Back to Journals » Infection and Drug Resistance » Volume 16
Rapid Identification of Carbapenemase-Producing Klebsiella pneumoniae Using Headspace Solid-Phase Microextraction Combined with Gas Chromatography-Mass Spectrometry
Authors Luo H, Hang Y, Zhu H , Zhong Q, Peng S , Gu S , Fang X, Hu L
Received 13 January 2023
Accepted for publication 14 April 2023
Published 1 May 2023 Volume 2023:16 Pages 2601—2609
DOI https://doi.org/10.2147/IDR.S404742
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Hong Luo,1 Yaping Hang,1 Hongying Zhu,2 Qiaoshi Zhong,1 Suqin Peng,1 Shumin Gu,1 Xueyao Fang,1 Longhua Hu1
1Jiangxi Provincial Key Laboratory of Medicine, Clinical Laboratory of the Second Affiliated Hospital of Nanchang University, Nanchang, Jiangxi, People’s Republic of China; 2Clinical Laboratory of Ganzhou People’s Hospital, Ganzhou, Jiangxi, People’s Republic of China
Correspondence: Longhua Hu, Email [email protected]
Background: Carbapenemase-producing Klebsiella pneumoniae is an unprecedented threat to public health, and its detection remains challenging. Analysis of microbial volatile organic compounds (VOCs) may offer a rapid way to determine bacterial antibiotic susceptibility.
Purpose: The aim of this study was to explore the VOCs released by carbapenemase-producing carbapenem-resistant Klebsiella pneumoniae (CRKP) using headspace solid-phase microextraction/gas chromatography-mass spectrometry (HS-SPME/GC-MS).
Methods: Test bacteria were incubated in trypticase soy broth to the end of exponential growth phase, and imipenem was added in the middle time. Headspace VOCs were concentrated and analyzed using HS-SPME/GC-MS.
Results: The compound 3-methyl-1-butanol was found to be a biomarker among the 26 bacterial isolates (10 KPC-positive, 10 NDM-positive, 2 IMP-positive, 2 carbapenemase-negative CRKP, and 2 carbapenem-susceptible K. pneumonoiae).
Conclusion: This study explored a promising new strategy for the screening of carbapenemase-producing CRKP strains. Further research with larger sample sizes will potentially accelerate the application of biomarkers in routine microbiology.
Keywords: carbapenem-resistantKlebsiella pneumoniae, volatile organic compounds, headspace solid-phase microextraction combined with gas chromatography-mass spectrometry
Corrigendum for this paper has been published.
Introduction
Klebsiella pneumoniae belongs to the genus Klebsiella, which is widespread in the environment and host-associated niches.1 K. pneumoniae has long been considered as opportunistic pathogen and remains the most common nosocomial pathogen worldwide.2,3 The treatment of K. pneumoniae infections has been complicated and limited by the emergence of antibiotic resistance, particularly to carbapenems. Carbapenem resistance in K. pneumoniae may be related to multiple mechanisms, including carbapenemase production and alterations in bacterial outer membrane permeability, combined with hyperproduction of other β-lactamases (eg, extended spectrum β-lactamase, AmpC β-lactamase).4–7 Notably, carbapenemase expression is the most common mechanism involved in CRKP, and the distribution of carbapenemase types carried by CRKP varied by country.8 In Europe, surveillance network data from 2013 to 2017 showed that among CRKP, K. pneunominae carbapenemase (KPC) producers were the most abundant, followed closely by oxacillinase (OXA)-48-like producers.8 In China, a multicenter genomic analysis of CRKP from 2015 to 2018 reported that KPC (98/147, 66.7%) and New Dehli metallo-β-lactamase (NDM; 27/147, 18.4%) were the main carbapenemases detected in CRKP isolates.9
Current evidence indicates that CRKP infections are associated with significant mortality rates,10,11 ranging from 23% to 75%.12 The average mortality rate will increase by 7.6% if administration of effective therapy in patients with septic shock is delayed per hour.13 The Treatment of carbapenem-resistant organism is complicated and often required combined therapy.14,15 Early detection of CRKP and its carbapenemase type is vital in providing a timely clinical therapy and thus resulting in lower mortality.16 However, current antibiotic susceptibility testing methods use either solid growth media or liquid broth and normally take at least 18–24 h after pathogen identification, which is time-consuming and significantly affects early clinical intervention. Therefore, new methods for the rapid identification of CRKP should be explored.
