Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 16
PSMC2 is a Novel Prognostic Biomarker and Predicts Immunotherapeutic Responses: From Pancreatic Cancer to Pan-Cancer
Authors Huang W, Qian Z, Shi Y, Zhang Z , Hou R, Mei J, Xu J , Ding J
Received 10 May 2023
Accepted for publication 1 August 2023
Published 9 August 2023 Volume 2023:16 Pages 747—758
DOI https://doi.org/10.2147/PGPM.S418533
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Wei Huang,1,* Zhengtao Qian,2,* Yuxin Shi,1,3,* Zheming Zhang,3 Rui Hou,1,3 Jie Mei,1,3 Junying Xu,1 Junli Ding1
1Department of Oncology, the Affiliated Wuxi People’s Hospital of Nanjing Medical University, Wuxi, 214023, People’s Republic of China; 2Department of Clinical Laboratory, Changshu Medicine Examination Institute, Changshu, 215500, People’s Republic of China; 3Wuxi School of Clinical Medicine, Nanjing Medical University, Wuxi, 214023, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Junli Ding; Junying Xu, Email [email protected]; [email protected]; [email protected]
Background: Proteasome 26S subunit ATPase 2 (PSMC2) is a part of the 19S regulatory complex, which catalyzes the unfolding and transport of substrates into the 20S proteasome. Our previous research demonstrated that PSMC2 participates in the tumorigenesis and progression of pancreatic cancer (PC). However, no systematic analysis has been conducted to conclude its expression pattern and correlation with tumor immunity.
Aim: To investigate the expression level of PSMC2 in PC, its prognostic value and its relationship with tumor immunity.
Methods: In numerous public and internal cohorts, the expression, prognostic significance, and immunological connections of PSMC2 in PC were investigated. Additionally, using data from The Cancer Genome Atlas (TCGA), a pan-cancer analysis was carried out to examine PSMC2’s immunological assocaition, and the predictive power of PSMC2 for immunotherapy was also evaluated in numerous public cohorts.
Results: PSMC2 was overexpressed in tumor tissues and linked to unfavorable prognosis in PC. PSMC2 was not only positively correlated with TIICs, also positively correlated with immune checkpoints in PC. In addition to PC, PSMC2 was expected to be an indicator of high immunogenicity in most cancer types. Importantly, PSMC2 could predict the immunotherapeutic responses in various cancer types, including urothelial carcinoma and breast cancer.
Conclusion: From PC to pan-cancer analysis, we report that PSMC2 is a novel prognostic biomarker in multiple cancer types. PSMC2 is related to the immuno-hot phenotype and predicts the outcome of immunotherapy. Therefore, the current study emphasizes that cancer patients with high PMSC2 expression should actively receive immunotherapy to improve their prognosis.
Keywords: PSMC2, tumor immunity, pancreatic cancer, biomarker, bioinformatics
Introduction
Pancreatic cancer (PC) is a common malignancy with poor prognosis, with the 5-year overall survival rate of about 10%.1 However, the majority of individuals with PC are determined to suffer from locally advanced or metastatic diseases.2 The 5-year survival rate (after resection) remains no more than 20%, and is only possible in 15%-20% of the diagnosed patients.3 Recently, immunotherapy, represented by the blocking antibodies against programmed cell death 1 (PD-1) or its ligand 1 (PD-L1), has become a viable hotspot treatment approach for PC.4 In fact, in addition to tumor cells, tumor tissue also contains immune cells, stromal cells, vascular networks, and several other cellular and non-cellular elements that help to create the tumor microenvironment (TME).5 Tumors could be classified as hot or cool tumors depending on the TME’s features. T cell infiltration and molecular signs of immune activation are markers of hot malignancies, whereas T cell exclusion or absence is conspicuously seen in cold tumors.6 Moreover, the TME contains a wide variety of potential targets for cancer treatment thanks to its several components. Hence, the identification of populations that can benefit from the immunotherapy necessitates the urgent need for possible biomarkers that could be utilized to determine tumor immunogenicity.
