Back to Journals » Biologics: Targets and Therapy » Volume 18
Recent Advancements in Reducing the Off-Target Effect of CRISPR-Cas9 Genome Editing
Authors Asmamaw Mengstie M , Teshome Azezew M, Asmamaw Dejenie T , Teshome AA , Tadele Admasu F , Behaile Teklemariam A , Tilahun Mulu A , Mekonnen Agidew M , Adugna DG , Geremew H , Abebe EC
Received 7 July 2023
Accepted for publication 16 January 2024
Published 18 January 2024 Volume 2024:18 Pages 21—28
DOI https://doi.org/10.2147/BTT.S429411
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Doris Benbrook
Misganaw Asmamaw Mengstie,1 Muluken Teshome Azezew,2 Tadesse Asmamaw Dejenie,3 Assefa Agegnehu Teshome,4 Fitalew Tadele Admasu,1 Awgichew Behaile Teklemariam,1 Anemut Tilahun Mulu,1 Melaku Mekonnen Agidew,1 Dagnew Getnet Adugna,5 Habtamu Geremew,6 Endeshaw Chekol Abebe1
1Department of Biochemistry, College of Medicine and Health Sciences, Debre Tabor University, Debre Tabor, Ethiopia; 2Department of Physiology, College of Medicine and Health Sciences, Debre Tabor University, Debre Tabor, Ethiopia; 3Department of Medical Biochemistry, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia; 4Department of Anatomy, College of Medicine and Health Sciences, Debre Tabor University, Debre Tabor, Ethiopia; 5Department of Anatomy, School of Medicine, College of Medicine and Health Sciences, University of Gondar, Gondar, Ethiopia; 6College of Health Sciences, Oda Bultum University, Chiro, Ethiopia
Correspondence: Misganaw Asmamaw Mengstie, Department of Biochemistry, College of Medicine and Health Sciences, Debre Tabor University, Debre Tabor, Ethiopia, Tel +25196715350, Email [email protected]
Abstract: The CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)) and the associated protein (Cas9) system, a young but well-studied genome-editing tool, holds plausible solutions to a wide range of genetic disorders. The single-guide RNA (sgRNA) with a 20-base user-defined spacer sequence and the Cas9 endonuclease form the core of the CRISPR-Cas9 system. This sgRNA can direct the Cas9 nuclease to any genomic region that includes a protospacer adjacent motif (PAM) just downstream and matches the spacer sequence. The current challenge in the clinical applications of CRISPR-Cas9 genome-editing technology is the potential off-target effects that can cause DNA cleavage at the incorrect sites. Off-target genome editing confuses and diminishes the therapeutic potential of CRISPR-Cas9 in addition to potentially casting doubt on scientific findings regarding the activities of genes. In this review, we summarize the recent technological advancements in reducing the off-target effect of CRISPR-Cas9 genome editing.
Keywords: CRISPR-Cas9, genome editing, off-target effect, recent advancements, review
Introduction
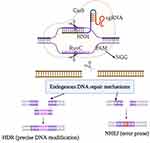
The CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) and its associated protein (Cas9) system, a new yet thoroughly researched genome-editing technology, holds viable solutions to a variety of genetic problems. It uses short guide RNAs (gRNAs) to direct Cas9, a DNA-cleaving enzyme, to its genomic target DNAs.1 As illustrated in Figure 1, Cas9 consists of the recognition lobe and the nuclease lobe, which are joined by a highly conserved arginine-rich helix to form contacts with the sgRNA.2 The recognition lobe and nuclease lobe each contain the RuvC and HNH domains. Eight to twelve base pairs (bps) upstream of the PAM are crucial for recognition by the Cas9-sgRNA complex, and the cutting site is three base pairs upstream of the PAM. The PAM is typically NGG.3 In medical research, CRISPR-Cas9 gene editing has proven to be a valuable tool. Its applications to a variety of diseases are nearly limitless, given its ability to add or silence genes in living hosts. Despite its enormous potential as a genome-editing tool, there are still several concerns that must be addressed during the application process. One of the most significant challenges in using CRISPR gene editing is off-target DNA cleavage, which occurs when the molecular scissor alters the wrong section of the host genome, resulting in a non-specific, unwanted /unexpected, and even adverse alteration to the genome.4 When these errors occur, the system produces genetic mutations instead of fixing them. To reduce off-target effects, varieties of methods have been developed such as sgRNA optimization, off-target detection, prime editing, Cas9 nuclease modification, and so on.5,6 This review focused on the recent advances to mitigate the off-target effect of CRISPR-Cas9 genome editing.
