Back to Journals » International Journal of General Medicine » Volume 15
Proviral Integration of Moloney Virus-2 (PIM-2) Expression Level as a Prognostic Marker in Patients with Acute Myeloid Leukemia
Authors Hamed G , Omar HM, Sarhan AM, Salah HE
Received 21 December 2021
Accepted for publication 22 March 2022
Published 20 April 2022 Volume 2022:15 Pages 4247—4258
DOI https://doi.org/10.2147/IJGM.S354092
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Gehad Hamed,1 Hisham M Omar,1 Abbas M Sarhan,2 Hossam E Salah1
1Department of Clinical Pathology, Faculty of Medicine, Zagazig University, Zagazig, Al-Sharkia, Egypt; 2Department of Clinical Oncology and Nuclear Medicine, Faculty of Medicine, Zagazig University, Zagazig, Al-Sharkia, Egypt
Correspondence: Gehad Hamed, Department of Clinical Pathology, Faculty of Medicine, Zagazig University, Zagazig, Al-Sharkia, 44519, Egypt, Tel +201092034529, Email [email protected]
Purpose: This study aimed to assess PIM-2 gene expression level as a prognostic marker in AML patients and to correlate the results with their clinical outcome.
Patients and Methods: This study was conducted on 50 de novo younger AML patients (median age 44). Quantitative real-time polymerase chain reaction (QRT-PCR) was used to assess the expression level of the PIM-2 gene. The transcription level of the target gene (PIM-2) was normalized to that of the reference gene (GAPDH). Twenty control samples were withdrawn from 20 age- and sex-matched individuals for the analysis of the results using the 2−ΔΔCT method. On day 28 following induction chemotherapy, patients’ bone marrow (BM) was examined for evaluation of their remission status.
Results: PIM-2 gene expression was higher among AML patients who did not achieve complete remission (CR); also, it was higher in patients in the intermediate and poor cytogenetic risk groups. A significant positive correlation was found between PIM-2 level and BM blasts on day 28. In AML patients, PIM-2 has been discovered to be an independent predictive factor for achieving CR following standard induction treatment. Receiver operating characteristic curve (ROC) and area under the curve (AUC) were performed for PIM-2 level at diagnosis to evaluate its role in achieving remission after induction. It was found that PIM-2 at cutoff ≤ 1.6 had an AUC (0.903) with a sensitivity (90.48%) and specificity (86.21%), P < 0.001.
Conclusion: Overexpression of the PIM-2 gene is associated with induction failure and low CR.
Keywords: AML, PIM-2 gene, QRT-PCR, induction chemotherapy, complete remission
Introduction
Acute myeloid leukemia (AML) is the most aggressive myeloid neoplasm triggered by the acquisition of genetic alterations that lead to the transformation of hematopoietic progenitor cells.1,2 In Egypt, leukemia comprises 10% of all malignancies, with AML representing 16.9% of the cases.3 Despite many AML patients responding to induction chemotherapy, the refractory disease is frequent, and relapse represents the principal cause of treatment failure, so identifying the precise clinical outcome indicators can aid in developing a tailored treatment for particular patients.4 One of the most important phenomena that leads to malignant cells’ accumulation in leukemia patients is apoptosis inhibition, and such finding guided the researchers to target the anti-apoptotic factor, Proviral Integration of Moloney virus-2 (PIM-2) which belongs to a family of serine/threonine kinase (PIM-1, PIM-2 and PIM-3).5
Several human cancers, particularly hematological malignancies, showed an increased expression of PIM kinases, where they aid the survival and proliferation of malignant cells.6 PIM kinases were found to be downstream effectors of vital oncoproteins like Janus Kinase 2 (JAK2), FMS-like tyrosine kinase 3 (FLT3) and Abelson (ABL)7; therefore, the PIM kinases have emerged as prospective pharmacological targets in a variety of hematological cancers with numerous Inhibitors demonstrating their efficacy in AML preclinical models.8 SGI-1776, a pan-PIM inhibitor has been shown to be effective in AML cell lines for inhibiting PIM-2 kinase and inducing apoptosis.9 PIM-2 plays a critical role in the survival, proliferation and differentiation of hematopoietic cells as many cytokines implicated in the growth and maturation of these cells (especially IL3) regulate the expression of the PIM-2 gene at both protein and mRNA levels.10 Over-expression of PIM-2 has been linked to the advancement of B-lymphoid malignancies as chronic lymphocytic leukemia (CLL), diffuse large B-cell lymphoma (DLBCL), Burkitt lymphoma (BL) and mantle cell lymphoma (MCL).11,12 Almost half of DLBCL cases revealed elevated PIM kinase expression, especially in the activated B-cell (ABC) subtype, because of constitutive activation of JAK/STAT3 signalling.13 A strong link between PIM kinase expression, STAT signaling, increased rates of proliferation, and progressive disease stages has been established.14
Several reports have revealed that PIM-2 is closely linked to myeloma cell growth and survival, so that it might be a promising therapeutic target.15 PIM-2 has also been reported to be persistently expressed in primary AML blasts and to have a role in protein translation through regulating 4E-BP1 phosphorylation.16 Elevated PIM-2 gene expression level was found to be linked to a low rate of complete remission (CR) in AML patients and their poor survival, suggesting another aspect of its possible prognostic significance.17 This study aimed to assess PIM-2 expression level in AML patients, correlate between its level, clinical and laboratory findings, response to treatment and patients’ outcome.
