Back to Journals » Journal of Hepatocellular Carcinoma » Volume 11
RAD21: A Key Transcriptional Regulator in the Development of Residual Liver Cancer
Authors Pang JS, Bai XM, Wan WJ , Kang T , Wen R, Li LP, Yin HH, Lu CM, Wen DY, He Y , Yang H
Received 10 November 2023
Accepted for publication 30 January 2024
Published 5 February 2024 Volume 2024:11 Pages 285—304
DOI https://doi.org/10.2147/JHC.S447915
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Laura A. Dawson
Jin-Shu Pang,1,* Xiu-Mei Bai,1,* Wei-Jun Wan,1 Tong Kang,1 Rong Wen,1 Li-Peng Li,1 Hai-Hui Yin,1 Chun-Miao Lu,2 Dong-Yue Wen,1 Yun He,1,* Hong Yang1,*
1Department of Medical Ultrasound, the First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, People’s Republic of China; 2Department of Experimental Research, the Affiliated Tumor Hospital of Guangxi Medical University, Nanning, 530021, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hong Yang; Yun He, Email [email protected]; [email protected]
Objective: Thermal ablation is a commonly used therapy for hepatocellular carcinoma (HCC). Nevertheless, inadequate ablation can lead to the survival of residual HCC, potentially causing rapid progression. The underlying mechanisms for this remain unclear. This study explores the molecular mechanism responsible for the rapid progression of residual HCC.
Methods: We established an animal model of inadequate ablation in BALB/c nude mice and identified a key transcriptional regulator through high-throughput sequencing. Subsequently, we conducted further investigations on RAD21. We evaluated the expression and clinical significance of RAD21 in HCC and studied its impact on HCC cell function through various assays, including CCK-8, wound healing, Transwell migration and invasion. In vitro experiments established an incomplete ablation model verifying RAD21 expression and function. Using ChIP-seq, we determined potential molecules regulated by RAD21 and investigated how RAD21 influences residual tumor development.
Results: High RAD21 expression in HCC was confirmed and correlated with low tumor cell differentiation, tumor growth, and portal vein thrombosis. Silencing RAD21 inhibited the migration, invasion, and proliferation significantly in liver cancer cells. Patients with high RAD21 levels showed elevated multiple inhibitory immune checkpoint levels and a lower response rate to immune drugs. Heat treatment intensified the malignant behavior of liver cancer cells, resulting in increased migration, invasion, and proliferation. After subjecting it to heat treatment, the results indicated elevated RAD21 levels in HCC. Differentially expressed molecules regulated by RAD21 following incomplete ablation were primarily associated with the VEGF signaling pathway, focal adhesion, angiogenesis, and hepatocyte growth factor receptor signaling pathway etc.
Conclusion: The upregulation of RAD21 expression after incomplete ablation may play a crucial role in the rapid development of residual tumors and could serve as a novel therapeutic target.
Keywords: residual hepatocellular carcinoma, incomplete ablation, RAD21, overexpression, progression
Introduction
Hepatocellular carcinoma (HCC) is the most prevalent form of liver cancer worldwide and the most commonly used treatments for early HCC include liver transplantation, radical surgical, and thermal ablation.1 Thermal ablation, including radiofrequency and microwave techniques, is widely recognized as the primary method for radical treatment and local control of HCC.2–5 Previous studies have shown that thermal ablation achieves efficacy comparable to surgery, offering advantages in terms of repeatability, safety, and minimal invasiveness.6,7 Several studies recommend thermal ablation techniques for treating medium- to large-sized HCCs. They have demonstrated their feasibility in treating solitary HCCs up to 5 cm or multiple HCCs smaller than 3 cm.8–11 Unfortunately, like hepatectomy, the high frequency of recurrence following ablation remains a significant factor negatively impacting the long-term survival of HCC patients.12–14 Incomplete ablation of the tumor periphery often leads to the recurrence of residual HCC, which, in turn, exhibits enhanced growth and invasiveness.15–17 However, the underlying mechanisms, to date, have not been extensively revealed for the increased malignant potential of residual HCC after ablation.
RAD21 encodes a DNA double-strand-break (DSB) repair protein that plays a crucial role in the cohesin complex. Additionally, it functions in various aspects of the mitosis process, including sister chromatid cohesion, preventing improper recombination, ensuring proper division, and post-replicative DNA repair.18 The roles of RAD21 in various diseases have been a subject of significant interest for decades, and its deregulation has been notably observed in human tumors. Studies have closely associated RAD21 expression levels with prognosis and metastatic behaviors, even though RAD21 mutations are rarely found in solid tumors.19 Notably, researchers have identified overexpression of RAD21 in epithelial breast cancer as linked to poor prognosis and resistance to drug treatment.20 Conversely, low RAD21 levels characterize metastasis and invasion in oral squamous cell carcinoma.21 Extensive studies with large sample sizes have observed positive RAD21 protein within the nuclei of colorectal cancer cells, and its presence has been linked to metastasis and reduced disease-specific survival.22
Furthermore, researchers have observed increased RAD21 expression in HCC compared to normal livers. In HCC patients with evaluated RAD21 expression, there is evidence of resistance to ionizing resistance, suggesting a connection between RAD21 levels and the tumor’s ability to protect itself.23 However, the precise mechanisms and function of RAD21 as a transcriptional regulator in residual HCC remain largely unclear. Understanding how deregulated RAD21 influences tumor behaviors in HCC is a topic that merits further exploration.