Mechanism of CRISPR/Cas9 Genome Editing
Adaptation, expression, and interference are the three phases of the CRISPR-Cas immune response.7 During the adaption step, an invasive DNA molecule is encountered by a complex of Cas proteins, which then attaches to it and induces two double-strand breaks. This motif is known as the short protospacer-adjacent motif (PAM). Interwoven between two CRISPR array repetitions, the released short DNA fragment of invasive phages or plasmids dubbed protospacer becomes a spacer.8 CRISPR expression and transcription into a lengthy precursor CRISPR RNA (pre-crRNA) take place during the expression stage of the gene. Pre-crRNA is processed by Cas proteins and auxiliary factors into short mature crRNA. The host cells are shielded from the infection during the interference step by the joint action of the Cas proteins and crRNA, which identify and mediate the cleavage of the foreign nucleic acid.9 The PAM sequence, which is conserved DNA with a length of two to five base pairs and varies in size depending on the type of bacteria, is located downstream of the cut location. The most widely used nuclease in the genome-editing tool, Cas9 protein, detects the PAM sequence at 5ʹ-NGG-3ʹ.6 Following its discovery of a target site with the proper PAM, Cas9 initiates local DNA melting, which leads to the creation of an RNA-DNA hybrid. Next, the Cas9 protein cleaving DNA produces a double-strand break. Ultimately, the host cellular machinery repairs the double-strand break using either homology-directed repair (HDR) pathways or non-homologous end joining (NHEJ).10,11
Off-Target Effect of Cas9 Nuclease
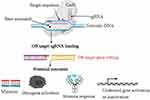
Off-target genome editing is a type of unintended and non-specific genetic modification that occurs when an engineered nuclease is used. Potential off-target cleavage activity could still occur on DNA sequences with even three to five base pair mismatches in the PAM-distal part of the sgRNA-guiding sequence, even though the frequency, nature, and location of this genetic modification were not explicitly stated.12 Noncanonical PAMs and a variety of distinct nucleotides from on-target sites are present in off-target sites. Mismatches are more easily tolerated by gRNAs at their 5′ end than at their 3′ end.13 A mismatch in the seed area can prevent Cas9 from activating if there are one or more of them. If there are three or more mismatches, DNA sequences bind to Cas9, obstructing HNH conformation and inhibiting cleavage. Off-target actions are typically influenced by the configurations and structures of gRNAs inside the system.2 Off-target genome editing confuses and diminishes the therapeutic potential of CRISPR-Cas9 in addition to potentially casting doubt on scientific findings regarding the activities of genes.14 As shown in Figure 2, this effect can end up with harmful events such as undesired DNA damage, immune response, and cytotoxicity.
