Back to Journals » International Journal of General Medicine » Volume 14
Prognostic Value of Long Noncoding RNA DLEU2 and Its Relationship with Immune Infiltration in Kidney Renal Clear Cell Carcinoma and Liver Hepatocellular Carcinoma
Authors Fu S, Gong B, Wang S, Chen Q, Liu Y, Zhuang C , Li Z, Zhang Z, Ma M, Sun T
Received 27 August 2021
Accepted for publication 26 October 2021
Published 11 November 2021 Volume 2021:14 Pages 8047—8064
DOI https://doi.org/10.2147/IJGM.S336428
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Shengqiang Fu,1,* Binbin Gong,1,* Siyuan Wang,1 Qiang Chen,1 Yifu Liu,1 Changshui Zhuang,2 Zhilong Li,1 Zhicheng Zhang,1 Ming Ma,1 Ting Sun1
1Department of Urology, The First Affiliated Hospital of Nanchang University, Nanchang, People’s Republic of China; 2Department of Urology, Union Shenzhen Hospital, Huazhong University of Science and Technology, Shenzhen, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ting Sun
Department of Urology, The First Affiliated Hospital of Nanchang University, Nanchang, 330000, Jiangxi Province, People’s Republic of China
Email [email protected]
Background: DLEU2 is a long noncoding RNA considered important in the progression of many cancers. However, correlations between DLEU2 and kidney renal clear cell carcinoma (KIRC) and liver hepatocellular carcinoma (LIHC) have rarely been reported.
Methods: We first analysed the expression of DLEU2 across cancers and the correlation between DLEU2 and the clinical features of KIRC and LIHC by using the “ggplot2” package in R and searched the Oncomine database and Timer website platform. We verified the expression of DLEU2 in the GEO dataset (GSE105261 and GSE45267). Receiver operating characteristic (ROC) curves were drawn using the “pROC” and “ggplot2” packages in R, and we constructed a DLEU2-based prognostic nomogram for KIRC and LIHC by using the “survival” and “rms” packages in R. Then, we analysed the correlation between DLEU2 expression and prognosis in R as well as the correlation between DLEU2 and immune cell infiltration in the TIMER database. Finally, we explored the causes of DLEU2 upregulation in the UCSC Xena and UALCAN databases.
Results: We found that DLEU2 was upregulated in many cancers, including KIRC and LIHC. Expression of DLEU2 is associated with tumour stage, grade, lymphatic metastasis, and distant metastasis in KIRC as well as alpha-fetoprotein (AFP), tumour stage, grade, lymphatic metastasis, and distant metastasis in LIHC. DLEU2 is an adverse factor for the prognosis of KIRC and LIHC. In addition, DLEU2 has moderate accuracy in diagnosing KIRC and LIHC and predicting their prognosis. Moreover, we found that expression of DLEU2 correlated positively with immune cell infiltration in KIRC and LIHC, and upregulation of DLEU2 in KIRC and LIHC suggests a poor prognosis based on immune cells analysis. Genetic and epigenetic analyses of DLEU2 indicate that copy number variations (CNVs) and methylation contribute to the upregulation of DLEU2.
Conclusion: The long noncoding RNA DLEU2 has the potential to predict the prognosis and immune infiltration of KIRC and LIHC.
Keywords: DLEU2, prognosis, immune infiltration, KIRC, LIHC
Introduction
Kidney renal clear cell carcinoma (KIRC) and liver hepatocellular carcinoma (LIHC) are two common solid tumours that are insensitive to traditional radiotherapy and chemotherapy.1,2 Immunotherapy, which has emerged in recent years, has become an optimized treatment option for many cancers, including KIRC and LIHC.3–5 Therefore, it is important to understand the relationship between cancer progression and immune infiltration. The tumour microenvironment (TME), which includes tumour cells, immunoinfiltrating cells, stromal cells and extracellular matrix, is pivotal for tumorigenesis and development. It has been reported that the interaction between tumour cells and infiltrating immune cells affects the progression of tumours.6 Immune infiltration is thought to play an important role in the progression of KIRC and LIHC.7,8 Previous studies have reported that the prognosis of KIRC is closely related to immune-infiltrating macrophages, CD4+ T cells, CD8+ T cells, and regulatory T cells.9,10 The treatment outcome of LIHC is believed to be correlated with infiltrating macrophages, B cells, CD4+ T cells, neutrophils, and dendritic cells.11 The immunoregulatory molecules PD1, PD-L1, PD-L2, and CTLA4 are believed to be involved in the malignant progression of KIRC,9 and PD-1 and CTLA-4 are believed to be associated with a poor prognosis of LIHC.12 These findings suggest that tumour-infiltrating immune cells may be a promising therapeutic target for KIRC and LIHC. Considerable progress has been achieved in the diagnosis, surgical treatment, and drug treatment of KIRC and LIHC, but their clinical outcomes are still unsatisfactory. Due to the heterogeneity of tumours, both KIRC and LIHC require further investigation to identify effective molecular targets for personalized therapy.