Materials and Methods
The institutional review board (IRB) of Zagazig University approved the study (approval number, ZU-IRB# 4240), and written informed consent was taken from all patients involved in the study. The study was conducted according to the ethical principles of the declaration of Helsinki. The study was carried out at Clinical Pathology Department, Zagazig University Hospitals. Fifty de novo AML patients admitted to Medical Oncology Department, Clinical Hematology Unit, Internal Medicine Department, Zagazig University Hospitals were included in this study between August 2019 and September 2020. Following informed permission and IRB approval, peripheral blood (PB) and bone marrow (BM) samples were taken when the diagnosis was made. Patients’ inclusion criteria include approval to participate in the study, newly diagnosed AML patients before receiving induction chemotherapy with no concurrent malignancy. Therapy-related AML and patients diagnosed with Acute Promyelocytic Leukemia (M3) were excluded. The patients were subjected to the following: Thorough history taking, clinical examination, routine lab investigations including Complete Blood Count (CBC) using Sysmex Xn (Sysmex, Japan), liver, kidney function tests and serum lactate dehydrogenase (LDH) using Cobas 6000 autoanalyzer (Roche Diagnostics, Germany), prothrombin time (PT) and activated partial thromboplastin time (APTT) using CS-1500 (Sysmex, Japan), erythrocyte sedimentation rate (ESR) using VISION-B (YHLO Biotech, China), BM aspiration and evaluation using Leishman and peroxidase stains, flow cytometry was used to determine immunophenotyping using FACSCanto II flow cytometer equipped with Diva software (Becton Dickinson, USA) and cytogenetic analysis (karyotyping) was performed with an image analyzer (Imstar, France) utilizing the G banding technique. Patients were categorized according to their cytogenetic risk stratification into favorable cytogenetic risk group (n=9 patients), Intermediate cytogenetic risk group (n=39 patients); 36 of them were with normal karyotyping, and the adverse cytogenetic risk group (n=2 patients). Radiological investigations such as Pavli-abdominal sonography, echocardiography and chest X-ray were done for the patients.