Microbial volatile organic compounds are carbon-based molecules thought to evolve as primary or secondary metabolites and may be used for bacterial identification.17–19 In the mVOC 2.0 database, approximately 2000 compounds from almost 1000 species were identified, which belong to a diverse group of chemical classes, including acids, alcohols, aldehydes, ketones, esters, hydrocarbons, and nitrogen- or sulfur-containing compounds.20
Numerous studies have analyzed the VOC profile of pathogens to differentiate between species.17,21,22 For instance, Escherichia coli and Pseudomonas aeruginosa can be rapidly identified by indole and 2-aminoacetophenone in culture media,23,24 while 2-phenylethyl alcohol, benzyl alcohol, and methyl mercaptan were identified as biomarkers for Streptococcus pneumoniae.25
However, to date, few studies have been conducted to differentiate strains according to their drug susceptibility and antibiotic resistance mechanisms. To the best of our knowledge, the production of drug hydrolysates is one of the most common mechanisms among resistant strains. Therefore, it is reasonable to suspect that hydrolysis or the type of hydrolase can influence the bacterial VOC profile after reacting with a specific antibiotic, which may become a new strategy for the rapid identification of CRKP.
Headspace solid-phase microextraction/gas chromatography-mass spectrometry (HS-SPME/GC-MS) is a sensitive analytical technique that can separate gas compounds and provide a chemical fingerprint for bacteria.17,19,26 SPME is a concentration technique that can be used to extract and concentrate various volatile and semi-volatile organic compounds from the sample headspace. The application of SPME is becoming increasingly common in various fields, mostly because SPME collection is rapid, simple, reproducible, and is available for transportation between laboratories and GC-MS analysis sites.26–28 Meanwhile, GC-MS with direct collection of gas constituents from the headspace possesses strong separation power, repeatable retention time (RT), and selective and sensitive mass detection.29 It has been regarded as the gold standard for VOCs analysis.30,31 Therefore, the aim of this study was to identify volatile compounds from the VOC profiles produced by CRKP using HS-SPME/GC-MS and provide a novel strategy for rapid identification of drug-resistant strains.
Materials and Methods
Bacterial Strains and Carbapenemase Detection
A total of 215 clinical CRKP isolates were collected from January 1, 2016 to December 31, 2021 at the Second Affiliated Hospital of Nanchang University. K. pneumoniae isolates were identified using matrix-assisted laser desorption/ionization-time of flight mass spectrophotometry (bioMérieux, France) or the Vitek 2 Compact system (bioMérieux). Antimicrobial susceptibility testing was conducted using the VITEK-2 Compact ASTGN16 (bioMérieux) or Kirby–Bauer test, and the minimum inhibitory concentrations of ertapenem and imipenem were determined according to the Clinical Laboratory Standards Institute criteria. For CRKP, the presence of carbapenemase was determined using the modified carbapenem inactivation method (mCIM), and EDTA-modified carbapenem inactivation method (eCIM) was used to determine whether the carbapenemase belongs to Ambler class B. Then, the type of carbapenemases were characterized using polymerase chain reaction (PCR).
According to the results of mCIM and eCIM, of the 215 CRKP isolates, 213 were carbapenemase-positive, of which 14 produced class B ß-lactamases. The results of PCR showed that among the 213 carbapenemase-positive strains, 199 produced KPC, 11 produced NDM, 2 produced imipenemase (IMP), and 1 produced NDM and IMP. Accordingly, we randomly selected 10 KPC-positive, 10 NDM-positive, 2 IMP-positive, and the 2 carbapenemase-negative strains for GC-MS analysis. Two strains of carbapenem-susceptible K. pneumonoiae (CSKP) were used as the control group.
Culture Conditions
Our early work showed that under the set culture conditions (microbial concentration: 107 colony forming units (CFU)/mL, culture: trypticase soy broth, temperature: 37 °C, agitation: 200 rpm) the growth rate of K. pneumoniae was the fastest at approximately 4 h and entered the end of exponential growth phase at around 7 h (Figure 1).
