Back to Journals » OncoTargets and Therapy » Volume 17
Prognostic Factors and Construction of Nomogram Prediction Model of Lung Cancer Patients Using Clinical and Blood Laboratory Parameters
Authors Zhang Y, Wan W, Shen R, Zhang B, Wang L, Zhang H, Ren X, Cui J, Liu J
Received 25 October 2023
Accepted for publication 31 January 2024
Published 21 February 2024 Volume 2024:17 Pages 131—144
DOI https://doi.org/10.2147/OTT.S444396
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Nagashree Seetharamu
Yamin Zhang,1 Wei Wan,1 Rui Shen,1 Bohao Zhang,1 Li Wang,1 Hongyi Zhang,2 Xiaoyue Ren,3 Jie Cui,4,* Jinpeng Liu1,*
1Department of Oncology, Xi’an International Medical Center Hospital, Xi’an, Shaanxi, 710100, People’s Republic of China; 2Department of Urology, The First Affiliated Hospital of Xi’an Medical University, Xi’an, Shaanxi, 710077, People’s Republic of China; 3College of Life Sciences, Northwest University, Xi’an, Shaanxi, 710069, People’s Republic of China; 4Department of Oncology, The First Affiliated Hospital of Xi’an Medical University, Xi’an, Shaanxi, 710077, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jie Cui, Department of Oncology, The First Affiliated Hospital of Xi’an Medical University, No. 48 Fenghao West Road, Lianhu District, Xi’an, Shaanxi, 710077, People’s Republic of China, Tel/Fax +86-15209277121, Email [email protected] Jinpeng Liu, Department of Oncology, Xi’an International Medical Center Hospital, No. 777, Xitai Road, Chang’an District, Xi’an, Shaanxi, 710100, People’s Republic of China, Tel/Fax +86-13772079179, Email [email protected]
Objective: This work aimed to explore the prognostic risk factors of lung cancer (LC) patients and establish a line chart prediction model.
Methods: A total of 322 LC patients were taken as the study subjects. They were randomly divided into a training set (n = 202) and a validation set (n = 120). Basic information and laboratory indicators were collected, and the progression-free survival (PFS) and overall survival (OS) were followed up. Single-factor and cyclooxygenase (COX) multivariate analyses were performed on the training set to construct a Nomogram prediction model, which was validated with 120 patients in the validation set, and Harrell’s consistency was analyzed.
Results: Single-factor analysis revealed significant differences in PFS (P< 0.05) between genders, body mass index (BMI), carcinoembryonic antigen (CEA), cancer antigen 125 (CA125), squamous cell carcinoma antigen (SCCA), treatment methods, treatment response evaluation, smoking status, presence of pericardial effusion, and programmed death ligand 1 (PD-L1) at 0 and 1– 50%. Significant differences in OS (P< 0.05) were observed for age, tumor location, treatment methods, White blood cells (WBC), uric acid (UA), CA125, pro-gastrin-releasing peptide (ProGRP), SCCA, cytokeratin fragment 21 (CYFRA21), and smoking status. COX analysis identified male gender, progressive disease (PD) as treatment response, and SCCA > 1.6 as risk factors for LC PFS. The consistency indices of the line chart models for predicting PFS and OS were 0.782 and 0.772, respectively.
Conclusion: Male gender, treatment response of PD, and SCCA > 1.6 are independent risk factors affecting the survival of LC patients. The PFS line chart model demonstrates good concordance.
Keywords: laboratory parameters, LC, nomogram prediction model, KM analysis, cox multivariate analysis
Introduction
Since the middle of the 20th century, the incidence and mortality of LC have grown rapidly, thus, LC has become one of the cancers with a high incidence worldwide and one of the cancers with the highest proportion of cancer-related deaths1. Although the incidence of LC in Europe and the United States has decreased to some extent in recent years, the incidence of LC in China is still very high, which is closely related to the high number of smokers in China.2 Many clinical studies have found that LC patients must achieve “three early”, that is, early detection, early diagnosis, and early treatment, in order to achieve good therapeutic results.3,4 However, the prognosis for most LC patients is still very poor. This is mainly because the early symptoms of LC are not obvious and have been overlooked by patients, while when patients enter the hospital, about 75% of patients are already in the late stage of LC, and the prognosis of the late stage is relatively poor.5 Therefore, the threat caused by LC to people’s lives and health has become a non-negligible problem in clinical practice, and improving the prognosis of patients with LC necessary.