26S proteasome, which is functioned as the executor of the ubiqutin-proteasome, plays the key role in protein degradation in the eukaryotic cells. A single- or double-capped (26S1 or 26S2) entire 26S proteasome is created when one or two 19S regulatory complexes unite with a single 20S catalytic complex, accordingly.7 Proteasome 26S subunit ATPase 2 (PSMC2), an important component of the 19S proteasome is encoded by a recently discovered gene on chromosome 7q22.1-q22.3.8 Several studies demonstrate that PSMC2 is crucial for controlling intracellular proteins in various human cancers, such as osteosarcoma,9 gastric cancer,7 PC.10 In our previous research, we discovered that PSMC2 was involved in the development of PC and was being considered as a possible therapeutic target.10 The potential relationship between PSMC2, the 26S proteasome’s regulatory complex, and immunological characteristics of PC has not yet been investigated.
The expression of PSMC2 and its immunological function in PC were first examined in the current investigation. PSMC2 was found to be increased in PC and to distinguish immuno-hot tumors, according to the current research. In addition, PSMC2 was also correlated with a majority of immunosuppressive molecules. Moreover, PSMC2 demonstrated close relationships with immunological variables in the majority of malignancies, and patients with high PSMC2 expression showed well immunotherapeutic response. Overall, PSMC2 was a promising biomarker for identifying high immunogenicity in not only PC and multiple solid cancer types.
Materials and Methods
Public Data Acquisition
The PC and pan-cancer normalized gene expression profiles, and clinical annotations of The Cancer Genome Atlas (TCGA) datasets were obtained from the online data portal UCSC Xena (https://xenabrowser.net/datapages/). Table S1 provides the abbreviations for various cancer types. Two PC datasets were downloaded to compare the PSMC2 expression in normal and tumor tissues, including GSE2873511 and GSE6245212 datasets. Two public datasets comprising RNA-seq data from patients receiving immunotherapy were downloaded, including IMvigor21013 and MEDI473614 datasets.
Enrichment Analysis of PSMC2 in PC
To identify PSMC2-related functions in PC, we first extracted correlated genes with PSMC2 in the TCGA dataset using the LinkedOmics tool.15 Then, for the enrichment analysis, the top 50 positively and negatively linked genes were submitted. Briefly, the most recent gene annotation for the KEGG pathway was retrieved and utilized as background (https://www.kegg.jp/kegg/rest/keggapi.html). Then, the KEGG analysis results of gene set enrichment were obtained using the R package clusterprofiler (version 3.14.3), which was used to execute the enrichment study. The minimum gene set was set to 5, the maximum gene was set to 5000 and one thousand re-samplings were set. P value < 0.05 was considered statistically significant.
TIMER Database Analysis
TIMER is a comprehensive resource from TCGA for the systematic investigation of immune infiltrates in pan-cancer (https://cistrome.shinyapps.io/timer/).16 Using gene expression profiles, TIMER uses a deconvolution method to estimate the quantity of tumor-infiltrating immune cells (TIICs).17 We analyzed correlations of PSMC2 expression with the abundance of immune infiltrates, including B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells (DCs). In addition, correlations between PSMC2 expression and immune checkpoints as well as markers of various subsets of immune cells were also assessed by the TIMER tool.
Pan-Cancer Analysis Using Sangerbox
The Sangerbox tool is a comprehensive and user-friendly clinical bioinformatics analysis platform.18 In this research, we examined PSMC2 expression and predictive values in pan-cancer using the Sangerbox tool. In addition, the Sangerbox tool was also used to assess correlations between PSMC2 expression and immunomodulators as well as TIICs in pan-cancer.
Clinical Samples
Outdo BioTech (Shanghai, China) provided the PC tissue microarray (TMA, Cat. HPanA120Su02). A total of 66 PaCa tissues and 54 pairs of neighboring normal tissues made up the HPanA120Su02 TMA. The Clinical Research Ethics Committee, Outdo BioTech, approved the research of the TMAs (No. YB-M-05-02) and Outdo BioTech provided detailed clinico-pathological cohort characteristics as well.