PIM-2 Quantitative Polymerase Chain Reaction (q PCR)
RNA was extracted from whole blood by RNA extraction kit (Gene JET RNA Purification Kit Catalog No. K0731 Thermo Scientific, USA). Its purity was determined using a Nanodrop2000 spectrophotometer (Thermo Scientific, USA). RNA was reverse transcribed to generate cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems Part Number: 4375575) through Veriti thermal cycler (Applied Biosystems, Japan). TaqMan gene expression assay (Thermo Fisher) was used to perform quantitative PCR with primers and hydrolysis probes for PIM2 (ref. Hs00179139_m1) and an internal control GAPDH (ref. Hs03929097_g1) (Applied Biosystems). The transcription level of the target gene (PIM-2) was normalized to that of the reference gene (GAPDH). Control samples were withdrawn from 20 age- and sex-matched individuals. They were selected from patients with hematological diseases to whom BM examination was one of the required investigations (hypersplenism and idiopathic thrombocytopenic purpura). Relative mRNA expression was calculated using the 2−ΔΔCT method.18
Treatment Plan and Follow-Up
Patients were treated by an induction regimen (7+3) that included a 7-day cytarabine infusion (100mg/m2) and 3 days of doxorubicin (25mg/m2) treatment. Low-intensity treatment: lower dose chemotherapy or hypomethylation agents were used to treat patients over 60 years old.19 Follow-up of the patients was done on day 28 after receiving induction chemotherapy by performing CBC and BM aspirate to evaluate their remission status. CR was defined as normocellular BM containing less than 5% blast cells and peripheral blood with at least 1×109/L neutrophils and ≥100×109/L platelets.20 Patients who achieved remission received three to four courses of high-dose cytarabine (1–2 g/ m2 every 12 h on days 1, 3 and 5; total, 12 g/ m2) as a consolidation.19
Statistical Analysis
The collected data were computerized and statistically analyzed using Statistical Package for Social Science program (SPSS version 24) (IBM; Armonk, New York, USA). The Shapiro–Wilk test was used to determine if the data had a normal distribution. Frequencies or relative percentages were used to express qualitative data. The difference between qualitative variables was calculated using the Chi-square test (χ2) and Fisher exact test. The median and range were used to express quantitative data. Mann–Whitney test was used to calculate the difference between quantitative variables in two groups. Kruskal–Wallis Test was used to calculate the difference between quantitative variables in more than two groups. Spearman correlation tests were used for correlating variables, respectively. Univariate and multivariate logistic regression analyses were performed with P<0.1 in univariate analysis. ROC curve was constructed to permit the selection of threshold values for test results and comparison of different testing strategies.
Results
Our research involved 50 de novo AML patients, with more than half of them being males (54%), M:F ratio 1.2, and their age ranged from 18 to 72 years with a median of 44 years. Bone ache and fatigue were the most frequent presentations (82%), followed by fever (64%), bleeding (60%), purpura (58%), gum hypertrophy (32%), splenomegaly (18%), hepatomegaly (12%) and finally lymphadenopathy (10%). According to FAB classification, the most frequent type was M4 (34%), followed by M2 (30%), while M1 and M5 constitute 18% for each. Normal cytogenetic was found in 72%, while abnormal cytogenetic was found in 28% (Table 1).
 |
Table 1 Baseline Patient Characteristics [Median (Range) or n (%)] |
PIM-2 level was assessed in different categories, and it was observed that PIM-2 level was significantly found at a lower level in the favorable cytogenetic risk group (median=1.06) versus other cytogenetic groups (median= 4) (p=0.006); also, a highly statistically significant lower PIM-2 level was observed in AML patients who achieved remission (p≤0.001). No statistically significant difference in PIM-2 level was found in patients who had early death than others, while a statistically significant higher PIM-2 level was observed in patients who suffered early relapse (p=0.02). The median of PIM-2 level in female AML patients was higher than male patients, but with no statistical significance. Regarding FAB, the median of PIM-2 expression was the highest in M1, followed by M5, then M2 and lastly M4, but with no statistical significance (Table 2).
 |
Table 2 PIM-2 Median (Range) Levels in Different Categories |
A highly significant positive correlation between PIM-2 level and BM blast on day 28 (p<0.001, r=0.855) (Figure 1), also a highly significant positive correlation was found between PIM-2 level and PB blast on day 28 (p<0.001, r=0.856), while no significant correlation was found between PIM-2 level, age, total leukocytic count (TLC), hemoglobin level (Hb), platelets count, ESR and LDH (Table 3).
 |
Table 3 Correlation Between PIM-2 Expression and Other Studied Parameters |
 |
Figure 1 Significant positive correlation between PIM-2 expression and BM blasts on day 28. |
The logistic regression was used to determine the effects of various parameters on the likelihood of achieving CR. The univariate analysis revealed that response was dependently associated with TLC, cytogenetics, and PIM-2. However, on multivariable analysis, PIM-2 level was independently associated with response to therapy (Table 4).