The purpose of this article is to comprehensively explore the function and significance of RAD21 in residual cancer. Given its difficulty to collect residual HCC specimens from clinical patients, we simulated residual HCC of incomplete ablation on the subcutaneous xenograft tumors and performed high-throughput sequencing for these specimens. The use of sequencing on residual tumors from animal models enables our study to conduct a detailed molecular analysis, which can reveal expression changes in the thousands of genes associated with the progression of residual HCC and the influence of RAD21. This information is vital for understanding the molecular pathways involved and for identifying potential therapeutic targets in residual HCC. Additionally, we also study the relationship between RAD21 and HCC clinicopathological parameters and the prognostic role of RAD21 through public big data and immunohistochemistry (IHC). Besides, we conducted in vivo experiments to further investigate the impact of RAD21 on the behaviors of HCC cells and further investigate its potential regulatory targets. Due to the gradual application of immune and adjuvant therapy in the treatment of liver cancer, our study also explored the potential of RAD21 in suggesting immune infiltration and immunotherapy response.12,24,25 To the best of our knowledge, our study is the first to focus on the role of RAD21 as a transcriptional regulator in the context of residual HCC development and progression.
Materials and Methods
Cell Culture
Liver tumor cell lines (HepG2, Huh7, MHCC97-H, MHCC97-L, Hep3B) were provided by Cellcook (Guangzhou, China) and FuHeng Biology (Shanghai, China). These HCC cells were cultured in an environment at 37 °C with 95% CO2 and 5% O2, using Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin solution. Subculture was then performed when the cells reached 80%–90% confluence in a 25 cm2 culture flask.
Animal Xenograft Experiment
The experiment observed a group of eight 6-week-old male immunodeficient mice (Balb/c nude mice) for a 3-day adaptation period. Subsequently, 100 µL (2 x 106 cells/mL) of MHCC97-H and HepG2 cells were subcutaneously injected on the lateral side of the spine. When the subcutaneous HCC xenografts reached a 1.0–1.5 cm size, the experimental group (N = 8) and the control group (N = 8) were randomly assigned. The Balb/c mice were anesthetized with sodium pentobarbital (0.1 mL / 20 g), and nude mice in the experimental group underwent incomplete ablation (power: 30 watts; time: 15s) using a Cool-tip™ (RFA) system. In contrast, the control mice only underwent a puncture injury using an ablation needle (power: 0 watts; time: 15s). After a 24-h obversion period, HCC xenografts were resected from Balb/c mice and frozen in –80 °C liquid nitrogen using RNAsafer stabilizer reagent for the subsequent high-throughput RNA-seq analysis. Differentially expressed genes were selected using a threshold of |fold-change |> 1.2 (and p < 0.05) following analysis using the Limma package in R software. The HepG2 xenografts have been uploaded into GEO (GSE234283).
Globally Bulk High-Throughput Cohorts and Microarrays
Globally multi-center bulk HCC RNA-seq data or microarrays, which included both HCC and non-HCC specimens, were obtained from various sources, such as TCGA, SRA, ArrayExpress, and GEO databases. The goal was to identify differentially expressed genes in HCC and non-tumor livers. The search query for these datasets was as follows: (HCC OR hepatocellular OR hepatic OR liver) AND (tumor OR cancer OR malignancy OR carcinoma OR neoplasm).
We also acquired transcription regulators including transcription factors (TFs) and transcription cofactors (ToFs), previously validated to play a regulatory role in transcriptional activity from the humanTFDB. The bulk RNA-seq data were then converted into standardized expression quantification using TPM, and all expression profiles underwent log (x + 1) normalization. For datasets obtained from the same platform, expression profiles were further processed into an aggregated expression matrix by the “sva” package in R software to eliminate the batch effect. In contrast to traditional methods for identifying differentially expressed genes (DEGs), we integrated global multicentric samples to assess genome-wide mRNA differences between HCC and non-HCC livers comprehensively.
Single-Cell RNA-Seq (scRNA-Seq) Analysis
We comprehensively used scRNA-seq data to reveal the disparities in gene expression and expression distribution within the GSE151530 dataset, comprising 17,164 malignant cells and 35,625 non-tumor cells collected from 30 samples. These cells were annotated and classified into seven primary categories: tumor cells, T cells, B cells, cancer-associated fibroblasts (CAFs), tumor-associated macrophages (CAFs), tumor-associated endothelial cells (TECs), and unclassified cells.
Immunohistochemical Staining and Proteomics Analysis
mRNA is the intermediary between genes and proteins, with proteins being the ultimate performers of genetic functions. While protein expression can often be inferred from mRNA levels, it is essential to note that mRNA expression levels do not always correlate strongly with protein levels. To address this, we extended our analysis by incorporating proteomics and IHC to evaluate protein levels when identifying differentially expressed genes (DEGs) at the mRNA level.
The IHC samples were obtained from tissue microarrays (LVC1601), purchased from Fanpu Biotech (Guilin, China). For the IHC assessment, intensity was defined as follows: negative (scored 0), weakly positive (scored 1), moderately positive (scored 2), and strongly positive (scored 3). Positive cell scores were defined based on the percentage of positive cell counts relative to the total number of cells, as follows: (1) the ratio of positive cells = 0%, scored 0; (2) 0% < positive cells < 25%, scored 1; (3) 25% < positive cells < 50%, scored 2; (4) 50%< positive cells < 75%, scored 3; (5) positive cells > 75%, scored 4. Finally, the IHC response scores were calculated by multiplying the staining intensity by the positive percentage scores.26
Proteomics data including 125 HCC and adjacent livers were downloaded from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) program. We used the KNN function of the “impute” package in the R software and set the number of neighbors to 10 to complete the missing data. We then analyzed the relations between RAD21 protein and clinical value as well as its prognostic significance.
Prognostic Value of RAD21 for HCC Patients
We employed Cox regression and Kaplan-Meier curves to evaluate the different survival conditions of key regulators on the overall survival (OS) and progression-free survival (PFS) of HCC patients. HCC patients from TCGA and CPTAC were divided by the median value of gene expression, and hazard ratio (HR) was used to quantify the OS and PFS prognosis of HCC patients.