DLEU2 is a long noncoding RNA aberrantly expressed in many cancers.13–20 Previous studies have reported that DLEU2 affects the progression of malignancies via multiple mechanisms.13,14,19,20 DLEU2 can mediate the progression of gastric cancer by regulating the EMT process and affecting the Akt signalling pathway and can promote the proliferation of cervical cancer cells by regulating the Notch signalling pathway.13,14 In addition, DLEU2 promotes the malignant progression of laryngeal squamous cell carcinoma by influencing the PI3K-Akt signalling pathway.20 Overall, DLEU2 is thought to play a crucial role in cancer progression.
Notably, DLEU2 was initially reported in immunocell-associated disease (diffuse large B-cell lymphoma),21 suggesting that DLEU2 may correlate with immune cell infiltration. KIRC and LIHC are common malignancies that have been reported to correlate closely with immune cell infiltration.7,8 However, whether DLEU2 contributes to the progression of KIRC and LIHC by affecting immune cell infiltration has not been reported.
In this study, we accessed the Oncomine database and the TIMER and Kaplan–Meier Plotter website platforms and analysed the expression of DLEU2 in KIRC and LIHC and the correlation between its expression and the prognosis of KIRC and LIHC. Then, we analysed correlation between DLEU2 expression and infiltrating immune cells on the TIMER website. We further explored the potential mechanism of dysregulated DLEU2 expression in KIRC and LIHC, such as DNA methylation, CNVs, and somatic cell mutations. Our findings identified an important role of DLEU2 in the prognosis of KIRC and LIHC. We also uncovered a mechanism by which DLEU2 can affect immune cell infiltration in the TME of KIRC and LIHC and affect the prognosis of KIRC and LIHC.
Methods
Oncomine Database Analysis
Oncomine (https://www.oncomine.org/resource/main.html) is the largest cancer gene chip database and integrated data search platform worldwide.22 Oncomine merges DNA and RNA-Seq data derived from TCGA, GEO and online publications. Oncomine contains comprehensive cancer mutation data, gene expression information and correlative clinical data. Using the Oncomine database, differential expression analyses, correlation analyses and coexpression analyses can be implemented to choose genes with differential content in certain cancers, identify target genes, and determine research directions. We analysed the differences in the DLEU2 content across cancers using the Oncomine database platform.
TIMER Database Analysis
TIMER (https://cistrome.shinyapps.io/timer/) is an online tool used to study tumour immunity led by Prof. Xiaobing Liu, a professor of immunoinformatics at Harvard University.23 TIMER uses RNA-Seq expression profile data to detect immune cell infiltration in tumour tissues and comprehensively analyses the correlation between gene expression and immune cell infiltration in various cancers. The TIMER algorithm was used to evaluate the abundance of six immune-infiltrating cells (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells). TIMER also allows users to analyse differences in gene expression between cancer and normal tissue in various cancers.23 We analysed the difference in the content of DLEU2 between various cancer specimens and matched normal specimens using the TIMER website. Then, we analysed the correlation between DLEU2 and 6 immune infiltrating cells and 16 immune cell molecular markers.
Kaplan–Meier Plotter Website Platform Analysis
The Kaplan–Meier Plotter (http://kmplot.com/analysis/) database combines microarray and RNA-SEq data from public databases, such as TCGA, GEO, and EGA, to evaluate the impact of tens of thousands of genes on survival in 21 cancers.24 We used the Kaplan–Meier Plotter database to evaluate the correlation between DLEU2 expression and survival in various cancers. We also used an online tool to perform a prognostic analysis of DLEU2 expression based on the abundance of different immune cells. The hazard ratios (HRs) and log-rank p-values of the 95% confidence intervals (CIs) were extracted.
UCSC Xena Database Analysis
The UCSC Xena database (http://xena.ucsc.edu/) is a cancer genomics data analysis platform that supports the visualization and analysis of multiple omics data from cancer samples.25 The platform contains numerous public datasets, including data from TCGA, GTEx, ICGC, TARGET and other large cancer research projects. The database provides information regarding gene expression, protein expression, copy numbers, methylation, and somatic mutations and clinical data, such as patient treatment and survival outcomes. We performed DLEU2 expression analysis, copy number variation (CNV) analysis, somatic mutation analysis and DNA methylation analysis using the UCSC Xena website platform.
UALCAN Database Analysis
UALCAN (http://ualcan.path.uab.edu/index.html) is a website that allows users to mine and analyse cancer data online.26 Its data are mainly derived from the TCGA database. The content of the website mainly includes expression analysis, survival analysis, methylation survival analysis, correlation analysis and pan-cancer view and can be linked to several other external databases for users to perform other analyses. We performed a methylation analysis of DLEU2 in KIRC and LIHC using this site.