 |
Table 4 Univariate and Multivariable Logistic Regression Analyses for Response to Therapy |
A highly statistically significant difference was observed between patients and the control samples as regard PIM-2 expression level (p<0.001) (Table 5). Receiver operating characteristic curve (ROC) and area under the curve (AUC) for PIM-2 at diagnosis for achieving CR after induction was calculated, and it was found that PIM-2 at cutoff < 1.6 had an AUC (0.903) with a sensitivity (90.48%) and specificity (86.21%), P= <0.001 (Figure 2 and Table 6).
 |
Table 5 Comparison Between PIM-2 Level (Median and Range) Among Patients and Control |
 |
Table 6 Receiver Operating Characteristic Curve (ROC) for PIM-2 at Diagnosis for Achieving Remission After Induction |
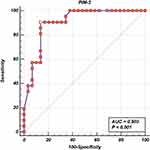 |
Figure 2 Receiver operating characteristic curve (ROC) and area under the curve (AUC) for PIM-2 at diagnosis (cut-off was 1.6). |
There was no statistically significant difference between AML patients with high PIM-2 level and those with low PIM-2 level regarding their clinical presentation (Table 7). Upon analysis of PIM-2 expression level among different cytogenetic risk groups, it was found that all AML patients in the favorable cytogenetic group had low PIM-2 level. In contrast, the intermediate-risk group included 39 patients (25 had high PIM-2 expression). The adverse cytogenetic risk group included only two patients. Both had high PIM-2 level and there was a statistically significant difference between the favorable cytogenetic risk group and other groups regarding their PIM-2 expression level (p= 0.003) (Table 8). A clinical outcome of patients based on PIM-2 expression was clarified in Table 9.
 |
Table 7 Clinical Presentation of Patients Based on PIM-2 Expression |
 |
Table 8 PIM-2 Expression Among Different Cytogenetic Risk Groups |
 |
Table 9 Clinical Outcomes of AML Group Based on PIM-2 Expression |
Discussion
AML is a group of heterogeneous hematological malignancies characterized by clonal expansion of myeloid precursors with diminished capacity for differentiation.21 It is triggered by genetic changes that cause hematopoietic progenitor cells to convert.1 Molecular abnormalities are the essential events in AML development despite cytogenetic changes being the most critical independent prognostic markers.22 Additional research into the involvement of fundamental genetic and epigenetic pathways in malignancy provides more knowledge about the mechanism of leukemogenesis in AML, suggests key prognostic standards and highlights the possible therapeutic targets.23,24
PIM-2 belongs to a serine/threonine kinase family, which includes three members that overlie in their function, compensate for each other and target the regulators of cell cycle and survival.25 PIM kinases are overexpressed in both hematologic malignancies, including lymphomas, CLL and acute leukemia and solid tumors such as prostate and colon cancer.12 HOXA9 overexpression and STAT activation in AML lead to PIM-2 upregulation as they can act as transcription factors.26,27
In this study, PIM-2 mRNA level was elevated in AML patients versus the control samples used to interpret our results. This agreed with what was reported by El Dahshan et al that AML patients had statistically significant higher PIM-2 expression level than their control group.28 Tamburini et al in their study reported an expression of PIM-2 in 23 AML cases compared to its deficiency in normal CD34 positive hematopoietic cells.16 Kapelko-Slowik et al showed that AML and ALL patients in their study had significantly higher PIM-2 expression level than the control group.17 Moreover, Mizuki et al compared the expression of PIM-2 mRNA in newly diagnosed AML patients’ BM samples to healthy BM samples and noticed that PIM-2 mRNA was considerably higher in AML.29
In this research, a positive correlation was detected between PIM-2 expression level and age but did not reach statistical significance. Kapelko-Slowik et al found a significant positive correlation between these two parameters,30 while El Dahshan et al reported a negative correlation between them but without statistical significance.28 Regarding the association between PIM-2 gene and different FAB subtypes, no statistically significant difference for PIM-2 gene was found among different FAB subtypes; this agreed with Abbassy et al31 and Kapelko-Slowik et al, who reported that no significant differences were observed between the subtypes stratified according to the FAB classification in AML.30
Regarding the correlation between PIM-2 gene and different lab parameters, our results showed positive correlations between PIM-2, TLC, initial BM blasts and initial PB blasts. Still, all of them did not reach a statistically significant level. At the same time, there were negative correlations between PIM-2 and both Hb level and platelet count, but none of them was statistically significant. This agreed with Kapelko-Slowik et al, who found that no considerable correlation was observed between PIM-2 expression and Hb level, platelet count and PB blasts.30 El Dahshan et al showed no correlation between PIM-2 gene level and Hb or BM blast percentage. A positive correlation was noticed between the expression of PIM-2 and TLC, while a negative correlation was observed between its expression and platelet count, but with no significance.28