Immunohistochemistry and Semi-Quantitative Scoring
Using conventional techniques, immunohistochemistry (IHC) staining was done directly on the HPanA120Su02 TMA. PSMC2 was detected using the primary antibody for PSMC2 (1:200 dilution, Cat. 14905-1-AP, ProteinTech, Wuhan, China), while PD-L1 was detected using the primary antibody for PD-L1 (Ready-to-use, Cat. GT2280, GeneTech, Shanghai, China). DAB and hematoxylin counterstain were used to visualize the antibody staining, and Aperio Digital Pathology Slide Scanners were utilized to scan the stained sections. Two pathologists independently evaluated the stained TMA. Then, PSMC2 and PD-L1 staining were semi-quantitatively evaluated using the H-score criterion.19
Single-Cell RNA-Seq (scRNA-Seq) Analysis
Tumor Immune Single-cell Hub (TISCH, http://tisch.comp-genomics.org/gallery/) is a scRNA-seq database focusing on TME and provides detailed cell-type annotation at the single-cell level.20 We used TISCH to analyze the cell subpopulation patterns of PSMC2 in the GSE162708 dataset. Default options were used for all parameters.
Statistical Analysis
R language 4.0.2 and Graphpad Prism 6.0 were used for statistical analysis and figure display. Depending on the circumstances, either the Student t test or the Mann–Whitney test was used to assess the statistical difference between the two groups for continuous variables. To assess the correlation between two variables under the relevant circumstances, the Pearson and Spearman correlation test was performed. Prognostic values of categorical variables were investigated by Log rank test. P value < 0.05 was regarded as statistically significant for all analyses.
Results
Expression and Prognostic Value of PSMC2 in PC
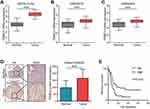
We first compared the expression of PSMC2 in normal and tumor tissues in PC. A total of three independent datasets were used. Notably, PSMC2 was highly expression in tumor tissues compared with normal tissues in GEPIA, GSE28735, and GSE62452 datasets (Figure 1A-C). To further validate the above results, a TMA of PC tissues was submitted for IHC staining. The majority of PSMC2’s immunoreactivity was localized to the cytoplasm, as shown in Figure 1D. Semi-quantitative analysis revealed that, in comparison to paired normal tissues, the IRS of PSMC2 in PC tissues was considerably higher (Figure 1D). Also, we discovered that PSMC2 high expression was significantly associated with poor differentiation and unfavorable clinical outcome (Table S2). Moreover, patients with higher PSMC2 expression had worse prognosis in term of overall survival (OS) than samples with lower PSMC2 expression (Figure 1E). Overall, we concluded that PSMC2 was overexpressed and related to poor clinical outcomes in PC.
Enrichment Analysis of PSMC2 in PC
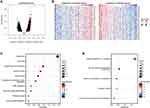
Next, the Linked Omics was utilized to look for the genes co-expressed with PSMC2 in the TCGA cohort in order to gain complete insights into the biological roles of PSMC2 in PC. The genes that were significantly linked to PSMC2 were shown in Figure 2A. The heat map displayed the top 50 significant genes positively and negatively related to PSMC2 (Figure 2B). In the term of KEGG analysis, the positively correlated genes were enriched in proteasome, cell cycle, and Epstein-Barr virus infection, and so on. The negatively positively correlated genes were enriched in Herpes simplex virus 1 infection, and metabolic pathways, such as glycine, serine and threonine metabolism and taurine and hypotaurine metabolism (Figure 2C and D). Taken together, PSMC2 was associated with multiple pathways, which may shed some insights into the molecular function of PSMC2 in PC
PSMC2 Was Related to an Inflamed TME in PC
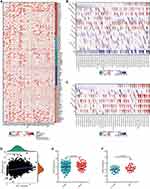
In light of the fact that PSMC2 was linked to Epstein-Barr virus infection, we next looked into PSMC2’s potential immunological function in PC. According to protocol to identify novel immunotherapy biomarker,21 we initially checked the expression pattern of PSMC2 expression in PC and found that PSMC2 was high expressed in tumor cells (Figure S1A-D). Then, the TIMER algorithm determined that PSMC2 expression was positively correlated with the infiltration levels of the majority of immune cells (Figure 3A). Next, we investigated the correlation between PSMC2 expression and subsets of TIICs based on the levels of immune marker expression in PC tissues using the TIMER tool. The correlation analysis was adjusted for purity since tumor purity in clinical samples affects the measurement of immune infiltration.22 The expression of marker genes from various TIICs was substantially linked with PSMC2 expression in PC tissues, including CD8+ T cells, general T cells, B cells, monocyte, M1/M2 macrophage, TAMs, neutrophils, NK cells, DCs, Th1 cells, Th2 cells, Tfh cells, Th17 cells, Treg cells, T cell exhaustion, according to analysis of the TIMER database (Table 1).