Statistical Analysis
DLEU2 expression analysis was performed with the ONCOMINE, TIMER, and R projects using the “ggplot2” package. ROC curves were drawn using the “pROC” and “ggplot2” package-derived R projects. A nomogram and calibration curves were drawn by manipulating the “survival” and “rms” packages in the R Project. The survival curve was drawn by combining the Kaplan–Meier Plotter database and “survival” package-derived R Project. The correlation analysis of gene expression in the TIMER database was performed by conducting a Spearman correlation analysis. Differences were considered statistically significant only when the P-value was less than 0.05.
Results
Expression of the Long Noncoding RNA DLEU2 in Various Cancers
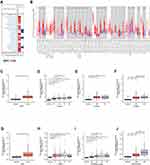
We searched for expression of DLEU2 in various cancers in the Oncomine database, and the results revealed that the abundance of DLEU2 expression in most cancer tissues was higher than that in the matched normal tissues (Figure 1A). The expression level of DLEU2 was then analysed in the TIMER database. The analysis results of the TIMER database revealed that DLEU2 was highly expressed in most human cancers compared with that in the matched normal tissues, especially KIRC and LIHC (Figure 1B). To obtain more convincing results, we used the “ggplot2” package in R Project to analyse the expression of DLEU2 in pan-cancer, and the results were consistent with those in the TIMER and ONCOMINE databases (S1). Next, we used the “ggplot” package in R to verify the findings in the Oncomine and TIMER databases and found that the expression of DLEU2 in KIRC and LIHC was significantly higher than that in their corresponding normal tissues (Figure 1C and G). In addition, we verified the expression of DLEU2 in the relevant datasets of KIRC (GSE105261) and LIHC (GSE45267) (S2), and the results were consistent with those obtained using the TIMER and Oncomine databases. Notably, DLEU2 expression is closely related to the primary tumour lesion (T), regional lymph node involvement (N), and distant metastasis (M) of KIRC in the TNM staging system (Figure 1D–F). Meanwhile, DLEU2 correlated significantly with the primary tumour lesion, histologic grade and alpha-fetoprotein level (AFP) of LIHC (Figure 1H–J).
DLEU2’s Efficiency in Predicting KIRC and LIHC
To explore the correlation between DLEU2 expression and KIRC and LIHC, we used the “pROC” package and “ggplot2” package in R to plot the receiver operating characteristic curve (ROC) of DLEU2 in KIRC and LIHC (Figure 2A and B). The graph shows that DLEU2 expression can predict KIRC (AUC=0.896, CI: 0.852–0.940) and LIHC (AUC=0.852, CI: 0.8 15–0.889) with a certain accuracy.
The Prognostic Value of DLEU2 and the Construction of a Prognostic Nomogram for KIRC and LIHC
To explore the factors influencing the prognosis of KIRC and LIHC, univariate and multivariate Cox hazard regression analyses were performed to identify factors that might affect the prognosis of KIRC and LIHC. The results of the univariate analysis showed that an age>60 years, right tumour, high T.N.M stage, high histologic grade, and high expression of DLEU2 affected the OS of KIRC (Table 1). The multivariate analysis showed that an age>60 years, high T.M stage, high histologic grade, and high expression of DLEU2 affected the OS of KIRC (Table 1). This finding implies that an age>60 years, high T and M stage, high histologic grade, and high expression of DLEU2 are independent prognostic factors of KIRC. Meanwhile, the univariate analysis and multivariate analysis of the factors that may affect the prognosis of LIHC showed that high histologic grades, tumours and high expression of DLEU2 were independent prognostic factors of LIHC (Table 2). Then, we extracted the independent prognostic factors of KIRC and LIHC and constructed nomograms for the prognosis of KIRC and LIHC by using the ‘rms’ package in R Project. As shown in Figure 3A, the nomogram revealed the relationship between age, T stage, M stage, and histologic grade and the DLEU2 expression level and the 1-, 2-, and 3-year survival probabilities of OS in KIRC. The nomogram for predicting the prognosis of LIHC showed the relations between the histologic grade, tumour status and DLEU2 expression and the 1-, 2-, and 3-year survival probabilities of OS in LIHC (Figure 3E). The DLEU2-based nomogram for both KIRC (C index=0.753) and LIHC (C index=0.710) showed good performance in terms of predicting the OS of KIRC and LIHC. The 1-, 2-, and 3-year calibration curves of KIRC (Figure 3B–D) and LIHC (Figure 3F–H) displayed consistency between our results and the predicted results, suggesting that the performance of the DLEU2-based nomogram in predicting the prognosis of KIRC and LIHC was satisfactory.