In the current study, evaluation of response to induction chemotherapy was carried out on day 28 due to BM aplasia on day 14 and poor general condition of the patients. In our study, there was a highly significant positive correlation between PIM-2 level and BM blasts and PB blasts on day 28; also, a highly statistically significant lower PIM-2 level was found in AML patients who achieved CR than those who did not achieve. This agreed with Kapelko-Slowik et al who reported that PIM-2 gene expression was significantly lower in AML patients who achieved CR compared to those who did not respond to induction therapy.30 El Dahshan et al noticed the same finding, but with no statistically significant difference.28 These results denote that high PIM-2 expression level is associated with resistance to induction chemotherapy as increased PIM-2 level promotes tumorgenesis through the phosphorylation of 4E-BP1.16 PIM-2 kinase also activates apoptosis inhibitor 5 (API-5), which is a downstream factor for Nuclear factor Kappa B (NF-KB),32 that target many genes associated with cell survival and apoptosis inhibition such as BCL-2 and BCL-XL.33
In our study, the univariate logistic regression analysis for response to therapy revealed that response was dependently associated with TLC, cytogenetics, and PIM-2. However, on multivariable analysis, PIM-2 level was independently related to response to treatment and hence PIM-2 expression is an independent prognostic marker for achieving CR after standard induction chemotherapy in AML patients. Kapelko-Slowik et al in their univariate analysis to assess the survival found that the overall survival of AML patients varied according to their PIM-2 expression level.30
ROC curve was done for PIM-2 gene expression at diagnosis to evaluate its role in achieving remission after induction, it was found that PIM-2 at cut off value >1.6 had an AUC of 0.903 with a sensitivity of 90.48% and a specificity of 86.21%. According to this cutoff value, AML patients were alienated into two groups: low PIM-2 expression (≤1.6) and high PIM-2 expression (>1.6). As regards clinical presentation of patients based on PIM-2 expression, our results showed that no statistically significant difference in the clinical presentation of patients with high PIM-2 level than those with low PIM-2 level, this agreed with El Dahshan et al who found that there was no association between the expression level of the gene and the existence of hepatosplenomegaly or lymphadenopathy.28
Our results showed that all AML patients in the favorable cytogenetic risk group have a statistically significant lower PIM-2 level than those in the intermediate and adverse groups. This data matches the findings of Kapelko-Slowik et al, who found that increased PIM-2 expression was linked to cytogenetic and molecular hazards in AML17 and Green et al who reported that PIM-2 was over-expressed in FLT3-ITD positive AML patients, PIM kinase inhibition reduces AML cell viability and PIM-2 knockdown blocks disease propagation in FLT3-ITD-positive AML xenografts.34
To investigate the impact of PIM-2 expression on the clinical outcome, our results showed that no significant association was found between PIM-2 level and mortality, while on following up the cases that achieved CR, 2 of them relapsed and it was found that they have a highly statistically significant higher PIM-2 level than those who did not relapse (p≤ 0.001). Abbassy et al had no mortality in their studied group.31 However, Kapelko-Slowik et al in their study showed that increased PIM-2 expression in AML patients’ blast cells is linked to a bad prognosis, while its lower expression resulted in prolongation of event-free survival (EFS).17
Finally, this study offers us an insight on PIM-2 gene, one of the PIM kinases that plays a critical role in the leukemogenesis process and resistance to chemotherapy, assesses its expression level in AML patients in order to evaluate its role as a prognostic marker for disease outcome.
Nevertheless, the current study had some inadequacies and limitations, such as the small sample size, that it was done in single centre, the short duration of patients’ follow-up and the lack of data about other genes that affect disease outcome in AML patients.
Conclusion
PIM-2 overexpression is associated with a low remission rate. PIM-2 is an independent prognostic marker for achieving CR after standard induction chemotherapy in AML patients. These results denote an association between the PIM-2 gene and poor outcome in AML. Further long-term clinical follow-up studies with a bigger sample size are recommended to make the statistical analysis more conclusive.