According to histopathological classification, LC is mainly divided into small cell LC (SCLC) and non-small cell LC (NSCLC) (including squamous cell carcinoma (SCC), adenocarcinoma, and large cell carcinoma). Among them, SCLC occurs in approximately 15% of all LC patients, is the most malignant form, and proliferates and metastasizes rapidly, and has a poor prognosis.6,7 NSCLC has a high incidence of approximately 85% among LC diseases, and although LC is less malignant and metastasizes later than SCLC, it still has a high mortality and recurrence rate.8,9 Therefore, both SCLC and NSCLC have the potential for a poor prognosis. At present, the TNM staging system is mainly used to evaluate the prognosis of LC patients in clinical practice,10 that is, the prognosis of LC patients is evaluated by evaluating the primary site of LC tumor (T stage), lymph node involvement (N stage), and whether distant metastasis occurs (M stage), and the application effect is good. Of course, in addition to oncological factors, other factors such as the patient’s treatment modality, sex, age, and laboratory parameters may also have an impact on the prognosis of patients with LC. Some experts have proposed that routine blood tests can predict the prognosis of nasopharyngeal carcinoma patients.11 Other studies have confirmed biomarkers for LC prognosis and risk stratification of leukocyte cholesterol 25-hydroxylase (CH25H).12 Although there have been many studies on prognostic factors in LC, there is still a lack of effective models to predict the prognosis of LC patients with LC. The proposed nomogram prediction model offers the possibility of solving this problem.13 A nomogram can be used to graphically display complex formulas quickly, intuitively, and accurately. It is often used in the medical field for the prediction of adverse events of diseases, such as the prediction of adverse events of chronic heart failure.14 At present, the nomogram prediction model has also been studied for the prediction of clinical metastasis of NSCLC,15 and the novel International Association for the Study of Lung Cancer (IASLC) grade of invasive lung adenocarcinoma,16 all of which reflect a good adjuvant effect in the treatment of patients.
Based on the above, the prognostic risk factors of patients were analyzed by collecting the LC of blood laboratory parameters of clinical patients as well as the prognosis of patients. A nomogram prediction model for LC prognosis was also constructed, and the Nomogram prediction model was internally validated to assess its practicability. It provides the possibility of improving the survival rate of patients by improving the treatment pertinence of clinical patients and providing a more effective means of disease evaluation in clinical practice.
Study Methods
Study Subjects
A total of 202 LC patients were diagnosed at the Xi’an International Medical Center Hospital between June 2019 and April 2022, and 120 patients from The First Affiliated Hospital of Xi’an Jiaotong University between October 2019 and April 2022 were retrospectively studied. The general clinical conditions of all patients (such as gender, age, body mass index (BMI), Eastern Cooperative Oncology Group (ECOG) score, smoking status, alcohol consumption, tumor conditions (including tumor location, pathological classification, and metastatic site), presence of PE/PF, etc.), treatment methods (immunotherapy alone/in combination with other therapies), number of treatment lines, adverse reactions during immunotherapy, clinical efficacy evaluation results, whether they died due to severe immune-related adverse events (irAEs), programmed death ligand 1 (PD-L1) test results, LC gene (epidermal growth factor receptor (EGFR)/anaplastic lymphoma kinase (ALK)/ Kirsten rat sarcoma viral oncogene homologue (KRAS)/TP53, etc.) expression, whether antibiotics are used and blood laboratory parameters (blood routine, liver and kidney function, coagulation, myocardial enzymes, thyroid, tumor markers, etc.) test results; through telephone or hospital follow-up to obtain the patient’s disease progression or death. A total of 202 patients from one center were included in the training set for statistical analysis and the construction of a Nomogram prediction model. Subsequently, the Nomogram prediction model was validated using a separate validation set of 120 patients from the other center. This study was conducted in accordance with the Declaration of Helsinki. As this study is a retrospective study, the data are anonymous and informed consent is not required. The Ethics Committee of Xi’an International Medical Center Hospital approved this retrospective study (GJYX-KY-2023-057).