 |
Table 1 Univariate and Multivariate Cox Regression Analysis of KIRC in TCGA |
 |
Table 2 Univariate and Multivariate Cox Regression Analysis of LIHC in TCGA |
Prognostic Significance of the Differential Expression of DLEU2 in Different Cancers
We searched the Kaplan–Meier Plotter database to determine the prognostic significance of DLEU2 expression in human cancers. As shown in Figure 4A–D, high DLEU2 expression predicted a poor outcome of KIRC, LIHC, UCEC, and KIRP. To obtain more intuitive results, we use the R project to analyse the correlation between the expression of DLEU2 and the prognosis of human cancer and the tumor sample size, HR, CI and P values were extracted to draw the forest plots. Forest plots showed that DLEU2 was a hazard for OS (HR = 1.89, P < 0.001), disease-specific survival (DSS) (HR = 1.97, P = 0.01) and progression-free interval (PFI) (HR = 1.37, P = 0.05) in KIRC and OS (HR = 1.56, P = 0.011), DSS (HR = 1.68, P = 0.024) (Figure 5A–C). These findings suggest that DLEU2 expression can predict a poor outcome of KIRC and LIHC.
DLEU2 Correlated with Immune Infiltration in KIRC and LIHC
It has been previously reported that tumour-infiltrated immune cells can be used to predict sentinel node involvement and the survival of cancer patients.27 Therefore, we analysed the correlation between DLEU2 expression and immune infiltration in various cancers in the TIMER database. The results showed that expression of DLEU2 in KIRC correlated significantly positively with the infiltration of B cells (r=0.133, p=4.30e−03), CD8+ T cells (r=0.286, p=1.15e−09), CD4+ T cells (r=0.276, p=1.67e−09), macrophages (r=0.182, p=1.03e−04), neutrophils (r=0.428, p=8.34e−22), and dendritic cells (r=0.268, p=6.17e−09) (Figure 6A). In addition, expression of DLEU2 correlated significantly positively with immune-infiltrated B cells (r=0.348, p=2.96e−11), CD8+ T cells (r=0.3, p=1.58e−08), CD4+ T cells (r=0.291, p=3.89e−08), macrophages (r=0.334, p=2.39e−10), neutrophils (r=0.269, p=3.77e−07), and dendritic cells (r=0.366, p=3.42e−12) in LIHC (Figure 6B).
DLEU2 Correlated with Specific Markers of Immune Cells
We searched the TIMER website platform to analyse the correlation between DLEU2 expression and molecular markers of infiltrating immune cells in KIRC and LIHC. Our results revealed that the expression of DLEU2 in KIRC correlated significantly with the immune markers of B cells, CD8+ T cells, dendritic cells, M2 macrophages, monocytes, neutrophils, T cell exhaustion, Tfh cells, Th1 cells, and Tregs (Table 3). In LIHC, DLEU2 expression correlated positively with the immune markers of B cells, CD8+ T cells, dendritic cells, monocytes, T cell exhaustion, TFH cells, Th1 cells, and Tregs (Table 3). Notably, we found that DLEU2 expression was significantly associated with molecular markers of various tumour-infiltrating immune cell subtypes, including CD8+ T cell markers, CD8A, CD8B, T cell (including general and exhausted) markers, CD3D, CD3E, CD2, GZMB, LAG-3, PD-1, Th (Th1 and Th2) cell markers, TBX21, STAT4, STAT1, IFNG, IL13, GATA3, STAT6, Treg markers, FOXP3, CCR8, STAT5B, TGFB1, B cell markers, CD19, CD79A, DC markers, ITGAX, NRP1, CD1C, HLA−DPA1, HLA−DRA, HLA−DQB1, neutrophil marker CCR7, ITGAM, CEACAM8, and HLA−DPB1 in KIRC. Similar to KIRC, the expression level of DLEU2 in LIHC also correlated positively with immune markers of various T-cell subtypes, such as CD8+ T cells, T cells (general), T cell exhaustion, Tfh cells, Th1 cells, Th2 cells, and Tregs. In addition, DLEU2 expression in LIHC was significantly associated with B cell, dendritic cell and monocyte immune markers. These findings suggest that DLEU2 expression possibly modulates immune infiltration in KIRC and LIHC.
 |
Table 3 Correlation Analysis of DLEU2 Expression with Molecular Markers of Immune Cells |
Prognostic Potential of DLEU2 in Different Cancers Based on Different Enriched Immune Cells
This study shows that high DLEU2 expression indicates a poor prognosis of KIRC and LIHC and is associated with immune cell infiltration in tumours. We hypothesized that DLEU2 influences the prognosis of KIRC and LIHC by, or at least partially by, influencing immune infiltration in the TME. We performed survival analysis of DLEU2 expression in KIRC and LIHC using the Kaplan–Meier Plotter website in the order of enrichment of B cells, CD4+ memory T cells, CD8+ T cells, macrophages, NK T cells, Treg T cells, Th1 cells, and Th2 cells. Surprisingly, high expression of DLEU2 in KIRC was associated with enriched B cells (p=0.0023), CD4+ memory T cells (p=4.5e-06), CD8+ T cells (p=2.7e-05), macrophages (p=1.1e-05), NK T cells (p=0.0011), Treg T cells (p=0.00054), and Th1 cells (p=0.013), and this cohort had a poor prognosis (Figure 7A–H). High expression of DLEU2 in enriched B cells (p=0.00028), CD4+ memory T cells (p=0.0037), CD8+ T cells (p=0.004), macrophages (p=0.039), NK T cells (p=0.038), Treg T cells (p=0.0069), Th1 cells (p=0.01), and Th2 cells (p=0.011) was also associated with a poor prognosis in LIHC (Figure 7I–P). These findings indicate that immune infiltration may, at least partially, influence the prognosis of KIRC and LIHC with high DLEU2 expression.