 |
Table 1 Correlation Analysis Between PSMC2 and Immune Cells Markers |
However, PSMC2 was also positively related to multiple immune checkpoints expression, including PD-L1, PD-L2, LAG3, CTLA4, PVR, and HAVCR2 (Figure 3B). Moreover, the positive correlation between PSMC2 and PD-L1 expression was also validated in the in-house cohort (Figure 3C and D). All the above results revealed that PSMC2, although was positively correlated with TIICs, also positively correlated with immune checkpoints, which perhaps explained the unfavorable clinical outcome of patients with high PSMC2 expression and the necessity of using immune checkpoint inhibitors in these patients.
Analysis of Expressions, Prognostic Values, and Immunological Correlations of PSMC2 in Pan-Cancer
All evidence revealed that PSMC2 was a significant functional gene in PC, thus we aimed to analyze its potential role in pan-cancer. As shown in Figure 4A, PSMC2 was significantly upregulated in almost all cancer types. In addition, PSMC2 was associated with poor OS and progression free survival (PFS) in all cancer patients from the TCGA database (Figure 4B and C). Analysis that was specific to various cancer types indicated PSMC2 was associated poor prognosis in LGG, GBM, UVM, and KICH (Figure 4D and E). Totally, these results showed that PSMC2 was a potential prognostic biomarker in multiple cancer types.
In addition, it was also unknown the immuno-correlation of PSMC2 in pan-cancer. Thus, the relationships between PSMC2 and chemokines, receptors, MHC, immunoinhibitors, and immunostimulators were then examined. PSMC2 was favorably linked with the expression levels of various immunomodulators, with the exception of a few cancer types. (Figure 5A). PSMC2 was positively related to TIICs levels in most cancer types (Figure 5B and C). Furthermore, PSMC2 was positively related to PD-L1 expression in pan-cancer (Figure 5D). Moreover, PSMC2 was highly expressed in tumors with well immunotherapeutic response in IMvigor210 (urothelial carcinoma) and MEDI4736 (breast cancer) cohorts (Figure 5E and F), and the the predictive values of PSMC2 and PD-L1 mRNA expression were similar (Figure S2A-B). Overall, the data suggested that PSMC2 was a pan-cancer biomarker for prognosis and immunotherapy except for a few tumor types.
Discussion
In eukaryotic cells, the ubiquitin-proteasome system (UPS) is a key mechanism for selective protein degradation.23 There are two key steps in the UPS. Ubiquitination, also known as the covalent attachment of ubiquitin(s) to a protein substrate, is the first step, and 26S proteasome degradation is the second.23 In eukaryotic cells, the 26S proteasome is the predominant nonlysosomal protease and is in charge of degrading 70% to 90% of all long-lived proteins in addition to all short-lived proteins. It follows that it is not surprising that the 26S proteasome is essential for a variety of biological processes, and that it is particularly involved in numerous regulatory pathways, such as cell proliferation and death. In fact, it has been suggested that the 26S proteasome could be a target for human cancer therapy.