Ethical Approval
All procedures involving human subjects were carried out in compliance with IRB’s ethical standards (ZU-IRB# 4240). The study was performed in accordance with the Helsinki Declaration and was approved by the IRB.
Informed Consent
Informed consent was obtained from all individuals.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Patel J, Gonen M, Figueroa M, et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366(12):1079–1089. doi:10.1056/NEJMoa1112304
2. Thol F, Heuser M, Damm F, Klusmann J, Reinhardt K, Reinhardt D. DNMT3A mutations are rare in childhood acute myeloid leukemia. Haematologica. 2011;96(8):1238–1240. doi:10.3324/haematol.2011.046839
3. Khorshid O, Diaa A, Abd El Moaty M, et al. Clinical features and treatment outcome of acute promyelocytic leukemia patients treated at Cairo National Cancer Institute in Egypt. Mediterr J Hematol Infect Dis. 2011;3(1):e2011060. doi:10.4084/mjhid.2011.060
4. Döhner H, Weisdorf D, Bloomfield C. Acute myeloid leukemia. N Engl J Med. 2015;373(12):1136–1152. doi:10.1056/NEJMra1406184
5. Brault L, Gasser C, Bracher F, Huber K, Knapp S, Schwaller J. PIM serine/threonine kinases in the pathogenesis and therapy of hematologic malignancies and solid cancers. Haematologica. 2010;95(6):1004–1015. doi:10.3324/haematol.2009.017079
6. Alvarado Y, Giles F, Swords R. The PIM kinases in hematological cancers. Expert Rev Hematol. 2012;5(1):81–96. doi:10.1586/ehm.11.69
7. Wernig G, Gonneville J, Crowley B, et al. The Jak2V617F oncogene associated with myeloproliferative diseases requires a functional FERM domain for transformation and for expression of the Myc and Pim proto-oncogenes. Blood. 2008;111(7):3751–3759. doi:10.1182/blood-2007-07-102186
8. Keeton E, McEachern K, Dillman K, et al. AZD1208, a potent and selective pan-pim kinase inhibitor, demonstrates efficacy in preclinical models of acute myeloid leukemia. Blood. 2014;123(6):905–913. doi:10.1182/blood-2013-04-495366
9. Chen LS, Redkar S, Taverna PM, Cortes JE, Gandhi V. Mechanisms of cytotoxicity to pim kinase inhibitor, SGI-1776 in acute myeloid leukemia. Blood. 2011;118(3):693–702. doi:10.1182/blood-2010-12-323022
10. Fox C, Hammerman P, Cinalli R, Master S, Chodosh L, Thompson C. The serine/threonine kinase Pim- 2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 2003;17(15):1841–1854. doi:10.1101/gad.1105003
11. Huttmann A, Klein-Hitpass L, Thomale J, et al. Gene expression signatures separate B-cell chronic lymphocytic leukaemia prognostic subgroups defined by ZAP-70 and CD38 expression status. Leukemia. 2006;20(10):1774–1782. doi:10.1038/sj.leu.2404363
12. Cohen A, Grinblat B, Bessler H, et al. Increased expression of the hPim-2 gene in human chronic lymphocytic leukemia and non-Hodgkin lymphoma. Leuk Lymphoma. 2004;45(5):951–955.