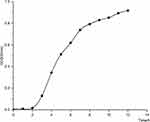 |
Figure 1 Growth curve of K. pneumoniae. |
Each test bacterium was cultured on blood agar plates at 37 °C overnight. Then, approximately 107 CFU/mL of each bacterium was inoculated into a 20 mL headspace vial (CNW Technologies GmbH, Germany) containing 6 mL trypticase soy broth (Solarbio, China) and then incubated at 37 °C with agitation at 200 rpm for 7 h (the end of exponential growth phase as early work about the growth curve of K. pneumoniae). Blank medium was used as a control. After 3.5 h, imipenem was added to each culture to a final concentration of 0.25 mg/mL (16μg/mL for carbapenemase-negative CRKP). The experiment was performed in triplicate for each strain.
According to the results of GC-MS, one 3-methyl-1-butanol producer (labeled as K198), one carbapenemase-negative CRKP (labeled as R1577), and one CSKP (labeled as S602) were randomly selected from the strains analyzed using GC-MS in order to explore the source of characteristic VOCs. Growth conditions were the same as described above; however, in the imipenem-free group, imipenem was not added during cultivation. Meanwhile, in the KPC inhibitor group, particularly for KPC-producing CRKP, avibactam sodium (final concentration: 1 mg/L; Solarbio, China) was added immediately after imipenem. Experiments were performed in triplicates.
Detection of Microbial VOCs
Headspace volatile metabolites were concentrated on a 50/30 μm divinylbenzene/carboxen/polydimethylsiloxane (DVB/CAR/PDMS) SPME fiber (Supelco, USA). After incubation at the inlet of the GC-MS at 250 °C for 60 min, the SPME was inserted into the headspace vial, which was equilibrated at 40 °C for 40 min. The fiber was exposed approximately 1 cm above the culture medium surface for 30 min at 40 °C prior to desorption in the GC injection port. Subsequently, the fiber was introduced into the inlet and desorbed for 5 min at 250 °C. The volatile molecules were separated using a DB-35MS column (30m × 0.25 mm, 0.25 μm, Agilent, America). The carrier gas was 99.999% pure helium at a constant flow rate of 1.0 mL/min.
The GC oven parameters were set as follows: initial temperature of 50 °C with a 3 min hold, ramped to 180 °C at 10 °C/min with no hold, and then a final ramp to 250 °C at a rate of 40 °C/min with a 5 min hold, with a total run time 23.2 min. The MS parameters were set as follows: the ion source temperature was 230 °C, ionization was performed via electron impact at an energy of 70 eV, and mass spectra were obtained over the range of 40 to 550m/z.
Data Analysis
Data acquisition and analysis were performed using the MassHunter GC/MS Acquisition software. The background signal was removed to avoid background interference and spectral overlap caused by the partial co-elution of compounds. An extracted ion chromatogram (EIC) within the range of 40–550m/z was derived for comparison. The Kruskal–Wallis H-test was applied to compare signal intensity in each group, and Bonferroni correction was applied to post hoc multiple comparisons.
To identify volatile molecules, the compound spectra were compared with the National Institute of Standards and Technology (NIST) 2017 mass spectral library. According to the NIST guidelines, match factor scores < 600 are considered a poor match, 700–800 as a fair match, 800–900 as a good match, and > 900 as an excellent match.32
Results
An EIC range of 40–550 m/z of VOCs were observed in the five different types of K. pneumoniae (KPC-producing, NDM-producing, and IMP-producing CRKP strains; carbapenemase-negative CRKP, and CSKP) (Figure 2). The characteristic peaks of KPC-producing and NDM-producing CRKP were observed at a retention time of 6.1 min, and the mass-charge ratios were 41, 42, 43, 55, and 70.
 |
Figure 2 Typical extracted ion chromatogram of the tested samples. |
The Kruskal–Wallis H-test showed that the signal intensity was significantly different among groups (p<0.001) when the mass-charge ratios were 41, 42, 43, 55, and 70. Furthermore, no significant difference was observed between the signal intensities of the KPC-positive and NDM-positive groups when the mass-to charge ratio was 41, 42, 43, 55, and 70 (Bonferroni-corrected p>0.05); however, the difference between the other groups was statistically significant (Bonferroni-corrected p<0.05) (Figure 3).
 |
Figure 3 Diagram showing the signal intensity of each strain. Note: Different letters indicate statistical difference (p<0.05). |
The compound spectra expressed at 6.1 min were imported into the NIST mass spectral library for identification, and the results showed that the characteristic VOC of KPC and NDM producers was 3-methyl-1-butanol, with a match factor score of 806 (Figure 4).