In vitro Experiments
siRNA Design and Transfection Efficacy
The siRNA targeting RAD21 was designed and provided by Sangon Biotech’s website. We have displayed the sequence of these RAD21-targeting siRNAs in Table 1. The reagents used for transfecting the siRNA were also procured from Sangon Biotech and employed according to the provided protocol. The qRT-PCR primers for detecting RAD21 mRNA are additionally provided in Table 1. The primary antibody (ab217678) used for RAD21 detection was sourced from the Abcam company, while the secondary rabbit antibody was obtained from Proteintech.
 |
Table 1 The Sequences of RAD21 siRNA and qRT-PCR Primer |
Quantitate Real-Time PCR and Cell Functional Assays
Once the RAD21 knockdown was successfully achieved, cell functional assays, which included CCK8 proliferation experiments, scratch test, and Transwell migration plus invasion experiments, were conducted as described in the provided protocol.
Candidate Transcriptional Targets of RAD21
Chromatin immunoprecipitation sequencing (ChIP-seq) is an antibody-based, high-throughput technique that selectively enriches proteins in specific regions of their DNA targets. We utilized ChIP-seq experiments to identify the putative targets regulated by RAD21 in the transcriptional process. Candidate targets were filtered with a cutoff score of ≥ 2.5. The final candidates influenced by RAD21 in residual HCC were selected from the overlap between HCC and residual HCC upregulated DEGs and the ChIP-seq targets. Gene Ontology and pathway analysis including KEGG, WikiPathways, Panther, and Reactome algorithms were performed based on WebGestalt tool.
RAD21 Levels and Tumor Immunity
The TIMER score algorithm was employed to estimate immune cell infiltration using RNA-sequencing profiles (level 3) involving six immune cells, including dendritic cells, neutrophils, B cells, CD8+ T, CD4+ T cells, and macrophages. Subsequently, Pearson correlation and survival analyses were conducted to establish connections between RAD21 levels and HCC immune infiltration. We then assessed the differential expression of immune checkpoint-related genes in patient groups with high and low RAD21 levels. Furthermore, we utilized the TCGA HCC samples to calculate the Tumor Immune Dysfunction and Exclusion (TIDE) score to predict the response to immune therapy response for HCC subtypes characterized by different RAD21 expressions. The TIDE algorithm, as provided by Jiang, P. et al27 employs a panel of checkpoint-related signatures to assess two types of immune escape mechanisms, including cytotoxic T lymphocyte (CTL) dysfunction and the rejection of CTLs by immunosuppression factors. High TIDE scores indicate a poor response and limited benefit from immune checkpoint blockade (ICB) therapy.
Statical Analysis
The expression values, whether mRNA or protein, were presented as mean ± standard deviation (SD). Statical differences between two or more groups were analyzed using independent Student’s t-test or analysis of variance (ANOVA). Globally large samples were then systematically evaluated for differential expression in HCC based on evidence-based algorithms. The standardized mean difference (SMD) was calculated to assess mRNA levels for each gene using the “meta” package in R software. A gene with an SMD > 0 (and p < 0.05) represented higher expression in HCC compared to non-HCC samples, while genes of an SMD < 0 (and p < 0.05) were considered to be under-expressed in HCC. Furthermore, a random-effects model would be more suitable for the SMD index when heterogeneity (I2) is ≥ 50%, while a fixed-effects model would be the optimal choice if I2 is < 50%. Begg’s and Egger’s tests were used together to detect publication bias, and sensitivity analysis was also performed to assess the possibility of heterogeneity if necessary.
Results
Key Regulatory Factors in Residual HCC
We compiled 2690 transcription regulators from the HumanTFDB database. Comprehensive RNA-seq and microarray analyses were conducted on 3443 tumors and 3941 non-cancer samples, revealing 8676 upregulated differentially expressed genes (DEGs) and 7125 downregulated DEGs in hepatocellular carcinoma (HCC) (Tables S1 and S2). Based on the in-house sequencing efforts, we observed 2554 overexpressed genes and 531 under-expressed genes in HepG2 xenografts subjected to imcomplete ablation (Figure 1A). Additionally, we found 1349 upregulated and 77 genes with dysregulated expression in MHCC97-H xenografts after imcomplete ablation (Figure 1B). The specimens and pathological images of MHCC97-H were showed in Figure S1.
Considering these findings, we ultimately identified 18 transcription regulators that were consistent across all four datasets (Figure 1C). Among these 18 candidates, ten genes (GTF2H1, ILF2, OTX1, PARP12, RNF10, PHF20, RAD21, ZMIZ2, PSEN1, and ETV3) exhibited statistically significant associations with clinical prognosis, particularly concerning OS (Figure 1D). Furthermore, eight genes (PHF20, RNF10, ILF2, ZMIZ2, GTF2H1, RAD21, PIAS1, and NCOR2) were valuable in predicting HCC recurrence or disease progression, as indicated by PFS analysis (Figure 1E).
Elevated RAD21 mRNA and Protein in HCC
As illustrated in Figure 2, 39 platforms, encompassing 3395 HCC samples and 3023 non-HCC samples, were included for a comprehensive evaluation of RAD21 mRNA expression. Among these, 30 cohorts showed a significant overexpression of RAD21 mRNA in HCC, with a p-value of < 0.05. Based on the random-effects model, the forest plot further demonstrates a notably high expression of RAD21 mRNA in HCC, with an SMD of 1.13 (p < 0.001, Figure 3A). Additionally, scRNA-seq analysis revealed that RAD21 was widely expressed in HCC cells but had limited expression in intratumor cells, except for T cells. This further confirms that malignant cells are primarily responsible for the elevated RAD21 expression in HCC (Figures 3B-D). Moreover, our proteomics analysis demonstrated a similar pattern of higher RAD21 protein levels in HCC compared to peritumor tissues (Figure 3E).
 |
Figure 2 The expression pattern of RAD21 mRNA in a single data set was analyzed by globally multicentric microarrays and bulk RNA-seq data. |
In-House IHC Verification
Representative IHC staining images of RAD21 protein in HCC are displayed in Figure 3F-I. Furthermore, in-house IHC experiments also revealed higher levels of RAD21 protein in HCC cells compared to non-tumor cells, consistent with the RAD21 mRNA trend observed in multicentric samples (Table 2). However, we discovered no significant relationship between RAD21 levels and HCC characteristics.