Advances in Reducing the off-Target Effect of CRISPR-Cas9 Genome Editing
Improving sgRNA Specificity
Various methods of guided RNA modification have been devised to minimize off-target effects, including GC content, sgRNA length, truncated sgRNA, and chemical modification.5 It was demonstrated that the area of GC content with sgRNA seeds correlates with the efficacy of sgRNA gene editing. Researchers discovered that GC content in the gRNA sequence between 40% and 60% increases on-target activity because higher GC content stabilizes the DNA: RNA duplex and destabilizes off-target binding.15 The specificity of sgRNA is likewise closely linked with its length depending on the specific Cas nuclease. Shorter sgRNA sequences, typically those with fewer than 20 nucleotides, can efficiently lessen the off-target effect without compromising the gene editing effect.16,17 Another easy approach to make sgRNA more specific is known as the “GG20” technique, which involves replacing the GX19 sgRNAs at the 5’ end of the sgRNA with two guanines (called ggX20 sgRNAs), which can significantly lessen the off-target effect and boost specificity.14 Studies have also shown that chemical modifications at particular locations in the guide sequences can significantly increase the specificity and flexibility of CRISPR-Cas systems for the precise editing of genes. For instance, a study revealed that a chemical modification (2′-O-methyl-3′-phosphonoacetate’) incorporated at specific sites in the ribose-phosphate backbone of sgRNAs can significantly reduce off-target cleavage activities while maintaining high on-target performance, as demonstrated in clinically relevant genes.18
Improved Cas Variants
Researchers argued that lowering non-specific Cas9/sgRNA binding to DNA, particularly the non-targeted DNA strand, could improve the fidelity of prototypic SpCas9 (Streptococcus pyogenes Cas9). The rational design of SpCas9 mutants like enhanced SpCas9 (eSpCas9) and SpCas9-HF1 (HF1 for high-fidelity variant-1) was prompted by this concept.19 This is due to the presence of a proofreading mechanism that trapped these mutants in an inactive state when bound to mismatched targets. So, in recent times, it has been advised to increase the usage of eSpCas9 and SpCas9-HF1 for effective and precise genome engineering.20,21 Scientists also compared the activity of mutants with SpCas9 at on-target sites using previously reported sgRNAs to determine the fidelity of these Cas9 derivatives. For instance, SpCas9-HF1 was found to retain on-target activity comparable to wild-type SpCas9 with >85% of sgRNAs tested in human cells.22
In addition to protein engineering, another effective method for reducing off-target effects is to employ CRISPR nickase to alter one nuclease domain in only one strand of DNA. Nickase is essential for producing nicks that are promptly repaired in the cell. Cas9 nickase, unlike normal Cas9, only breakdown one strand of DNA. As a result, editing genes with Cas9 nickase reduces further damage in the target DNA and plays a significant role in reducing off-targeting.23 In general, the main advantage of the Cas9-paired nickase over Cas9 is its ability to reduce undesired mutations. The off-target effect can also be decreased by discovering novel Cas9 homologs with rarer PAM sequences, which have a lower probability of binding on non-targeted genomic DNA.12 For example, in contrast to SpCas9, which uses a relatively common PAM sequence of 5’-NGG-3’, the SaCas9 produced from Staphylococcus aureus requires a more difficult PAM sequence of 5’-NGGRRT-3.24 SaCas9 is said to be effective for a range of genome engineering purposes because of its longer PAM sequence, small size for delivery, and paired nickase approach, thus minimizing off-target effects.25
Use of Prime Editors
The CRISPR-Cas9 systems’ off-target effect mainly results from the generation of DNA double-strand breaks (DBSs) at target genomic loci, the requirement to include a donor DNA molecule, and the activation of DNA repair mechanisms.26 Prime editing is a search-and-replace genome-editing technique that does not require donor DNA or a double-strand break and enables all 12 conceivable base-to-base conversions as well as insertions and deletions.27 Prime editing has several benefits, but its most notable advantage is removing the need for a DNA donor to carry out gene editing.28 Prime editing guided RNA (PegRNA), a reverse transcriptase (RT) enzyme, and an engineered Cas9 nickase (nCas9) is the three main molecular components involved in prime editing.29 As opposed to Cas9 employed in typical CRISPR-Cas9-based genome editing, nCas9 can only cut single-strand DNA that is complementary to the template RNA. The RT enzyme is an RNA-dependent DNA polymerase that can use an RNA template to create a complementary DNA strand, whereas the PegRNA acts as a guide RNA for base-complementary recognition of target DNA locations.30 In addition to a prime binding site (PBS) region and the sequence that will be introduced to the target gene, PegRNA also comprises a spacer that is complementary to one DNA strand.31 The primer-binding site (PBS) region on the DNA is bound to the 5′ of the pegRNA, exposing the non-complimentary strand. The PAM-containing strand’s unbound DNA is nicked by nCas9, which produces a primer for the RT enzyme that is linked to nCas9. By using the inside of the pegRNA as a template to extend the nicked PAM strand, the RT then modifies the target region in a programmed way.32,33 Numerous researchers have successfully used this gene-editing toolbox in diverse systems for various genome-editing applications. Recent studies further revealed that the prime editor did not cause any detectable off-target mutations in transfected human cells, demonstrating its excellent specificity.34