CNV, Somatic Mutation, and Methylation Analyses of DLEU2
This study revealed that DLEU2 is aberrantly expressed in KIRC and LIHC. We further explored the causes of this dysregulation of DLEU2 expression in KIRC and LIHC. Previous studies have reported that CNVs, somatic mutations, and DNA methylation play a crucial role in genetic and epigenetic regulation, especially DNA methylation and CNVs, and correlate closely with oncogenesis and progression.28–30 We searched for CNV levels, somatic mutations and DNA methylation of DLEU2 in KIRC and LIHC using the UCSC Xena website platform. The heat map (Figure 8) results showed that expression of DLEU2 correlated closely with CNVs and DNA methylation, but not with somatic mutations, in both KIRC and LIHC. In addition, we analysed methylation levels of DLEU2 in KIRC and LIHC samples compared with their normal samples in the UALCAN database, and the results showed that the aberrant expression of DLEU2 was significantly related to KIRC (p=0.004) and LIHC (p=0.01) methylation (S3). In summary, CNVs and methylation may contribute to abnormal DLEU2 expression.
Discussion
DLEU2 is a long noncoding RNA initially reported to be deleted in lymphocytic leukaemia.31 DLEU2 has been reported to regulate the evolution or progression of several types of cancer.32 This study is the first to analyse the expression of DLEU2 and its correlation with clinically relevant indicators, prognosis, immune infiltration, and gene variation in KIRC and LIHC.
In this study, we found that the expression of DLEU2 was higher in KIRC and LIHC samples than in matched normal samples and that the expression of DLEU2 was associated with the TNM stage of KIRC (Figure 1C–F). In addition, DLEU2 expression correlated with the T stage, histological grade, and AFP level of LIHC (Figure 1G–J). ROC curve analysis (Figure 2) shows that DLEU2 has a certain accuracy in predicting KIRC and LIHC. The prognostic nomogram and 1-, 2-, and 3-year calibration curves for KIRC and LIHC revealed the satisfactory performance of the DLEU2-based nomogram (Figure 3). These findings show that DLEU2 is closely related to the progression of KIRC and LIHC and that DLEU2 correlates with the prognosis of KIRC and LIHC and has the potential to be a significant diagnostic and prognostic marker of KIRC and LIHC.
The prognosis analyses of DLEU2 suggested that high expression of DLEU2 was associated with a poor prognosis of KIRC and LIHC (Figures 4 and 5). These results are consistent with previous reports showing that DLEU2 can promote malignant phenotypes, such as tumour growth, migration and invasion. Dong et al reported that DLEU2 could be a competitive RNA to influence the EMT and glycolysis in endometrial cancer.33 Han et al reported that DLEU2 could promote malignant behaviours in gastric cancer cells.13 Our results demonstrate the potential of DLEU2 to predict the outcomes of KIRC and LIHC.
DLEU2 has been previously reported to affect the proliferation and differentiation of immune cells.34 The correlation analysis between DLEU2 and immune cells showed that DLEU2 expression correlated positively with 6 types of immune infiltrating cells in KIRC and LIHC (Figure 6A and B). These results suggest that DLEU2 mediates immune cell infiltration in KIRC and LIHC.
Then, we analysed the association between DLEU2 and immune cell markers in KIRC and LIHC (Table 3). After the cell purity calibration, we found that DLEU2 was positively associated with several immune cell molecular markers. This finding further supports the correlation between DLEU2 and immune infiltration in KIRC and LIHC. Notably, our results show that DLEU2 correlates positively with markers of Treg and T cell exhaustion, such as TGFB1 and CTLA4. TGFB1 and CTLA4 have been shown to play important immunoregulatory roles in immune microloops. CTLA4 has been reported to mediate T cell apoptosis and inhibit T cell proliferation and cytokine release to mediate tumour immune escape, while TGFB1 enhances this effect by increasing the expression of CTLA4.12 It has been previously reported that T cells play an important role in immune regulation in the tumour microenvironment, such as the recruitment of regulatory T cells (Tregs) in the immune microenvironment by limiting the signal transduction of effector T cells and their killing effect on tumour cells.35 Yang et al reported that a high proportion of DC cells and Treg cells and T cell exhaustion promote the tumour microenvironment in bladder cancer.36 Our results also show that the expression of DLEU2 was associated with immune markers of CD8+ T cells, Tfh cells, Th1 cells, and Th2 cells. These findings shed light on the potential mechanism by which DLEU2 mediates immune cell infiltration in KIRC and LIHC. Therefore, DLEU2 may contribute to a poor prognosis of KIRC and LIHC by recruiting or regulating the infiltration of immune cells in the TME.