 |
Table 2 Immunohistochemistry Shows the Expression of RAD21 in HCC and Adjacent Cancer and Its Relationship with Clinicopathological Parameters |
Clinical Significance of RAD21 in HCC
Based on a large dataset of samples, the high expression of RAD21 in HCC suggests superior performance in differentiating HCC from non-HCC livers, with a summarized ROC (sROC) value of 0.89 (Figure 4A). The pooled sensitivity, specificity, DLR negative, and positive were 0.92, 0.72, 3.24, and 0.11, respectively (Figures S1 and S2). Concerning HCC prognosis, we discovered that patients with high RAD21 levels exhibited a more favorable prognosis regarding both OS and PFS (Figures 4B and C). Additionally, patients with high RAD21 protein levels experienced worse OS, although the predictive performance of RAD21 for PFS patients was less apparent in HCC patients, as indicated by proteomics samples (Figures 4D and E). Regarding clinicopathological features from 371 RNA-seq samples, HCC patients with advanced stages and larger tumors exhibited higher RAD21 levels. Furthermore, high RAD21 levels in HCC were associated with a poorly differentiated status, ranging from grade 1 to grade 3 (Table 3). Similarly, proteomics data also indicated that high RAD21 protein levels in HCC were closely related to a poor differentiation grade and the presence of portal thrombus (Table 4).
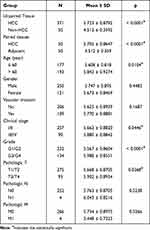 |
Table 3 The Relationship Exploration Based on Transcriptomics Between RAD21 Expression and HCC Clinicopathological Parameters |
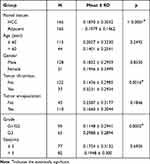 |
Table 4 The Relationship Exploration Based on the Proteomics Between RAD21 Protein Expression and HCC Clinicopathological Parameters |
RAD21 is a Helpful Marker in Predicting Immune Checkpoints and Resistance to ICB Therapy
Patients with high RAD21 expression levels exhibit significantly elevated levels of CD274, CTLA4, HAVCR2, PDCD1, and TIGIT, which are well-known inhibitory immune checkpoints, in comparison to patients with low RAD21 levels (Figure 5A). Importantly, patients with high RAD21 scores also show increased TIDE scores, indicating a lower likelihood of responding to immune checkpoint blockade (ICB) therapy than patients with low RAD21 scores (Figure 5B).
Knockdown of RAD21 Suppressed HCC Proliferation, Migration, and Invasion
Following a 48-h transfection of RAD21 siRNA, we observed a significant downregulation of RAD21 mRNA and protein through qRT-PCR and WB, thus confirming the successful knockdown of RAD21 expression in Huh7 and Hep3B cells (Figure 6). Loss-of-function arrays revealed that RAD21 knockdown significantly inhibited the viability of HCC cells, affecting proliferation, migration, and invasion (Figures 7 and 8).
Aggressive Behavior of Sublethal Heat-Treated HCC Cells
Both Transwell migration and invasion experiments indicated that Huh7 and Hep3B HCC cells exhibited stronger migration and invasion abilities than the control group after sublethal heat treatment (Figures 9A-E). CCK-8 arrays also suggested that Huh7 and Hep3B cells displayed enhanced proliferation and reduced apoptosis in response to heat stress (Figures 9C and F). We observed increased RAD21 mRNA and protein levels in sublethal HCC cells through qRT-PCR and WB analysis, respectively (Figures 9G-J).
Transcriptional Mechanism Regulated by RAD21
RAD21 transcriptional targets were identified by summarizing data from nine ChIP-seq datasets, and 4603 candidates validated by at least five experiments were selected. Among these candidates, a final set of 184 genes exhibited significantly elevated expression in both HCC and residual HCC (Figure 10A). Based on GO analysis (Figure 10B), these 184 RAD21 targets were found to be involved in various biological processes, including cellular localization, organelle organization, and symbiont process (biological process, BP). Regarding cellular components (CC), RAD21 targets were distributed in the cytosol, adherents and anchoring junctions, and focal adhesion. Regarding molecular function (MF), the functions of proteins encoded by RAD21 targets were mainly categorized as phosphatase binding, cadherin binding, and oxygen sensor activity. Additionally, the pathway enrichment analysis (Figure 10C-F) indicated that the RAD21 targets in residual HCC were associated with multiple HCC-related pathways, including the VEGF signaling pathway, focal adhesion, angiogenesis, and hepatocyte growth factor receptor signaling pathway.
Discussion
Aberrant epigenetic features, such as gene mutations and expression patterns, can manifest in certain cases. These abnormalities suggest that cells may produce abnormal or excessive protein products, leading to uncontrolled cancer growth and metastasis, which are hallmark characteristics of cancer.28,29 Within the context of cancer, oncogenes are genes that affect the development and progression of cancer when mutated or overexpressed. When comparing cancer cells to normal cells, genes exhibiting significantly higher expression in cancer cells are more likely to be oncogenes. This is because they play essential roles in cellular processes dysregulated in cancer. These processes include promoting cell proliferation, inhibiting apoptosis, enhancing angiogenesis, or activating signaling pathways that drive tumor metastasis and progression.28,29 Identifying differentially highly expressed genes in cancer cells is crucial in understanding the potential mechanisms underlying cancer development and progression.
TF and ToF are both proteins involved in gene transcription, but they exhibit differences in function and mechanisms of action. TFs regulate chromatin transcription by recognizing specific DNA sequences and recruiting coactivators, forming a complex system to control gene expression in the genome.30 When combined with TFs, ToFs further stabilize the structure of the transcription complex and enhance gene transcription.31 Additionally, ToFs can mediate signal transduction pathways that influence gene transcription regulation.32 The occurrence and progression of cancer involve a series of changes in these transcription regulators.33–36 However, current research on residual cancer has not yet addressed the elucidation of transcription factor regulators. Therefore, we propose the following question: Does the process of residual cancer following ablation involve alterations and regulation of critical transcription regulators?