 |
Figure 4 Mirror plot. |
The detection rates of 3-methyl-1-butanol in KPC-positive and NDM-positive groups were 100.0% (10/10), while those in other groups were 0.0% (0/2) (Table 1). Moreover, 3-methyl-1-butanol could be detected in samples K198, R1577, and S602 when drugs were not added. Interestingly, 3-methyl-1-butanol was not detected in KPC inhibited by avibactam sodium (Table 2).
 |
Table 1 Detection of 3-Methyl-1-Butanol in Each Group |
 |
Table 2 Explore the Source of 3-Methyl-1-Butanol |
Discussion
This study is the first to use HS-SPME-GCMS for the analysis of VOCs produced by CRKP. Our results showed that 3-methyl-1-butanol could act as a biomarker for the identification of KPC- and NDM-positive CRKP after the addition of imipenem.
Bacteria can release various volatile compounds in different combinations and quantities; however, it is difficult to quantify and characterize each compound from a complex VOC profile. Consequently, the identification and analysis of biomarkers represent a better choice for bacterial identification. In recent years, a growing body of research has successfully performed volatile compound analysis in discriminating bacteria at the species or strain level. Our early research indicated that indole synthesized by E. coli can act as a biomarker for the identification of this organism.21 In addition, both 3-methyl-butanal and 3-methyl-butanoic acid were unique targets for S. aureus.17 The success of microbial VOCs in identification of bacterial species driving its application to antibiotic susceptibility testing. In a sensor-based study, VOCs produced by oxacillin-sensitive and oxacillin-resistant S. aureus as well as ampicillin-sensitive and oxacillin-resistant E. coli were monitored using Ion-molecule Reaction- Mass Spectrometry. Methanethiol was found to be a valid target for the growth of both strains and has been recommended as an indicator of antibiotic susceptibility after six hours of incubation.33 Smart et al34 utilized thermal desorption-GC-MS and detected nine compounds that distinguished cephalexin-sensitive and-resistant isolates (p<0.05). The addition of cephalexin induced more differences between the two groups, which may be attributed to bacterial cell wall lysis, thereby releasing cell components into the culture media. In the present study, 3-methyl-1-butanol was not detected when the activity of KPC was inhibited by avibactam sodium, suggesting that this biomarker may be released from carbapenem hydrolysis. However, further analysis showed 3-methyl-1-butanol could be produced by KPC-producing CRKP, carbapenemase-negative CRKP, and CSKP during metabolism and without the interference of imipenem indicating that the biomarker was not the produced from imipenem hydrolysis. Thus, this compound is a volatile metabolite released by K. pneumoniae during its natural metabolism process.
Previous studies have observed that 3-methyl-1-butanol can be produced by E. coli,24 K. pneumoniae,34,35 and S. aureus.17 The decomposition of amino acids can produce several compounds, such as 3-methyl-1-butanol, 3-methylbutanoic acid, or 3-methylbutanal.36 K. pneumoniae metabolizes leucine via the Ehrlich pathway and synthesizes short-chain branched alcohols, such as 3-methyl-1-butanol. The chemical reaction starts with the deamination of 2-amino-4-methylpentanoic acid, generating 4-methyl-2-oxopentanoic acid, which decarboxylates to 3-methyl-1-butanal. Finally, 3-methyl-1-butanal is reduced to 3-methyl-1-butanol.37 Therefore, based on our results, imipenem may interfere with the Ehrlich pathway, which affects 3-methyl-1-butanol production and the degree of interference may be related to the mechanism of bacterial resistance, however, the detailed mechanism remains to be fully elucidated. According to our data, 3-methyl-1-butanol was not produced in IMP-positive strains. Several studies found that IMP-4 positive CRKP confers reduced susceptibility to carbapenems, and the MICs to imipenem was much reduced (0.064 mg/liter).38,39 In addition, high-level resistance of IMP-positive CRKP to imipenem may be due to the combination of IMP and deficiency of outer membrane proteins.38 Therefore, it is reasonable to speculate that IMP enzyme hydrolyzing imipenem is inferior to KPC and NDM enzymes, the defect is more evident in the environment of high concentration of imipenem (0.25mg/L), which leads to a great effect of imipenem on the Ehrlich pathway of IMP-producers and the non-release of 3-methyl-1-butanol from IMP-producing CRKP.