The results obtained using the Kaplan–Meier plotter website platform showed that high expression of DLEU2 enriched in several immune cell cohorts of KIRC and LIHC was associated with a poor prognosis (Figure 7). Tregs can limit the immune system’s immunity to tumours and increase the body’s immune tolerance to tumours, leading to the immune escape of tumours.35 DCs can lead to tumour immunosuppression and promote tumour growth and metastasis by increasing Tregs and activating CD8+ T cells.37 Moreover, M2 tumour-associated macrophages can increase the activity of tumour cells and mediate the EMT process in tumours.38 TGFB1 is an important immune marker of Tregs, and expression of DLEU2 correlates positively with TGFB1. Bao et al showed that TGFB1 induces immune escape in LIHC by increasing the expression of PD-1 and CTLA-4 on T cells.12 Moreover, several studies have reported that macrophages, Tregs, and CD8+ T cells in the KIRC microenvironment can promote tumour growth and immune escape.39–41 These results suggest that DLEU2 may influence the prognosis of KIRC and LIHC by regulating immune cell infiltration in the TME.
CNVs, gene mutations and DNA methylation are important mechanisms that mediate carcinogenesis.42 In our study, the heat map (Figure 8) showed that DLEU2 expression was associated with CNVs and DNA methylation but not somatic mutations. DLEU2 methylation leads to the dysregulation of its expression, which mediates disease progression.43 CNVs are an important marker of carcinogenesis and can lead to aberrant copy numbers, including amplification, insertion, loss, and deletion. Moreover, studies have shown that CNVs play an important role in the progression of cancers, such as lung adenocarcinoma, breast cancer, and ovarian cancer.29,44,45
Conclusion
DLEU2 is upregulated in various cancers, especially KIRC and LIHC, and its aberrantly upregulated expression is associated with poor clinicopathologic features, a poor prognosis, and high immune cell infiltration in KIRC and LIHC. Copy number variations and DNA methylation may contribute to the high expression of DLEU2 in KIRC and LIHC. Furthermore, our study revealed that DLEU2 expression correlates with the poor outcome of KIRC and LIHC by influencing immune infiltration in the TME. Therefore, this study provides a new approach to the immunotherapy of KIRC and LIHC.
Abbreviations
AFP, Alpha-fetoprotein; CNV, Copy number variations; CI, Confidence intervals; DSS, Disease specific survival; KIRC, Kidney renal clear cell carcinoma; LIHC, Liver hepatocellular carcinoma; OS, Overall survival; PFI, Progression free interval; ROC, Receiver operating characteristic; TME, Tumour microenvironment.
Data Sharing Statement
All data or materials involved in the manuscript are freely available from the open database The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/).
Ethics Statement
Since all patient data in this study were obtained from public databases, informed consent was obtained from the patients before data disclosure, and literature related to the database was quoted when we invoked data in the database; thus, an ethical review of this study was exempted from the Ethics Committee of the First Affiliated Hospital of Nanchang University.
Funding
There is no funding to report.
Disclosure
The authors have no conflicts of interest or competing financial interests in this study to declare.
References
1. Federico P, Petrillo A, Giordano P, et al. Immune checkpoint inhibitors in hepatocellular carcinoma: current status and novel perspectives. Cancers. 2020;12(10):3025. doi:10.3390/cancers12103025
2. Xu T, Gao S, Ruan H, et al. METTL14 acts as a potential regulator of tumor immune and progression in clear cell renal cell carcinoma. Front Genet. 2021;12:609174. doi:10.3389/fgene.2021.609174
3. Thouvenin J, Martínez Chanzá N, Alhalabi O, et al. Efficacy of immune checkpoint inhibitors in upper tract urothelial carcinomas: current knowledge and future directions. Cancers. 2021;13(17):4341. doi:10.3390/cancers13174341
4. Yu EM, Linville L, Rosenthal M, Aragon-Ching JB. A contemporary review of immune checkpoint inhibitors in advanced clear cell renal cell carcinoma. Vaccines. 2021;9(8):919. doi:10.3390/vaccines9080919
5. Zheng X, Jin W, Wang S, Ding H. Progression on the roles and mechanisms of tumor-infiltrating T lymphocytes in patients with hepatocellular carcinoma. Front Immunol. 2021;12:729705. doi:10.3389/fimmu.2021.729705
6. Nishikawa H, Koyama S. Mechanisms of regulatory T cell infiltration in tumors: implications for innovative immune precision therapies. J Immunother Cancer. 2021;9(7):e002591.