In this study, we employed evidence-based random-effects and fixed-effects models, integrating 39 publicly available datasets to calculate the genome-wide mRNA expression patterns. This approach allowed us to identify highly reliable differentially expressed genes specific to liver cancer. Compared to previous single-center, small-sample, and incomplete coverage studies, our research benefits from a larger and more accurate dataset and advanced analytical methods. Furthermore, we integrated two in-house sequencing datasets of residual HCC and human transcriptional regulators to explore potential transcriptional regulators that may exacerbate malignant behavior in incompletely ablated HCC. We identified 18 molecules, and 10 genes (GTF2H1, ILF2, OTX1, PARP12, RNF10, PHF20, RAD21, ZMIZ2, PSEN1, and ETV3) exhibited a strong predictive capability for OS in HCC patients. Additionally, eight genes (PHF20, RNF10, ILF2, ZMIZ2, GTF2H1, RAD21xPIAS1, and NCOR2) demonstrated significant statistical value in predicting HCC recurrence risk. After reviewing relevant literatures, the biological function of RAD21 may be related to the development of liver cancer, namely its key role in DNA repair, cell cycle regulation, and various aspects of the mitosis process, therefore making it a relatively attractive research target. Finally, we focused our attention on RAD21 for an in-depth investigation.
RAD21 is an essential nuclear protein in chromosome meiosis and DNA repair during cell mitosis. It is not a novel protein, as research in the 1990s already indicated its association with DNA repair.18 Recently, an increasing body of evidence suggests that abnormal expression of RAD21 is implicated in the initiation, progression, and metastasis of various cancers. RAD21 is found to be highly expressed in multiple human cancers, and its overexpression is associated with a poor prognosis.37 Furthermore, overexpression of RAD21 may lead to metastasis and invasion of breast cancer.38,39 This study has also detected elevated expression of RAD21 in ovarian cancer, and it has correlated its amplification with a poor prognosis and immune escape in ovarian cancer.40
We conducted comprehensive data curation and analysis on 3395 cases of HCC tissue and 3024 cases of non-HCC tissue to confirm the overexpression of RAD21 in HCC. The single-cell analysis further revealed relatively low expression of RAD21 in non-malignant cells, indicating that the elevated RAD21 in HCC originates from malignant cells rather than infiltrating cells from other tumors. Subsequently, we analyzed the correlation between RAD21 levels and the prognosis and clinicopathological characteristics of 371 RNA-seq HCC patients and 125 proteomic HCC patients. Specifically, the discovery of elevated RAD21 levels in tissues from late-stage HCC patients and larger tumor tissues suggests that RAD21 expression may be involved in tumor growth and progression. The association between high RAD21 levels and the low differentiation status of cancer cells indicates that RAD21 may play a role in dedifferentiation, often associated with more aggressive tumor behavior. The finding of a correlation between high RAD21 levels and the possibility of portal vein thrombosis indicates that RAD21 may be involved in the development of HCC-related vascular complications and tumor metastasis, both of which are crucial factors influencing patient prognosis.
Cell function experiments showed that heat-treated liver cancer cells exhibited enhanced abilities to proliferate, migrate, and invade after regaining viability from sublethal damage. Furthermore, after subjecting them to sublethal heat treatment, we observed an upregulation of RAD21 expression levels in the liver cancer cells. On the other hand, knocking down RAD21 inhibited the proliferation, migration, and invasion abilities of liver cancer cells, suggesting an association between RAD21 expression and these capabilities in liver cancer cells. Specifically, the increase in RAD21 expression after heat treatment appears to strengthen liver cancer cells’ proliferation, migration, and invasion abilities while knocking down RAD21 suppresses these abilities. This phenomenon might be attributed to RAD21’s involvement in DNA damage repair, promoting cancer cell survival and invasive capabilities. This indicates that RAD21 is one of the critical molecules necessary for the continuous growth and progression of HCC. Targeting RAD21 could be a potential therapeutic strategy for treating incompletely ablated residual liver cancer.
To explore the complexity of the upregulated RAD21 in influencing the malignant behavior of residual cancer cells, we conducted an analysis using ChIP-seq to identify potential transcription targets affected by RAD21 at the whole-genome level. By comparing the ChIP-seq candidate targets with the upregulated genes in both HCC and ablated residual HCC, we identified 184 genes shared among the three arrays, suggesting that RAD21 might influence these genes in residual liver cancer. The utilization of ChIP-seq and high-throughput data provided robust support for investigating the transcriptional regulatory mechanisms of RAD21 and its potential role in residual cancer.
The GO analysis and pathway enrichment results indicated that the 184 genes regulated by RAD21 play various roles in cell biology, mediating the progression of residual cancer. These roles include cell localization, organelle assembly, and symbiosis. Factors such as cell localization, organelle assembly, and symbiosis can directly or indirectly affect the malignant behavior of tumors. For instance, tumors may alter the localization of proteins or structures to better adapt to their needs of growth and spreading, as observed with the maspin protein.41 Certain tumors may regulate the formation of ribosomes to facilitate cell transformation and metastasis.42 Tumor cells may form symbiotic relationships with surrounding immune cells to evade immune attacks and promote cell proliferation and metastasis.43 Pathway enrichment analysis indicated that RAD21 and its targets are involved in several HCC-related signaling pathways, including the VEGF signaling pathway, cell adhesion plaques, angiogenesis, and hepatocyte growth factor receptor signaling pathway (HGF receptor signaling pathway). Existing research has confirmed that these pathways play critical roles in cancer progression by promoting cell proliferation, migration, invasion, and angiogenesis. The VEGF signaling pathway is a crucial regulator of angiogenesis, forming new blood vessels to support tumor growth.44 Cell adhesion plaques connect cells to the extracellular matrix (ECM) and are essential for cell adhesion and migration.45 Upregulated RAD21 and its targets may promote angiogenesis and tumor growth by stimulating the production of VEGF and other angiogenic factors. They can also enhance tumor cell migration and invasion by increasing interactions between cancer cells and the ECM.