The UPS is crucial for cell immunity, particularly in myeloid cells that control essential aspects of innate and adaptive immunity.23 Moreover, ubiquitylation and substrate degradation through E3-ligases/UPS govern the networks of the innate and adaptive immune systems. Inhibitors of apoptosis proteins (IAPs), which work downstream of TLR, TNFR, and NOD2, are also known to control NF-B/MAPK signaling through substrate ubiquitylation.24 Moreover, UPS controls the immune system’s adaptive cells. According to a recent discovery, the E3 ubiquitin ligase Cul5 encourages CD4+ T cells to become Treg by degrading Phospho Janus Kinase 1.25 However, it has not been determined if these proteins are connected to TME and antitumor immunity.
PSMC2 is a component of the 26S proteasome’s 19S regulatory complex, and its expression is necessary for the assembly of both the 19S and 26S proteasomes. The top CYCLOPS gene, PSMC2, was identified by Nijhawan et al as a distinct class of cancer-specific vulnerabilities brought on by genomic instability.26 The study found that among 3131 tumors representing a wide range of cancer types, the probability of partial genomic deletion of PSMC2 was 0.10, resulting in a significant dependence of cancer cells on the remaining PSMC2.26 This finding further showed that PSMC2 might be a useful target for cancer therapy.
In our previous research, we focused on the biological role of PSMC2 in pancreatic cancer and found that knockdown of PSMC2 in pancreatic cancer cells inhibited proliferation and enhanced apoptosis, suggesting that PSMC2 might be involved in the progression of pancreatic cancer and may serve as a potential therapeutic target.10 Using large-scale RNA-seq data and the verified cohort, we examined PSMC2 expression and prognostic significance in PC in this work. The findings suggested that PSMC2 may play a predictive function in PC. Another significant finding was the correlation between the levels of immunological checkpoints and TIICs in PC and the expression of PSMC2. All of the above results may contribute to the poor prognosis of patients with high PSMC2 expression and the requirement for the use of immune checkpoint inhibitors in these patients.
Last but not least, we performed a pan-cancer analysis, and the results exhibited that PSMC2 was remarkably upregulated in almost all cancer types and associated with unfavorable prognosis in total cancer patients from the TCGA dataset. All results suggested that PSMC2 is an oncogene in multiple cancers. Interestingly, the oncogenic role of PSMC2 has been confirmed in ovarian cancer,27 breast cancer,28 glioma,29 gastric cancer,7 and skin cutaneous melanoma.30 In addition, PSMC2 was positively related to most immunomodulators and TIICs in most cancer types. Moreover, despite the unfavorable prognosis of patients with high PSMC2 expression, the better efficacy of immune checkpoint inhibitors in such patients, indicating the need for immune checkpoint inhibitors to improve the prognosis. Although the predictive values of PSMC2 and PD-L1 mRNA expression were similar, the presence of glycosylation modification of PD-L1 can hinder the antibody detection rate of PD-L1.31,32 PSMC2 is a cytosolic protein without glycosylation modification, so PSMC2 may be an important supplementary detection biomarker for immunotherapy. Overall, PSMC2 was a pan-cancer biomarker for prognosis and immunotherapy except for a few tumor types.
Conclusion
Our study reports that PSMC2 is overexpressed and associated with the inflamed TME in PC, which may affect the immunotherapeutic responses. Additionally, PSMC2 showed tight correlations with immunological factors in most cancers. Overall, PSMC2 might be a potential biomarker for assessing patients’ prognosis, tumor immunogenicity, and guiding immunotherapy in multiple cancers.
Abbreviation
PC, Pancreatic cancer; PD-1, programmed cell death 1; TME, tumor microenvironment; PSMC2, Proteasome 26S subunit ATPase 2; TCGA, The Cancer Genome Atlas; TIIC, tumor-infiltrating immune cell; IHC, immunohistochemistry; OS, overall survival; PFS, progression free survival; UPS, ubiquitin-proteasome system.
Data Sharing Statement
All data human data used in study is publicly available. Data and materials can be provided upon reasonable request to the corresponding authors.
Ethics Approval and Consent to Participate
Ethical approval (No. YB-M-05-02) for the use of TMA was granted by the Clinical Research Ethics Committee in Outdo Biotech (Shanghai, China).