7. Peng B, Yan Y, Xu Z. The bioinformatics and experimental analysis of AlkB family for prognosis and immune cell infiltration in hepatocellular carcinoma. PeerJ. 2021;9:e12123. doi:10.7717/peerj.12123
8. Wang B, Chen D, Hua H. TBC1D3 family is a prognostic biomarker and correlates with immune infiltration in kidney renal clear cell carcinoma. Mol Ther Oncolytics. 2021;22:528–538. doi:10.1016/j.omto.2021.06.014
9. Wang S, Ma X, Ying Y, et al. Upregulation of ARNTL2 is associated with poor survival and immune infiltration in clear cell renal cell carcinoma. Cancer Cell Int. 2021;21(1):341. doi:10.1186/s12935-021-02046-z
10. Jacquier A, Lambert T, Delattre J-F, et al. Tumor infiltrating and peripheral CD4ILT2 T cells are a cytotoxic subset selectively inhibited by HLA-G in clear cell renal cell carcinoma patients. Cancer Lett. 2021;519:105–116. doi:10.1016/j.canlet.2021.06.018
11. Xue C, Gao P, Cui X, et al. ASRGL1 correlates with immune cell infiltration in hepatocellular carcinoma and can serve as a prognostic biomarker. Front Oncol. 2021;11:680070. doi:10.3389/fonc.2021.680070
12. Bao S, Jiang X, Jin S, Tu P, Lu J. TGF-β1 induces immune escape by enhancing PD-1 and CTLA-4 expression on T lymphocytes in hepatocellular carcinoma. Front Oncol. 2021;11:694145. doi:10.3389/fonc.2021.694145
13. Han S, Qi Y, Xu Y, et al. lncRNA DLEU2 promotes gastric cancer progression through ETS2 via targeting miR-30a-5p. Cancer Cell Int. 2021;21(1):376. doi:10.1186/s12935-021-02074-9
14. He M, Wang Y, Cai J, et al. LncRNA DLEU2 promotes cervical cancer cell proliferation by regulating cell cycle and NOTCH pathway. Exp Cell Res. 2021;402(1):112551. doi:10.1016/j.yexcr.2021.112551
15. Yang J, Huang Y, Dong B, Dai Y. Long noncoding RNA DLEU2 drives the malignant behaviors of thyroid cancer through mediating the miR-205-5p/TNFAIP8 axis. Endocr Connect. 2021;10(4):471–483. doi:10.1530/EC-21-0046
16. Guo Y, Bai M, Lin L, et al. LncRNA DLEU2 aggravates the progression of hepatocellular carcinoma through binding to EZH2. Biomed Pharmacother. 2019;118:109272. doi:10.1016/j.biopha.2019.109272
17. Salerno D, Chiodo L, Alfano V, et al. Hepatitis B protein HBx binds the DLEU2 lncRNA to sustain cccDNA and host cancer-related gene transcription. Gut. 2020;69(11):2016–2024. doi:10.1136/gutjnl-2019-319637
18. Chen Z, Zhang J, Zhang Z, et al. The putative tumor suppressor microRNA-30a-5p modulates clear cell renal cell carcinoma aggressiveness through repression of ZEB2. Cell Death Dis. 2017;8(6):e2859. doi:10.1038/cddis.2017.252
19. Li G, Zhang Z, Chen Z, Liu B, Wu H. LncRNA DLEU2 is activated by STAT1 and induces gastric cancer development via targeting miR-23b-3p/NOTCH2 axis and Notch signaling pathway. Life Sci. 2021;277:119419. doi:10.1016/j.lfs.2021.119419
20. Li X, Xu F, Meng Q, et al. Long noncoding RNA DLEU2 predicts a poor prognosis and enhances malignant properties in laryngeal squamous cell carcinoma through the miR-30c-5p/PIK3CD/Akt axis. Cell Death Dis. 2020;11(6):472. doi:10.1038/s41419-020-2581-2
21. Mian M, Scandurra M, Chigrinova E, et al. Clinical and molecular characterization of diffuse large B-cell lymphomas with 13q14.3 deletion. Ann Oncol. 2012;23(3):729–735. doi:10.1093/annonc/mdr289
22. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi:10.1016/S1476-5586(04)80047-2
23. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi:10.1093/nar/gkaa407
24. Hou GX, Liu P, Yang J, Wen S. Mining expression and prognosis of topoisomerase isoforms in non-small-cell lung cancer by using Oncomine and Kaplan-Meier plotter. PLoS One. 2017;12(3):e0174515. doi:10.1371/journal.pone.0174515
25. Wang S, Xiong Y, Zhao L, et al. UCSCXenaShiny: an R/CRAN package for interactive analysis of UCSC xena data. Bioinformatics. 2021. doi:10.1093/bioinformatics/btab561
26. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
27. Azimi F, Scolyer RA, Rumcheva P, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol. 2012;30(21):2678–2683. doi:10.1200/JCO.2011.37.8539