Inclusion and Exclusion Criteria
(1) Inclusion criteria: All patients were diagnosed by lung biopsy; complete blood routine, liver and kidney function, tumor markers, and other test data of patients before treatment; patients had at least once, or a course of treatment; patients received subsequent follow-up investigation, and follow-up data were complete.
(2) Exclusion criteria: Patients with pneumonia, advanced liver disease, and severely abnormal liver function before treatment; patients with severe blood and immune system diseases before treatment and patients with other tumor diseases.
Grouping of Training Set
(1) Grouping according to general clinical data
Gender: male patients (1); female patients (0).
Age: ≤ 64 years (0); > 64 years (1).
BMI: ≤ 22.3 kg/m2 (0); > 22.3 kg/m2 (1).
ECOG score: 0 ~ 1 point (0); 2 points (1).
Smoking or not: smokers (1); non-smokers (0).
Alcohol drinking or not: drinkers (1); non-drinkers (0).
Pathological types of tumors: patients with SCLC (0), SCC (1), adenocarcinoma (2), and other pathological types (3).
Tumor metastasis: non-metastatic (0), metastasis to one organ (1), metastasis to two organs (2), and metastasis to three or more organs (3).
Treatment: Patients treated with immune monotherapy (0); patients treated with immunotherapy combined with radiotherapy/chemotherapy/bevacizumab (1).
Clinical efficacy evaluation: Patients with complete response (CR), partial response (PR), stable disease (SD) (0); and patients with progressive disease (PD) (1).
Immune adverse reactions: presence of irAE (1); absence of irAE (0).
Death due to irAEs: patients who died due to irAEs (1); other patients (0).
PD-L1 assay results:17 patients with TPS 0 (0); 1% ≤ patients with TPS < 50% (1); patients with TPS >50% (2).
LC gene detection: patients with EGFR/ALK/KRAS/TP53 results of 0 (0); patients with other EGFR/ALK/KRAS/TP53 results (1).
(2) Blood laboratory index examination data
These mainly include routine blood tests, liver function, renal function, myocardial enzymes, inflammatory index, coagulation function, tumor markers, and thyroid function tests.
Routine blood tests included the neutrophil/lymphocyte ratio (NLR), platelet/lymphocyte ratio (PLR), monocyte/lymphocyte ratio (MLR), eosinophil/lymphocyte ratio (ELR), systemic immune inflammatory index (SII), prognostic nutritional index (PNI), acute lung injury (ALI), White blood cell count (WBC), neutrophil count (NEUT), hemoglobin (HB), platelet count (PLT), lymphocyte count (LY), eosinophil (EO), basophil (BASO), and monocyte ratio (MON).
Liver and kidney function tests included gamma-glutamyl transferase (GGT), alkaline phosphatase (ALP), serum prealbumin (PA), albumin (ALB), and blood uric acid (UA).
Myocardial enzyme examination revealed, creatine kinase MB (CK-MB), lactate dehydrogenase (LDH).
Inflammatory markers: C-reactive protein (CRP), and procalcitonin (PCT).
Coagulation function tests: D-dimer (D-D), fibrinogen (FIB), and fibrin degradation products (FDP).
Tumor marker examination revealed, carcinoembryonic antigen (CEA), carbohydrate antigen 125 (CA125), pro-gastrin-releasing peptide (ProGRP), squamous cell carcinoma antigen (SCCA), cytokeratin fragment 21 (CYFRA21).