Astonishing progress has been made from the initial discovery and characterization of the prime-editing system to potential applications in reducing the off-target effect of CRISPR-Cas9 in biomedical sciences. Nevertheless, prime editing is still in its infancy and there are a number of issues that need to be resolved, most importantly the issue of low efficiency.33
Anti-CRISPR Proteins
Anti-CRISPR (Acr) proteins are naturally occurring inhibitors of the CRISPR-Cas system that are encoded by a variety of mobile genetic elements and inhibit CRISPR-Cas immune function at different stages.35 More than 50 anti-CRISPR proteins have so far been found to interact with CRISPR-Cas systems including Cascade-Cas3, Cas9, Cas12, and Cas13. While certain Acr proteins are exclusive to a certain Cas protein, others can block CRISPR enzymes from several bacterial species.36 The proteins in the Acr family are numbered in order of discovery and are named for the type of CRISPR-Cas system they block. For instance, AcrIIA4 denotes the fourth Acr protein that has been shown to disrupt type II-A CRISPR-Cas systems. The majority of anti-CRISPR proteins inhibit CRISPR action in one of three ways: by inhibiting DNA binding, inhibiting crRNA loading, or inhibiting DNA cleavage.37,38 More frequently, Acr binds directly to the CRISPR-Cas complex and prevents it from binding to DNA. This is known as DNA binding inhibition. Blocking the recognition of the PAM site is another method of preventing DNA binding.39 In doing so, they lower the concentration of the CRISPR-Cas effector complex, which enhances target selectivity. Acr can be employed in conjunction with the CRISPR-Cas system to neutralize and counteract the excessive activity induced by the CRISPR-Cas effector complex and reduce the frequency of off-target effects. Furthermore, Acrs can be delivered to tissues or cells to avoid undesired DNA cleavage caused by the CRISPR-Cas system.40 Acrs essentially mimic DNA, binding in the region where the cutting enzyme Cas9 typically binds the target DNA before it cuts and then never let it go. In general, using Acrs to turn off Cas9 or decrease its activity provides a safeguard against undesirable off-target consequences. However, Acr is still a relatively recent addition to the ever-expanding CRISPR toolbox, and scientists are working to identify other varieties along with understanding its biology and possible applications.
Discovery of SuperFi-Cas9
While various attempts have been made to modify Cas9 to prevent off-target interactions, all of them have suffered from reduced rates of on-target DNA cleavage. However, scientists at the University of Texas at Austin have recently redesigned Cas9, a key component of the CRISPR gene-editing tool.2 The new version has been titled as SuperFi-Cas9 and is 4000 fold less likely to cut off target sites. Normally, the Cas9 protein searches the DNA code for a specific sequence of 20 letters, but if it finds one with 18 of the 20 letters matching its target, it may proceed with the edit. The team used cryo-electron microscopy to observe what Cas9 does when it interacts to figure out why this happens. They came across an unusual finger-like structure that had never been seen before. This finger reached out and stabilized the DNA sequence, allowing the protein to continue editing. The team tweaked this finger after discovering this mechanism, causing it to no longer stabilize the DNA and instead push away from it. Cas9 is unable to edit the undesirable sequence as a result of this. The researchers created a 7D mutant (called SuperFi-Cas9) by changing the seven mismatch-stabilizing residues to aspartic acid. The competition assay revealed that SuperFi-Cas9 had a 6.3-fold preference for on-target DNA vs DNA with 18–20 mismatches, whereas wild-type SpCas9 only had a 1.55-fold preference ratio. This research showed that SuperFi-Cas9 was capable of distinguishing between on- and off-target substrates.41 A recent lab-based study also showed that superFi-Cas9 demonstrated a high activity rate and high fidelity exhibiting features in mammalian cells compared to previously used Cas9.42 Another study on the nematode Caenorhabditis elegans to assess the genome editing potential of SuperFi-Cas9 found that the rates of gene mutagenesis induced by SuperFi-Cas9 through either non-homologous end joining or homology-directed repair were much lower than those induced by wild-type Cas9, indicating that Super-Cas9 had very low in vivo DNA cleavage activity.43 SuperFi-Cas9 has so far only been tested on DNA in test tubes, but the researchers hope to use it to edit genes in living cells in the near future. Table 1 summarizes the current strategies to reduce the off-target effect of CRISPR-Cas9 genome editing.