The hepatocyte growth factor (HGF) receptor signaling pathway, also known as the Met signaling pathway, is another essential factor mediating liver cancer progression.46 HGF is a ligand that binds to the Met receptor, a transmembrane receptor tyrosine kinase. HGF activates the Met receptor, activating downstream signaling pathways, including PI3K/Akt, Ras/MAPK, and STAT pathways.47,48 These pathways ultimately stimulate cancer cell proliferation, leading to tumor enlargement, and can protect cancer cells from apoptosis, allowing them to survive and continue to increase. Activation of the Met receptor can also enhance cancer cell migration and invasion, facilitating the spread of cancer cells to other parts of the body.48
Moreover, the HGF/Met signaling pathway can activate the VEGF signaling pathway, promoting the migration and proliferation of endothelial cells, thereby stimulating the formation of new blood vessels to provide oxygen and nutrients for growing tumors.49 Therefore, upregulated RAD21 in residual cancer promotes the expression of downstream targets involved in multiple HCC-related signaling pathways, facilitating tumor growth and metastasis in residual cancer. Understanding the molecular mechanisms underlying these pathways and their interactions with RAD21 can provide valuable insights for developing novel therapeutic strategies for residual HCC. However, further research is needed to confirm this and to understand the mechanisms involved in RAD21.
However, this study also has some limitations. HCC progression is greatly influenced by tumor microenvironment including immune cell infiltration and its spatial distribution. We used immunodeficient nude mice to construct the incomplete ablation model, which might not capture HCC complexity partly. Additionally, while we have conducted extensive research on the functions of RAD21, the exact mechanisms of RAD21 in residual HCC are still largely unknown and need more investigation. The exploration of RAD21 targets was based on ChIP-seq, which is also a high-throughput method. The validation of RAD21 downstream targets requires more precise experiments such as WB and dual-luciferase experiments.
In summary, we think our research represents a pioneering effort to understand the role of RAD21 as a transcriptional regulator in residual HCC. Since RAD21’s function in residual HCC is not well understood, this research might open a novel insight into cancer biology and provide a potential therapeutic target for residual HCC. More importantly, the relationship between RAD21 levels and HCC clinicopathological parameters indicated by our study may offer predictive insights, helping clinicians better assess prognosis and treatment strategies. Due to difficulties in collecting residual HCC specimens from clinical patients, our research uses subcutaneous xenografts in animal models, enabling detailed molecular analysis through high-throughput sequencing, which also partly reveals the other progressive mechanisms of residual HCC. In the next years, we could try to develop new targeted therapies against residual HCC from these findings, particularly focusing on RAD21. With a deeper understanding of the prognostic role of RAD21, personalized treatment plans based on individual RAD21 expression profiles might become a reality, improving treatment outcomes.
Conclusions
These results indicate that RAD21 is a key molecule in the proliferation, migration, and invasion of liver cancer cells. The upregulation of RAD21 after incomplete ablation may be a significant factor exacerbating the malignant behavior of residual cancer cells. The upregulated RAD21 participates in various biological processes such as cell localization, organelle assembly, and symbiosis by promoting the expression of downstream genes. It also activates the VEGF signaling pathway, cell adhesion plaques, angiogenesis, and the hepatocyte growth factor receptor signaling pathway, further promoting the proliferation, migration, and invasion of tumor cells and worsening the tumor behavior of residual cancer after incomplete ablation.
Ethical Declarations
The research including animal experiment and clinical samples followed the Guide for the Care and Use of Laboratory Animals by the National Research Council committee of the USA and received ethical approval for from the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University.
Acknowledgments
We thank the Laboratory of Guangxi Zhuang Autonomous Region Engineering Research Center for Artificial Intelligence Analysis of Multimodal Tumor Images, Key Laboratory of Ultrasonic Molecular Imaging and Artificial Intelligence, Guangxi Key Laboratory of Early Prevention and Treatment for Regional High-Frequency Tumor/Key Laboratory of Early Prevention and Treatment for Regional High-Frequency Tumor (Guangxi Medical University), and Ministry of Education for their support of this study, as well as thank the language editing of Home for Researchers editorial team (www.home-for-researchers.com).
Funding
This study was funded by the National Natural Science Foundation of China (81960329, 82160350, and 82160336), the Natural Science Foundation of Guangxi (2023GXNSFDA026013 and 2020GXNSFDA238005).
Disclosure
The authors declare no conflicts of interest regarding the publication of this paper.