28. Zhou F, Shen D, Xiong Y, et al. CTHRC1 is a prognostic biomarker and correlated with immune infiltrates in kidney renal papillary cell carcinoma and kidney renal clear cell carcinoma. Front Oncol. 2020;10:570819. doi:10.3389/fonc.2020.570819
29. Li N, Zeng Y, Tai M, et al. Analysis of the prognostic value and gene expression mechanism of in lung adenocarcinoma. Front Mol Biosci. 2021;8:688274. doi:10.3389/fmolb.2021.688274
30. Koelsche C, Benhamida JK, Kommoss FKF, et al. Intimal sarcomas and undifferentiated cardiac sarcomas carry mutually exclusive MDM2, MDM4, and CDK6 amplifications and share a common DNA methylation signature. Mod Pathol. 2021. doi:10.1038/s41379-021-00874-y
31. Liu Y, Corcoran M, Rasool O, et al. Cloning of two candidate tumor suppressor genes within a 10 kb region on chromosome 13q14, frequently deleted in chronic lymphocytic leukemia. Oncogene. 1997;15(20):2463–2473. doi:10.1038/sj.onc.1201643
32. Xu W, Wang B, Cai Y, Guo C, Liu K, Yuan C. DLEU2: a meaningful long noncoding RNA in oncogenesis. Curr Pharm Des. 2021;27(20):2337–2343. doi:10.2174/1381612826666201026150857
33. Dong P, Xiong Y, Konno Y, et al. Long non-coding RNA DLEU2 drives EMT and glycolysis in endometrial cancer through HK2 by competitively binding with miR-455 and by modulating the EZH2/miR-181a pathway. J Exp Clin Cancer Res. 2021;40(1):216. doi:10.1186/s13046-021-02018-1
34. Klein U, Lia M, Crespo M, et al. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17(1):28–40. doi:10.1016/j.ccr.2009.11.019
35. Poli A, Abdul-Hamid S, Zaurito AE, et al. PIP4Ks impact on PI3K, FOXP3, and UHRF1 signaling and modulate human regulatory T cell proliferation and immunosuppressive activity. Proc Natl Acad Sci U S A. 2021;118(31):e2010053118. doi:10.1073/pnas.2010053118
36. Yang C, Wu S, Mou Z, et al. Transcriptomic analysis identified ARHGAP family as a novel biomarker associated with tumor-promoting immune infiltration and nanomechanical characteristics in bladder cancer. Front Cell Dev Biol. 2021;9:657219. doi:10.3389/fcell.2021.657219
37. Sawant A, Hensel JA, Chanda D, et al. Depletion of plasmacytoid dendritic cells inhibits tumor growth and prevents bone metastasis of breast cancer cells. J Immunol. 2012;189(9):4258–4265. doi:10.4049/jimmunol.1101855
38. Xing Z, Zhang M, Liu J, Liu G, Feng K, Wang X. LINC00337 induces tumor development and chemoresistance to paclitaxel of breast cancer by recruiting M2 tumor-associated macrophages. Mol Immunol. 2021;138:1–9. doi:10.1016/j.molimm.2021.07.009
39. Xu W, Liu WR, Xu Y, et al. Hexokinase 3 dysfunction promotes tumorigenesis and immune escape by upregulating monocyte/macrophage infiltration into the clear cell renal cell carcinoma microenvironment. Int J Biol Sci. 2021;17(9):2205–2222.
40. Zhu Q, Cai MY, Weng DS, et al. PD-L1 expression patterns in tumour cells and their association with CD8 tumour infiltrating lymphocytes in clear cell renal cell carcinoma. J Cancer. 2019;10(5):1154–1161. doi:10.7150/jca.29052
41. Qi Y, Xia Y, Lin Z, et al. Tumor-infiltrating CD39+CD8+ T cells determine poor prognosis and immune evasion in clear cell renal cell carcinoma patients. Cancer Immunol Immunother. 2020;69(8):1565–1576. doi:10.1007/s00262-020-02563-2
42. Cui K, Wu X, Gong L, et al. Comprehensive characterization of integrin subunit genes in human cancers. Front Oncol. 2021;11:704067. doi:10.3389/fonc.2021.704067
43. Morenos L, Chatterton Z, Ng JL, et al. Hypermethylation and down-regulation of DLEU2 in paediatric acute myeloid leukaemia independent of embedded tumour suppressor miR-15a/16-1. Mol Cancer. 2014;13:123. doi:10.1186/1476-4598-13-123
44. Graf RP, Eskander R, Brueggeman L, Stupack DG. Association of copy number variation signature and survival in patients with serous ovarian cancer. JAMA Netw Open. 2021;4(6):e2114162. doi:10.1001/jamanetworkopen.2021.14162
45. Pariyar M, Johns A, Thorne RF, Scott RJ, Avery-Kiejda KA. Copy number variation in triple negative breast cancer samples associated with lymph node metastasis. Neoplasia. 2021;23(8):743–753. doi:10.1016/j.neo.2021.05.016
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.