 |
Table 1 Summary of Recent Strategies to Reduce the Off-Target Effect of CRISPR-Cas9 |
Conclusion
In order to reduce the off-target effects of CRISPR-Cas9, several innovative strategies have been developed, including prime editors, improved Cas variants, optimized sgRNA, and anti-CRISPR proteins. In an effort to improve therapeutic gene editing, researchers are working rapidly to develop modified Cas9 variations and novel gene-targeting methods in mammalian cells that have minuscule off-target effects. With few off-target consequences on human cells, the primary editing technique now under development holds great potential for treating genetic illnesses in the future. However, detecting off-target sites in a highly sensitive and comprehensive manner, on the other hand, remains a major challenge in the field of gene editing.
Ethical Approval
Since this is a review article, ethical review and approval were not required.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Kato-Inui T, Takahashi G, Hsu S, Miyaoka Y. Clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 with improved proof-reading enhances homology-directed repair. Nucleic Acids Res. 2018;46(9):4677–4688. doi:10.1093/nar/gky264
2. Bravo JPK, Liu M-S, McCool RS, Jung K, Johnson KA, Taylor DW. Structural basis for mismatch surveillance by CRISPR/Cas9. bioRxiv. 2021;603:343–347.
3. Zheng N, Li L, Wang X. Molecular mechanisms, off-target activities, and clinical. Clin Transl Med. 2020;10(1):412–426. doi:10.1002/ctm2.34
4. Kotagama OW, Jayasinghe CD, Abeysinghe T. Era of genomic medicine: a narrative review on CRISPR technology as a potential therapeutic tool for human diseases. Biomed Res Int. 2019;201:1–15. doi:10.1155/2019/1369682
5. Naeem M, Majeed S, Hoque MZ, Ahmad I. Latest developed strategies to minimize the off-target effects in CRISPR-cas-mediated genome editing. Cells. 2020;9(7):1–23. doi:10.3390/cells9071608
6. Mengstie MA, Wondimu BZ. Mechanism and applications of crispr/ cas-9-mediated genome editing. Biol Targets Ther. 2021;15:353–361. doi:10.2147/BTT.S326422
7. Janik E, Niemcewicz M, Ceremuga M, Krzowski L. Various aspects of a gene editing system — CRISPR – cas9. Int J Mol Sci. 2020;2020:1–20.
8. Xue C, Sashital DG, State I. Mechanisms of type I-E and I-F CRISPR-Cas systems in Enterobacteriaceae. EcoSal Plus. 2020;8(2):1–38.
9. Hille F, Charpentier E. CRISPR-Cas: biology, mechanisms and relevance. Philos Trans R Soc B Biol Sci. 2016;371(1707):20150496. doi:10.1098/rstb.2015.0496