The concept of using mVOCs as a new method for rapid detection of antibiotic-resistant strains remains a big hope, but our results suggest the approach is promising. However, the data in our study should be interpreted carefully from several aspects. Firstly, previous studies have suggested that VOCs produced by different strains have species specificity, but the production is strongly influenced by culture conditions, such as nutrients, temperature, pH, incubation time, and headspace volume.17,37,40–42 Moreover, test results of VOCs were strongly dependent on instrumental techniques, such as sampling techniques and instrument model.35,43 Consequently, differences in the environment of bacteria growth and instrumental techniques between studies make inter-study comparisons difficult. Secondly, the sample size and the type of carbapenemases are limited in our study, further work is required to confirm these findings on a more comprehensive and larger data set.
In conclusion, this study demonstrated that 3-methyl-1-butanol can act as a biomarker for the identification of KPC- and NDM-positive CRKP, which promises to open new frontiers in rapid diagnosis of bacterial infection. Further studies should focus on expanding the sample size and enzyme type to accelerate the application of VOC for the rapid identification of bacterial strains.
Abbreviations
VOCs, volatile organic compounds; HS-SPME/GC-MS, Headspace solid-phase microextraction/gas chromatography-mass spectrometry; CRKP, carbapenem-resistant K. pneumoniae; CSKP, carbapenem-susceptible K. pneumonoiae; KPC, Klebsiella pneunominae carbapenemase; NDM, New Dehli metallo-β-lactamase; OXA, oxacillinase; IMP, imipenemase; RT, retention time; EIC, extracted ion chromatogram.
Ethics Statement
As the Klebsiella pneumoniae clinical isolation in this study was the routine hospital laboratory procedure, we have confirmed that the isolation has no identifiable patient data, the Second Affiliated Hospital of Nanchang University Medical Research Ethics Committee exempted this research for review.
Funding
This work was supported by the National Natural Science Foundation of China (91200316), the Natural Science Foundation of Jiangxi Province (No. 20202BAB216021), the Health Commission of Jiangxi Province (No. 20201034).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bagley ST. Habitat association of Klebsiella species. Infect Control. 1985;6(2):52–58. doi:10.1017/s0195941700062603
2. Pendleton JN, Gorman SP, Gilmore BF. Clinical relevance of the ESKAPE pathogens. Expert Rev Anti Infect Ther. 2013;11(3):297–308. doi:10.1586/eri.13.12
3. Hansen DS, Gottschau A, Kolmos HJ. Epidemiology of Klebsiella bacteraemia: a case control study using Escherichia coli bacteraemia as control. J Hosp Infect. 1998;38(2):119–132. doi:10.1016/S0195-6701(98)90065-2
4. Walsh TR, Toleman MA, Poirel L, Nordmann P. Metallo-beta-lactamases: the quiet before the storm? Clin Microbiol Rev. 2005;18(2):306–325. doi:10.1128/cmr.18.2.306-325.2005
5. Bush K. Past and present perspectives on β-lactamases. Antimicrob Agents Chemother. 2018;62(10). doi:10.1128/aac.01076-18
6. Jacoby GA, Mills DM, Chow N. Role of beta-lactamases and porins in resistance to ertapenem and other beta-lactams in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48(8):3203–3206. doi:10.1128/aac.48.8.3203-3206.2004
7. Doumith M, Ellington MJ, Livermore DM, Woodford N. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother. 2009;63(4):659–667. doi:10.1093/jac/dkp029
8. Kazmierczak KM, Jonge BL, Stone GG, Sahm DF. Longitudinal analysis of ESBL and carbapenemase carriage among Enterobacterales and Pseudomonas aeruginosa isolates collected in Europe as part of the International Network for Optimal Resistance Monitoring (INFORM) global surveillance programme, 2013-17. J Antimicrob Chemother. 2020;75(5):1165–1173. doi:10.1093/jac/dkz571
9. Cienfuegos-Gallet AV, Zhou Y, Ai W, Kreiswirth BN, Yu F, Chen L. Multicenter genomic analysis of carbapenem-resistant Klebsiella pneumoniae from bacteremia in China. Microbiol Spectr. 2022;10(2):e0229021. doi:10.1128/spectrum.02290-21
10. Mathers AJ, Peirano G, Pitout JDD. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev. 2015;28(3):565–591. doi:10.1128/cmr.00116-14
11. Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant Enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(suppl_1):S28–S36. doi:10.1093/infdis/jiw282
12. Pitout JDD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae, a key pathogen set for global nosocomial dominance. Antimicrob Agents Chemother. 2015;59(10):5873–5884. doi:10.1128/aac.01019-15
13. Yu J, Liu J, Li Y, et al. Rapid detection of carbapenemase activity of Enterobacteriaceae isolated from positive blood cultures by MALDI-TOF MS. Ann Clin Microbiol Antimicrob. 2018;17(1):22. doi:10.1186/s12941-018-0274-9
14. Katip W, Uitrakul S, Oberdorfer P. Acinetobacter baumannii a comparison of colistin versus colistin plus meropenem for the treatment of carbapenem-resistant in critically ill patients: a propensity score-matched analysis. Antibiotics. 2020;9(10):647. doi:10.3390/antibiotics9100647
15. Katip W, Oberdorfer P, Kasatpibal N. Acinetobacter baumannii effectiveness and nephrotoxicity of loading dose colistin-meropenem versus loading dose colistin-imipenem in the treatment of carbapenem-resistant infection. Pharmaceutics. 2022;14(6):1266. doi:10.3390/pharmaceutics14061266
16. Wang X, Wang Q, Cao B, et al. EnterobacteriaceaeRetrospective observational study from a Chinese network of the impact of combination therapy versus monotherapy on mortality from carbapenem-resistant bacteremia. Antimicrob Agents Chemother. 2019;63(1):e01511–e01518. doi:10.1128/aac.01511-18
17. Chen J, Tang J, Shi H, Tang C, Zhang R. Characteristics of volatile organic compounds produced from five pathogenic bacteria by headspace-solid phase micro-extraction/gas chromatography-mass spectrometry. J Basic Microbiol. 2017;57(3):228–237. doi:10.1002/jobm.201600505
18. Drabinska N, Costello B, Hewett K, Smart A, Ratcliffe N. From fast identification to resistance testing: volatile compound profiling as a novel diagnostic tool for detection of antibiotic susceptibility. Trends Analyt Chem. 2019;115:1–12. doi:10.1016/j.trac.2019.03.019
19. Timm CM, Lloyd EP, Egan A, Mariner R, Karig D. Direct growth of bacteria in headspace vials allows for screening of volatiles by gas chromatography mass spectrometry. Front Microbiol. 2018;9:491. doi:10.3389/fmicb.2018.00491
20. Lemfack MC, Gohlke B-O, Toguem SMT, Preissner S, Piechulla B, Preissner R. mVOC 2.0: a database of microbial volatiles. Nucleic Acids Res. 2018;46(D1):D1261–D1265. doi:10.1093/nar/gkx1016
21. Zhong Q, Cheng F, Liang J, et al. Profiles of volatile indole emitted by Escherichia coli based on CDI-MS. Sci Rep. 2019;9(1):13139. doi:10.1038/s41598-019-49436-y
22. Rees CA, Franchina FA, Nordick KV, Kim PJ, Hill JE. Expanding the Klebsiella pneumoniae volatile metabolome using advanced analytical instrumentation for the detection of novel metabolites. J Appl Microbiol. 2017;122(3):785–795. doi:10.1111/jam.13372
23. Ratiu I-A, Ligor T, Bocos-Bintintan V, Buszewski B. Mass spectrometric techniques for the analysis of volatile organic compounds emitted from bacteria. Bioanalysis. 2017;9(14):1069–1092. doi:10.4155/bio-2017-0051
24. Ratiu I-A, Ligor T, Bocos-Bintintan V, et al. The effect of growth medium on an Escherichia coli pathway mirrored into GC/MS profiles. J Breath Res. 2017;11(3):036012. doi:10.1088/1752-7163/aa7ba2
25. Cox CD, Parker J. Use of 2-aminoacetophenone production in identification of Pseudomonas aeruginosa. J Clin Microbiol. 1979;9(4):479–484. doi:10.1128/jcm.9.4.479-484.1979
26. Bahroun NHO, Perry JD, Stanforth SP, Dean JR. Use of exogenous volatile organic compounds to detect Salmonella in milk. Anal Chim Acta. 2018;1028:121–130. doi:10.1016/j.aca.2018.03.065
27. Fitzgerald S, Duffy E, Holland L, Morrin A. Multi-strain volatile profiling of pathogenic and commensal cutaneous bacteria. Sci Rep. 2020;10(1):17971. doi:10.1038/s41598-020-74909-w
28. Huang Z, Zhang J, Zhang P, Wang H, Pan Z, Wang L. Analysis of volatile organic compounds in pleural effusions by headspace solid-phase microextraction coupled with cryotrap gas chromatography and mass spectrometry. J Sep Sci. 2016;39(13):2544–2552. doi:10.1002/jssc.201600279
29. Koek MM, Muilwijk B, Werf M, Hankemeier T. Microbial metabolomics with gas chromatography/mass spectrometry. Anal Chem. 2006;78(4):1272–1281.