Author Contributions
All listed authors must have made a significant scientific contribution to the research in the manuscript approved its claims and agreed to be an author. All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was supported by the Wuxi Municipal Health Commission Foundation (No. M202108), Beijing Hengji Health Management and Development Foundation (HJ-HX-ZLXD-202209-002) and the Wuxi Science and Technology Bureau Foundation (No. N20202018).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Park W, Chawla A, O’Reilly EM. Pancreatic cancer: a review. JAMA. 2021;326(9):851–862. doi:10.1001/jama.2021.13027
2. Yeo D, Giardina C, Saxena P, Rasko JEJ. The next wave of cellular immunotherapies in pancreatic cancer. Mol Ther Oncolytics. 2022;24:561–576. doi:10.1016/j.omto.2022.01.010
3. Eibl G, Rozengurt E. Metformin: review of epidemiology and mechanisms of action in pancreatic cancer. Cancer Metastasis Rev. 2021;40(3):865–878. doi:10.1007/s10555-021-09977-z
4. Wang X, Li X, Wei X, et al. PD-L1 is a direct target of cancer-FOXP3 in pancreatic ductal adenocarcinoma (PDAC), and combined immunotherapy with antibodies against PD-L1 and CCL5 is effective in the treatment of PDAC. Signal Transduct Target Ther. 2020;5(1):38. doi:10.1038/s41392-020-0144-8
5. Duan Q, Zhang H, Zheng J, Zhang L. Turning cold into hot: firing up the tumor microenvironment. Trends Cancer. 2020;6(7):605–618. doi:10.1016/j.trecan.2020.02.022
6. Cai Y, Ji W, Sun C, et al. Interferon-induced transmembrane protein 3 shapes an inflamed tumor microenvironment and identifies immuno-hot tumors. Front Immunol. 2021;12:704965. doi:10.3389/fimmu.2021.704965
7. Liu T, Zhang J, Chen H, et al. PSMC2 promotes the progression of gastric cancer via induction of RPS15A/mTOR pathway. Oncogenesis. 2022;11(1):12. doi:10.1038/s41389-022-00386-7
8. Smith DM, Fraga H, Reis C, Kafri G, Goldberg AL. ATP binds to proteasomal ATPases in pairs with distinct functional effects, implying an ordered reaction cycle. Cell. 2011;144(4):526–538.
9. Song M, Wang Y, Zhang Z, Wang S. PSMC2 is up-regulated in osteosarcoma and regulates osteosarcoma cell proliferation, apoptosis and migration. Oncotarget. 2017;8(1):933–953. doi:10.18632/oncotarget.13511
10. Qin J, Wang W, An F, Huang W, Ding J. PSMC2 is up-regulated in pancreatic cancer and promotes cancer cell proliferation and inhibits apoptosis. J Cancer. 2019;10(20):4939–4946. doi:10.7150/jca.27616
11. Zhang G, Schetter A, He P, et al. DPEP1 inhibits tumor cell invasiveness, enhances chemosensitivity and predicts clinical outcome in pancreatic ductal adenocarcinoma. PLoS One. 2012;7(2):e31507. doi:10.1371/journal.pone.0031507
12. Yang S, He P, Wang J, et al. A Novel MIF Signaling Pathway Drives the Malignant Character of Pancreatic Cancer by Targeting NR3C2. Cancer Res. 2016;76(13):3838–3850. doi:10.1158/0008-5472.CAN-15-2841
13. Necchi A, Joseph RW, Loriot Y, et al. Atezolizumab in platinum-treated locally advanced or metastatic urothelial carcinoma: post-progression outcomes from the Phase II IMvigor210 study. Ann Oncol. 2017;28(12):3044–3050. doi:10.1093/annonc/mdx518
14. Blenman KRM, Marczyk M, Karn T, et al. Predictive Markers of Response to Neoadjuvant Durvalumab with Nab-Paclitaxel and Dose-Dense Doxorubicin/Cyclophosphamide in Basal-Like Triple-Negative Breast Cancer. Clin Cancer Res. 2022;28(12):2587–2597. doi:10.1158/1078-0432.CCR-21-3215
15. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–D63. doi:10.1093/nar/gkx1090
16. Li T, Fan J, Wang B, et al. TIMER: a Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017;77(21):e108–e10. doi:10.1158/0008-5472.CAN-17-0307
17. Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi:10.1186/s13059-016-1028-7
18. Shen W, Song Z, Zhong X, et al. Sangerbox: a comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta. 2022;1(3):e36. doi:10.1002/imt2.36
19. Mei J, Fu Z, Cai Y, et al. SECTM1 is upregulated in immuno-hot tumors and predicts immunotherapeutic efficacy in multiple cancers. iScience. 2023;26(2):106027. doi:10.1016/j.isci.2023.106027
20. Sun D, Wang J, Han Y, et al. TISCH: a comprehensive web resource enabling interactive single-cell transcriptome visualization of tumor microenvironment. Nucleic Acids Res. 2021;49(D1):D1420–D30. doi:10.1093/nar/gkaa1020
21. Mei J, Cai Y, Xu R, et al. Protocol to identify novel immunotherapy biomarkers based on transcriptomic data in human cancers. STAR Protocols. 2023;4(2):102258. doi:10.1016/j.xpro.2023.102258
22. Mei J, Wang R, Xia D, et al. BRCA1 Is a Novel Prognostic Indicator and Associates with Immune Cell Infiltration in Hepatocellular Carcinoma. DNA Cell Biol. 2020;39(10):1838–1849. doi:10.1089/dna.2020.5644
23. Cetin G, Klafack S, Studencka-Turski M, Kruger E, Ebstein F. The Ubiquitin-Proteasome System in Immune Cells. Biomolecules. 2021;11(1):60. doi:10.3390/biom11010060
24. Lalaoui N, Vaux DL. Recent advances in understanding inhibitor of apoptosis proteins. F1000Res. 2018;7:1889. doi:10.12688/f1000research.16439.1
25. Kumar B, Field NS, Kim DD, et al. The ubiquitin ligase Cul5 regulates CD4(+) T cell fate choice and allergic inflammation. Nat Commun. 2022;13(1):2786. doi:10.1038/s41467-022-30437-x
26. Nijhawan D, Zack TI, Ren Y, et al. Cancer vulnerabilities unveiled by genomic loss. Cell. 2012;150(4):842–854. doi:10.1016/j.cell.2012.07.023
27. Zhu D, Huang J, Liu N, Li W, Yan L. PSMC2/CCND1 axis promotes development of ovarian cancer through regulating cell growth, apoptosis and migration. Cell Death Dis. 2021;12(8):730. doi:10.1038/s41419-021-03981-5
28. Wang Y, Zhu M, Li J, et al. Overexpression of PSMC2 promotes the tumorigenesis and development of human breast cancer via regulating plasminogen activator urokinase (PLAU). Cell Death Dis. 2021;12(7):690. doi:10.1038/s41419-021-03960-w
29. Zheng X, Wang Y, Wang D, et al. PSMC2 is overexpressed in glioma and promotes proliferation and anti-apoptosis of glioma cells. World J Surg Oncol. 2022;20(1):84. doi:10.1186/s12957-022-02533-1
30. Yang Y, Qi F, Wei C, et al. PSMC2 knockdown suppressed tumor progression of skin cutaneous melanoma. Cell Death Discov. 2021;7(1):323. doi:10.1038/s41420-021-00727-2
31. Lee HH, Wang YN, Xia W, et al. Removal of N-Linked Glycosylation Enhances PD-L1 Detection and Predicts Anti-PD-1/PD-L1 Therapeutic Efficacy. Cancer Cell. 2019;36(2):168–78 e4. doi:10.1016/j.ccell.2019.06.008
32. Mei J, Xu J, Yang X, et al. A comparability study of natural and deglycosylated PD-L1 levels in lung cancer: evidence from immunohistochemical analysis. Mol Cancer. 2021;20(1):11. doi:10.1186/s12943-020-01304-4
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.