10. Chavez M, Chen X, Finn PB, Qi LS. Advances in CRISPR therapeutics. Nat Rev. 2023;19(9):1–14.
11. Richter H, Charpentier E, Randau L, Randau L. DNA and RNA interference mechanisms by CRISPR-Cas surveillance complexes. FEMS Microbiol Rev. 2015;39:442–463. doi:10.1093/femsre/fuv019
12. Zhang XH, Tee LY, Wang XG, Huang QS, Yang SH. Off-target effects in CRISPR/Cas9-mediated genome engineering. Mol Ther Nucleic Acids. 2015;4(11):e264. doi:10.1038/mtna.2015.37
13. Corsi GI, Qu K, Alkan F, Pan X, Luo Y, Gorodkin J. CRISPR/Cas9 gRNA activity depends on free energy changes and on the target PAM context. Nat Commun. 2022;13(1):3006. doi:10.1038/s41467-022-30515-0
14. Guo C, Ma X, Gao F, Guo Y, Wang K. Off-target effects in CRISPR / Cas9 gene editing. Front Bioeng Biotechnol. 2023;11:1–11.
15. Javaid N, Choi S. CRISPR / cas system and factors affecting its precision and ef fi ciency. Front Cell Dev Biol. 2021;9(November):1–25.
16. Tycko J, Myer VE, Hsu PD, Jolla L. Methods for optimizing CRISPR-Cas9 genome editing specificity. Mol Cell. 2017;63(3):355–370. doi:10.1016/j.molcel.2016.07.004
17. Baghini SS, Gardanova ZR, Zekiy AO, Shomali N, Tosan F, Jarahian M. Optimizing sgRNA to improve CRISPR / Cas9 knockout ef fi ciency: special focus on human and animal. Front Bioeng Biotechnol. 2021;9:775309. doi:10.3389/fbioe.2021.775309
18. Ryan DE, Taussig D, Steinfeld I, et al. Improving CRISPR – cas specificity with chemical modifications in single-guide RNAs. Nucleic Acids Res. 2018;46(2):792–803. doi:10.1093/nar/gkx1199
19. Zuo Z, Babu K, Ganguly C, Zolekar A, Newsom S. Rational engineering of CRISPR-Cas9 nuclease to attenuate position-dependent off-target effects. Cris J. 2022;5(2):329–340. doi:10.1089/crispr.2021.0076
20. Zhang D, Zhang H, Li T, Chen K, Qiu J, Gao C. Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases. Genome Biol. 2017;18(191):1–7. doi:10.1186/s13059-017-1325-9
21. Kulcsár PI, Tálas A, Huszár K, et al. Crossing enhanced and high fidelity SpCas9 nucleases to optimize specificity and cleavage. Genome Biol. 2017;18(109):1–17. doi:10.1186/s13059-017-1318-8
22. Kleinstiver BP, Pattanayak V, Prew MS, et al. High-fidelity CRISPR-Cas9 variants with undetectable genome- wide off-targets. Nature. 2016;529(7587):490–495. doi:10.1038/nature16526
23. Rasul MF, Hussen BM, Salihi A, Ismael BS, Jalal PJ. Strategies to overcome the main challenges of the use of CRISPR / Cas9 as a replacement for cancer therapy. Mol Cancer. 2022;24(64):1–30.
24. Topkar VV, Zheng Z, Joung JK. Broadening staphylococcus aureus Cas9 targeting range by modifying PAM recognition. Nature Biotechnol. 2016;33(12):1293–1298.
25. Friedland AE, Baral R, Singhal P, et al. Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16(257):1–10. doi:10.1186/s13059-015-0817-8
26. Xue C, Greene EC, Biophysics M. DNA repair pathway choices in CRISPR-Cas9 mediated genome editing. Trends Genet. 2022;37(7):639–656. doi:10.1016/j.tig.2021.02.008
27. Anzalone AV, Randolph PB, Davis JR, et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2020;576(7785):149–157. doi:10.1038/s41586-019-1711-4
28. Matsoukas IG. Prime editing: genome editing for rare genetic diseases without double-strand breaks or donor DNA. Front Genet. 2020;11(528):1–6. doi:10.3389/fgene.2020.00528
29. Schole J, Harrison PT, Harrison PT. Prime editing-an update on the field. Gene Ther. 2021;28(7):396–401. doi:10.1038/s41434-021-00263-9
30. Lee J, Lim K, Chung E, Cho S, Kim A, Mok YG. Prime editing with genuine Cas9 nickases minimizes unwanted indels. Nat Commun. 2023;14(1):1786. doi:10.1038/s41467-023-37507-8