13. Bachmann M, Möröy T. The serine/threonine kinase Pim-1. Int J Biochem Cell Biol. 2005;37(4):726–730. doi:10.1016/j.biocel.2004.11.005
14. Brault L, Menter T, Obermann E, et al. PIM kinases are progression markers and emerging therapeutic targets in diffuse large B-cell lymphoma. Br J Cancer. 2012;107(3):491–500. doi:10.1038/bjc.2012.272
15. Lu J, Zavorotinskaya T, Dai Y, et al. Pim2 is required for maintaining multiple myeloma cell growth through modulating TSC2 phosphorylation. Blood. 2013;122(9):1610–1620. doi:10.1182/blood-2013-01-481457
16. Tamburini J, Green A, Bardet V, et al. Protein synthesis is resistant to rapamycin and constitutes a promising therapeutic target in acute myeloid leukemia. Blood. 2009;114(8):1618–1627. doi:10.1182/blood-2008-10-184515
17. Kapelko-Slowik K, Owczarek T, Grzymajlo K, et al. Elevated PIM2 gene expression is associated with poor survival of patients with acute myeloid leukemia. Leuk Lymphoma. 2016;13:1–10. doi:10.3109/10428194.2015.1124991
18. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
19. Tefferi A, Letendre L. Going beyond 7 + 3 regimens in the treatment of adult acute myeloid leukemia. J Clin Oncol. 2012;30(20):2425–2428. doi:10.1200/JCO.2011.38.9601
20. Marcucci G, Mrózek K, Ruppert AS, et al. Abnormal cytogenetics at date of morphologic complete remission predicts short overall and disease-free survival, and higher relapse rate in adult acute myeloid leukemia: results from Cancer and Leukemia Group B study 8461. J Clin Oncol. 2004;22(12):2410–2418. doi:10.1200/JCO.2004.03.023
21. Estey E, Döhner H. Acute myeloid leukaemia. Lancet. 2006;368(9550):1894–1907. doi:10.1016/S0140-6736(06)69780-8
22. Renneville A, Roumier C, Biggio V, et al. Cooperating gene mutations in acute myeloid leukemia: a review of the literature. Leukemia. 2008;22(5):915–931. doi:10.1038/leu.2008.19
23. Conway O’Brien E, Prideaux S, Chevassut T. The epigenetic landscape of acute myeloid leukemia. Adv Hematol. 2014;2014:103175. doi:10.1155/2014/103175
24. Mikkers H, Allen J, Knipscheer P, et al. High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat Genet. 2002;32(1):153–159. doi:10.1038/ng950
25. Konietzko U, Kauselmann G, Scafidi J, et al. Pim kinase expression is induced by LTP stimulation and required for the consolidation of enduring LTP. EMBO J. 1999;18(12):3359–3369. doi:10.1093/emboj/18.12.3359
26. Benekli M, Baumann H, Wetzler M. Targeting signal transducer and activator of transcription signaling pathway in leukemias. J Clin Oncol. 2009;27(26):4422–4432. doi:10.1200/JCO.2008.21.3264
27. Hu YL, Passegué E, Fong S, Largman C, Lawrence H. Evidence that the Pim1 kinase gene is a direct target of HOXA9. Blood. 2007;109(11):4732–4738. doi:10.1182/blood-2006-08-043356
28. El Dahshan D, Hammam A, Ahmed A, El Samra M. Expression of Proviral Integration of Moloney Virus-2 (PIM2) gene in patients with acute myeloid leukemia and its clinical significance. IJBC. 2020;12(1):6–11.
29. Mizuki M, Schwable J, Steur C, et al. Suppression of myeloid transcription factors and induction of STAT response genes by AML-specific Flt3 mutations. Blood. 2003;101(8):3164–3173. doi:10.1182/blood-2002-06-1677
30. Kapelko-Słowik K, Urbaniak-Kujda D, Wołowiec D, et al. Increased expression of PIM-2 and NF-κB genes in patients with acute myeloid leukemia (AML) and acute lymphoblastic leukemia (ALL) is associated with complete remission rate and overall survival. Postepy Hig Med Dosw. 2013;67:553–559. doi:10.5604/17322693.1052449
31. Abbassy H, Donia H, Ghallab O. Association of PIM2 and NF-KB with poor clinical outcome in Egyptian patients with acute myeloid leukemia. Int J Curr Res. 2016;8(07):34399–34402.
32. Ren K, Zhang W, Shi Y, Gong J. pim-2 activates API-5 to inhibit the apoptosis of hepatocellular carcinoma cells through NF-KB pathway. Pathol Oncol Res. 2010;16(2):229–237. doi:10.1007/s11253-009-9215-4
33. Li Q, Verma IM. NF-KB regulation in the immune system. Nat Rev Immunol. 2002;2(10):725–734. doi:10.1038/nri910
34. Green A, Maciel T, Hospital MA, et al. Pim kinases modulate resistance to FLT3 tyrosine kinase inhibitors in FLT3-ITD acute myeloid leukemia. Sci Adv. 2015;1(8):e1500221. doi:10.1126/sciadv.1500221
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