References
1. Rizzo A, Ricci AD, Brandi G. Trans-Arterial Chemoembolization Plus Systemic Treatments for Hepatocellular Carcinoma: an Update. J Pers Med. 2022;12(11). doi:10.3390/jpm12111788
2. Kamal A, Elmoety AAA, Rostom YAM, Shater MS, Lashen SA. Percutaneous radiofrequency versus microwave ablation for management of hepatocellular carcinoma: a randomized controlled trial. J Gastrointest Oncol. 2019;10(3):562–571. doi:10.21037/jgo.2019.01.34
3. Munoz NM, Dupuis C, Williams M, et al. Molecularly targeted photothermal ablation improves tumor specificity and immune modulation in a rat model of hepatocellular carcinoma. Commun Biol. 2020;3(1):783. doi:10.1038/s42003-020-01522-y
4. Chen J, Peng K, Hu D, et al. Tumor Location Influences Oncologic Outcomes of Hepatocellular Carcinoma Patients Undergoing Radiofrequency Ablation. Cancers (Basel). 2018;10(10). doi:10.3390/cancers10100378
5. Radosevic A, Quesada R, Serlavos C, et al. Microwave versus radiofrequency ablation for the treatment of liver malignancies: a randomized controlled Phase 2 trial. Sci Rep. 2022;12(1):316. doi:10.1038/s41598-021-03802-x
6. Maeda M, Saeki I, Sakaida I, et al. Complications after Radiofrequency Ablation for Hepatocellular Carcinoma: a Multicenter Study Involving 9411 Japanese Patients. Liver Cancer. 2020;9(1):50–62. doi:10.1159/000502744
7. Wang J, Liang P, Yu J, et al. Clinical outcome of ultrasound-guided percutaneous microwave ablation on colorectal liver metastases. Oncol Lett. 2014;8(1):323–326. doi:10.3892/ol.2014.2106
8. Sutter O, Fihri A, Ourabia-Belkacem R, Sellier N, Diallo A, Seror O. Real-Time 3D Virtual Target Fluoroscopic Display for Challenging Hepatocellular Carcinoma Ablations Using Cone Beam CT. Technol Cancer Res Treat. 2018;17:1533033818789634. doi:10.1177/1533033818789634
9. Wang Z, Liu M, Zhang DZ, et al. Microwave ablation versus laparoscopic resection as first-line therapy for solitary 3-5-cm HCC. Hepatology. 2022;76(1):66–77. doi:10.1002/hep.32323
10. Cui R, Wang XH, Ma C, et al. Comparison of Microwave Ablation and Transarterial Chemoembolization for Single-Nodule Hepatocellular Carcinoma Smaller Than 5cm: a Propensity Score Matching Analysis. Cancer Manag Res. 2019;11:10695–10704. doi:10.2147/CMAR.S213581
11. Benson AB, D’Angelica MI, Abbott DE, et al. Hepatobiliary Cancers, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2021;19(5):541–565. doi:10.6004/jnccn.2021.0022
12. Rizzo A, Ricci AD, Brandi G. Systemic adjuvant treatment in hepatocellular carcinoma: tempted to do something rather than nothing. Future Oncol. 2020;16(32):2587–2589. doi:10.2217/fon-2020-0669
13. Qin Z, Xiang C, Zhong F, et al. Transketolase (TKT) activity and nuclear localization promote hepatocellular carcinoma in a metabolic and a non-metabolic manner. J Exp Clin Cancer Res. 2019;38(1):154. doi:10.1186/s13046-019-1131-1
14. Li N, Zheng D, Xue J, et al. Cidan inhibits liver cancer cell growth by reducing COX-2 and VEGF expression and cell cycle arrest. Exp Ther Med. 2015;9(5):1709–1718. doi:10.3892/etm.2015.2351
15. Ren Y, Cao Y, Ma H, et al. Improved clinical outcome using transarterial chemoembolization combined with radiofrequency ablation for patients in Barcelona clinic liver cancer stage A or B hepatocellular carcinoma regardless of tumor size: results of a single-center retrospective case control study. BMC Cancer. 2019;19(1):983. doi:10.1186/s12885-019-6237-5
16. Kong J, Yao C, Ding X, et al. ATPase Inhibitory Factor 1 Promotes Hepatocellular Carcinoma Progression After Insufficient Radiofrequency Ablation, and Attenuates Cell Sensitivity to Sorafenib Therapy. Front Oncol. 2020;10:1080. doi:10.3389/fonc.2020.01080
17. Su T, Huang M, Liao J, et al. Insufficient Radiofrequency Ablation Promotes Hepatocellular Carcinoma Metastasis Through N6-Methyladenosine mRNA Methylation-Dependent Mechanism. Hepatology. 2021;74(3):1339–1356. doi:10.1002/hep.31766
18. Zhang BN, Liu Y, Yang Q, et al. rad21 Is Involved in Corneal Stroma Development by Regulating Neural Crest Migration. Int J Mol Sci. 2020;21(20). doi:10.3390/ijms21207807
19. Mintzas K, Heuser M. Emerging strategies to target the dysfunctional cohesin complex in cancer. Expert Opin Ther Targets. 2019;23(6):525–537. doi:10.1080/14728222.2019.1609943
20. Xu H, Yan M, Patra J, et al. Enhanced RAD21 cohesin expression confers poor prognosis and resistance to chemotherapy in high grade luminal, basal and HER2 breast cancers. Breast Cancer Res. 2011;13(1):R9. doi:10.1186/bcr2814
21. Yamamoto G, Irie T, Aida T, Nagoshi Y, Tsuchiya R, Tachikawa T. Correlation of invasion and metastasis of cancer cells, and expression of the RAD21 gene in oral squamous cell carcinoma. Virchows Arch. 2006;448(4):435–441. doi:10.1007/s00428-005-0132-y
22. Deb S, Xu H, Tuynman J, et al. RAD21 cohesin overexpression is a prognostic and predictive marker exacerbating poor prognosis in KRAS mutant colorectal carcinomas. Br J Cancer. 2014;110(6):1606–1613. doi:10.1038/bjc.2014.31
23. Wang J, Zhao H, Yu J, et al. MiR-320b/RAD21 axis affects hepatocellular carcinoma radiosensitivity to ionizing radiation treatment through DNA damage repair signaling. Cancer Sci. 2021;112(2):575–588. doi:10.1111/cas.14751