31. Marzec M, Br A. Prime editing: a new way for genome editing. Trends Cell Biol. 2020;xx(xx):1–3.
32. Kantor A, Mcclements ME, MacLaren R. CRISPR-Cas9 DNA base-editing and prime-editing. Int J Mol Sci. 2020;21(6240):1–21. doi:10.3390/ijms21176240
33. Huang Z, Liu G. Current advancement in the application of prime editing. Front Bioeng Biotechnol. 2023;11:1039315. doi:10.3389/fbioe.2023.1039315
34. Godbout K, Tremblay JP. Prime editing for human gene therapy: where are we now ? Cell. 2023;12(4):536. doi:10.3390/cells12040536
35. Marino ND, Pinilla-redondo R, Csörgő B, Bondy-denomy J. Anti-CRISPR protein applications: natural brakes for CRISPR-Cas technologies. Nat Methods. 2021;17(5):471–479. doi:10.1038/s41592-020-0771-6
36. Forsberg KJ. Anti-CRISPR discovery: using magnets to find needles in haystacks. J Mol Biol. 2023;435(7):167952. doi:10.1016/j.jmb.2023.167952
37. Pinilla-redondo R, Shehreen S, Marino ND, et al. Discovery of multiple anti-CRISPRs highlights anti-defense gene clustering in mobile genetic elements. Nat Commun. 2020;11(1):5652. doi:10.1038/s41467-020-19415-3
38. Zhang F, Song G, Tian Y. Anti ‐ CRISPRs: the natural inhibitors for CRISPR ‐ Cas systems. Anim Model Exp Med. 2019;2(2):69–75. doi:10.1002/ame2.12069
39. Sun W, Zhao X, Wang J, et al. Anti-CRISPR AcrIIC5 is a dsDNA mimic that inhibits type II-C Cas9 effectors by blocking PAM recognition. Nucleic Acids Res. 2023;51(4):1984–1995. doi:10.1093/nar/gkad052
40. Zhang Y, Marchisio MA, Marchisio MA. Type II anti-CRISPR proteins as a new tool for synthetic biology. RNA Biol. 2021;18(8):1085–1098. doi:10.1080/15476286.2020.1827803
41. Nuclease C, Huang X, Yang D, Zhang J, Xu J, Chen YE. Recent advances in improving gene-editing specificity through CRISPR–Cas9 nuclease engineering. Cells. 2022;11(14):2186. doi:10.3390/cells11142186
42. Kulcsár PI, Tálas A, Welker E. SuperFi-Cas9 exhibits extremely high fidelity but reduced activity in mammalian cells. bioRxiv. 2022;2022:493683.
43. Ferdousy SCB. An assessment of genome-editing efficiency of a newly developed Cas9 in. microPublication Biol. 2022;10:17912.
44. Matson AW, Hosny N, Swanson ZA, Hering BJ, Burlak C. Optimizing sgRNA length to improve target specificity and efficiency for the GGTA1 gene using the CRISPR / Cas9 gene editing system. PLoS One. 2019;14:12.
45. Hahn F, Nekrasov V. CRISPR / Cas precision: do we need to worry about off-targeting in plants ? Plant Cell Rep. 2019;38(4):437–441. doi:10.1007/s00299-018-2355-9
46. Aquino-jarquin G. Current advances in overcoming obstacles of CRISPR / Cas9 off-target genome editing. Mol Genet Metab. 2021;134(1–2):77–86. doi:10.1016/j.ymgme.2021.08.002
47. Jain S, Xun G, Abesteh S, et al. Precise regulation of Cas9-mediated genome engineering by an anti-CRISPR based inducible CRISPR controllers. ACS Synth Biol. 2021;10(6):1320–1327. doi:10.1021/acssynbio.0c00548
48. Casini A, Olivieri M, Petris G, et al. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol. 2018;36(3):265–271. doi:10.1038/nbt.4066
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.