30. Phillips M, Basa-Dalay V, Blais J, et al. Point-of-care breath test for biomarkers of active pulmonary tuberculosis. Tuberculosis. 2012;92(4):314–320. doi:10.1016/j.tube.2012.04.002
31. Zhang J, Fang A, Wang B, et al. iMatch: a retention index tool for analysis of gas chromatography-mass spectrometry data. J Chromatogr A. 2011;1218(37):6522–6530. doi:10.1016/j.chroma.2011.07.039
32. Jordi Labs. NIST/EPA/NIH mass spectral library compound scoring: match factor, reverse match factor, and probability; 2019. Available from: https://jordilabs.com/wp-content/uploads/2017/07/Whitepaper-NISTEPA-NIH-Mass-Spectral-Library-Compound-Scoring.pdf.
33. Wiesnera K, Jaremekb M, Pohlea R, Sicarda O, Stuetza E. Monitoring of bacterial growth and rapid evaluation of antibiotic susceptibility by headspace gas analysis. Procedia Eng. 2014;87:332–335. doi:10.1016/j.proeng.2014.11.750
34. Smart A, Costello B, White P, et al. Sniffing out resistance - rapid identification of urinary tract infection-causing bacteria and their antibiotic susceptibility using volatile metabolite profiles. J Pharm Biomed Anal. 2019;167:59–65. doi:10.1016/j.jpba.2019.01.044
35. Tait E, Perry JD, Stanforth SP, Dean JR. Identification of volatile organic compounds produced by bacteria using HS-SPME-GC-MS. J Chromatogr Sci. 2014;52(4):363–373. doi:10.1093/chromsci/bmt042
36. Moller JT, Cluitmans P, Rasmussen LS, et al. Long-term postoperative cognitive dysfunction in the elderly ISPOCD1 study. ISPOCD investigators. International study of post-operative cognitive dysfunction. Lancet. 1998;351(9106):857–861. doi:10.1016/s0140-6736(97)07382-0
37. Sutton S. Measurement of microbial cells optical density. J Valid Technol. 2011;17:46–49.
38. Yu F, Ying Q, Chen C, et al. Outbreak of pulmonary infection caused by Klebsiella pneumoniae isolates harbouring blaIMP-4 and blaDHA-1 in a neonatal intensive care unit in China. J Med Microbiol. 2012;61:984–989. doi:10.1099/jmm.0.043000-0
39. Liu Y, Zhang B, Cao Q, Huang W, Shen L, Qin X. Two clinical strains of Klebsiella pneumoniae carrying plasmid-borne blaIMP-4, blaSHV-12, and armA isolated at a pediatric center in Shanghai, China. Antimicrob Agents Chemother. 2009;53(4):1642–1644. doi:10.1128/aac.01325-08
40. Zareian M, Silcock P, Bremer P. Effect of medium compositions on microbially mediated volatile organic compounds release profile. J Appl Microbiol. 2018;125(3):813–827. doi:10.1111/jam.13908
41. Misztal PK, Lymperopoulou DS, Adams RI, et al. Emission factors of microbial volatile organic compounds from environmental bacteria and fungi. Environ Sci Technol. 2018;52(15):8272–8282. doi:10.1021/acs.est.8b00806
42. Schulz-Bohm K, Martín-Sánchez L, Garbeva P. Microbial volatiles: small molecules with an important role in intra- and inter-kingdom interactions. Front Microbiol. 2017;8:2484. doi:10.3389/fmicb.2017.02484
43. Ratiu I-A, Bocos-Bintintan V, Monedeiro F, Milanowski M, Ligor T, Buszewski B. An optimistic vision of future: diagnosis of bacterial infections by sensing their associated volatile organic compounds. Crit Rev Anal Chem. 2020;50(6):501–512. doi:10.1080/10408347.2019.1663147
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