24. Santoni M, Rizzo A, Mollica V, et al. The impact of gender on The efficacy of immune checkpoint inhibitors in cancer patients: the MOUSEION-01 study. Crit Rev Oncol Hematol. 2022;170:103596. doi:10.1016/j.critrevonc.2022.103596
25. Mollica V, Rizzo A, Marchetti A, et al. The impact of ECOG performance status on efficacy of immunotherapy and immune-based combinations in cancer patients: the MOUSEION-06 study. Clin Exp Med. 2023. doi:10.1007/s10238-023-01159-1
26. Liu CZ, Guo WP, Peng JB, et al. Clinical significance of CCNE2 protein and mRNA expression in thyroid cancer tissues. Adv Med Sci. 2020;65(2):442–456. doi:10.1016/j.advms.2020.09.001
27. Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. doi:10.1038/s41591-018-0136-1
28. Yang Y, Ren P, Liu X, et al. PPP1R26 drives hepatocellular carcinoma progression by controlling glycolysis and epithelial-mesenchymal transition. J Exp Clin Cancer Res. 2022;41(1):101. doi:10.1186/s13046-022-02302-8
29. Hu C, Xin Z, Sun X, et al. Activation of ACLY by SEC63 deploys metabolic reprogramming to facilitate hepatocellular carcinoma metastasis upon endoplasmic reticulum stress. J Exp Clin Cancer Res. 2023;42(1):108. doi:10.1186/s13046-023-02656-7
30. Lambert SA, Jolma A, Campitelli LF, et al. The Human Transcription Factors. Cell. 2018;172(4):650–665. doi:10.1016/j.cell.2018.01.029
31. Alerasool N, Leng H, Lin ZY, Gingras AC, Taipale M. Identification and functional characterization of transcriptional activators in human cells. Mol Cell. 2022;82(3):677–695 e7. doi:10.1016/j.molcel.2021.12.008
32. Haberle V, Arnold CD, Pagani M, Rath M, Schernhuber K, Stark A. Transcriptional cofactors display specificity for distinct types of core promoters. Nature. 2019;570(7759):122–126. doi:10.1038/s41586-019-1210-7
33. Song J, Xie C, Jiang L, et al. Transcription factor AP-4 promotes tumorigenic capability and activates the Wnt/beta-catenin pathway in hepatocellular carcinoma. Theranostics. 2018;8(13):3571–3583. doi:10.7150/thno.25194
34. An T, Dong T, Zhou H, et al. The transcription factor Kruppel-like factor 5 promotes cell growth and metastasis via activating PI3K/AKT/Snail signaling in hepatocellular carcinoma. Biochem Biophys Res Commun. 2019;508(1):159–168. doi:10.1016/j.bbrc.2018.11.084
35. Zhang L, Wang K, Deng Q, Li W, Zhang X, Liu X. Identification of Key Hydroxymethylated Genes and Transcription Factors Associated with Alpha-Fetoprotein-Negative Hepatocellular Carcinoma. DNA Cell Biol. 2019;38(11):1346–1356. doi:10.1089/dna.2019.4689
36. Ji X, Chen X, Zhang B, et al. T-box transcription factor 19 promotes hepatocellular carcinoma metastasis through upregulating EGFR and RAC1. Oncogene. 2022;41(15):2225–2238. doi:10.1038/s41388-022-02249-2
37. Cheng H, Zhang N, Pati D. Cohesin subunit RAD21: from biology to disease. Gene. 2020;758:144966. doi:10.1016/j.gene.2020.144966
38. Cui R, Chen P, Wang Y, et al. Cohesin RAD21 Gene Promoter Methylation Correlated with Better Prognosis in Breast Cancer Patients. Cytogenet Genome Res. 2022;162(3):109–118. doi:10.1159/000524735
39. Atienza JM, Roth RB, Rosette C, et al. Suppression of RAD21 gene expression decreases cell growth and enhances cytotoxicity of etoposide and bleomycin in human breast cancer cells. Mol Cancer Ther. 2005;4(3):361–368. doi:10.1158/1535-7163.MCT-04-0241
40. Deng P, Wang Z, Chen J, et al. RAD21 amplification epigenetically suppresses interferon signaling to promote immune evasion in ovarian cancer. J Clin Invest. 2022;132(22). doi:10.1172/JCI159628
41. Sakabe T, Wakahara M, Shiota G, Umekita Y. Role of cytoplasmic localization of maspin in promoting cell invasion in breast cancer with aggressive phenotype. Sci Rep. 2021;11(1):11321. doi:10.1038/s41598-021-90887-z
42. Harold CM, Buhagiar AF, Cheng Y, Baserga SJ. Ribosomal RNA Transcription Regulation in Breast Cancer. Genes. 2021;12(4). doi:10.3390/genes12040502
43. Zhu J, Wang Y, Li D, Zhang H, Guo Z, Yang X. Interleukin-35 promotes progression of prostate cancer and inhibits anti-tumour immunity. Cancer Cell Int. 2020;20:487. doi:10.1186/s12935-020-01583-3
44. Morse MA, Sun W, Kim R, et al. The Role of Angiogenesis in Hepatocellular Carcinoma. Clin Cancer Res. 2019;25(3):912–920. doi:10.1158/1078-0432.CCR-18-1254
45. Shang N, Wang H, Bank T, et al. Focal Adhesion Kinase and beta-Catenin Cooperate to Induce Hepatocellular Carcinoma. Hepatology. 2019;70(5):1631–1645. doi:10.1002/hep.30707
46. Zhao Y, Ye W, Wang YD, Chen WD. HGF/c-Met: a Key Promoter in Liver Regeneration. Front Pharmacol. 2022;13:808855. doi:10.3389/fphar.2022.808855
47. Han T, Zheng H, Zhang J, et al. Downregulation of MUC15 by miR-183-5p.1 promotes liver tumor-initiating cells properties and tumorigenesis via regulating c-MET/PI3K/AKT/SOX2 axis. Cell Death Dis. 2022;13(3):200. doi:10.1038/s41419-022-04652-9
48. Yu J, Xia X, Dong Y, et al. CYP1A2 suppresses hepatocellular carcinoma through antagonizing HGF/MET signaling. Theranostics. 2021;11(5):2123–2136. doi:10.7150/thno.49368
49. Vimalraj S. A concise review of VEGF, PDGF, FGF, Notch, angiopoietin, and HGF signalling in tumor angiogenesis with a focus on alternative approaches and future directions. Int J Biol Macromol. 2022;221:1428–1438. doi:10.1016/j.ijbiomac.2022.09.129
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.









