskorte A, Block D, Drescher M, et al. Inactivation of thyA in Staphylococcus aureus attenuates virulence and has a strong impact on metabolism and virulence gene expression. mBio. 2014;5(4):e01447–14. doi:10.1128/mBio.01447-14
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.