Thyroid function tests included serum total triiodothyronine (T3), free thyroxine (T4), thyroid-stimulating hormone (TSH).
Follow-Up
All patients were followed up via telephone at the hospital or home, or consultation at the hospital. The follow-up cutoff was July 2022 or the patient died. PFS was assessed from the time of treatment initiation until disease recurrence or death, and OS was assessed from the time of treatment initiation until death. The PFS and OS of patients in the above groups were analyzed to evaluate the relationship between their clinical conditions, blood laboratory parameters, PFS, and OS.
Statistical Methods
All data were imported into Excel software and subjected to statistical analysis using SPSS 19.0. Descriptive statistics for continuous variables were expressed as mean ± standard deviation, and t-tests were employed for analysis. Multivariate analysis was conducted using Cox regression to identify independent prognostic factors. Independent risk factors influencing patient prognosis were determined, and a significance level of P<0.05 was considered statistically significant. The Nomogram prediction model was constructed using R software based on the selected variables. The model was then validated using a validation set consisting of 120 lung cancer (LC) patients. The predictive value of the column line chart model was evaluated using the Harrell concordance index.18
Results
Analysis of Patient Survival
In this work, the PFS time for 202 LC patients ranged from 1 to 32 months, with a median PFS time of 7 months. The OS time ranged from 2 to 39 months, with a median OS time of 12 months. The overall survival status of all patients is illustrated in Figure 1.
 |
Figure 1 Survival analysis. (A): PSF time, (B) OS time). |
Statistical Situation of PFS in Each Group
Statistical results of PFS in the General Case Group
As indicated in Table 1, significant statistical differences in PFS were observed among different genders, BMI levels, treatment methods, treatment response, smoking status, and the presence of pericardial effusion, as well as between TPS 0 and 1% versus TPS 1% to 50% (P<0.05). However, no significant differences were observed with other indicators (P>0.05).
 |
Table 1 Comparison of PFS Under General Clinical Indicators Group |
Statistical Results of PFS Under Blood Routine Index Grouping
As can be observed from Figure 2, there was no significant statistical difference in PFS among different levels of NLR, PLR, MLR, ELR, SII, PNI, ALI, WBC, NEUT, HB, PLT, LY, EO, BASO, and MON (P>0.05).
 |
Figure 2 The statistical analysis of PFS under the grouping of routine blood parameters. (A–O) NLR, PLR, MLR, ELR, SII, PNI, ALI, WBC, NEUT, HB, PLT, LY, EO, BASO, and MON). |
PFS Statistical Results of Liver, Kidney, Myocardium, Inflammation, and Coagulation Indexes
From Figure 3, it can be observed that there are no significant statistical differences in progression-free survival (PFS) among different levels of GGT, ALP, PA, ALB, UA, CK-MB, LDH, CRP, PCT, D-D, FIB, and FDP (P>0.05).
 |
Figure 3 The statistical analysis of PFS under the grouping of liver, kidney, cardiac, inflammation, and coagulation indicators. (A–L) GGT, ALP, PA, ALB, UA, CK-MB, LDH, CRP, PCT, D-D, FIB, and FDP). |
Statistical Results of PFS in Tumor Markers and Thyroid Function Groups
From Figure 4, it can be observed that there are no significant statistical differences in progression-free survival (PFS) among different levels of ProGRP, CYFRA21, T3, T4, and TSH (P>0.05). However, there are significant statistical differences in PFS among different levels of CEA, CA125, and SCCA (P<0.05).
OS Statistics of Patients in Each Group
General OS Statistics in a Group
As outlined in Table 2, significant statistical differences in OS were observed among different age groups, tumor locations, treatment methods, and smoking status (P<0.05). However, there were no significant differences with other indicators (P>0.05).
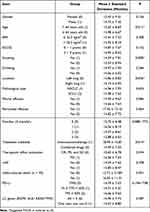 |
Table 2 Comparison of OS Under the Group of General Clinical Indicators |
The Statistical Results of OS Under the Grouping of Blood Routine Indexes
As illustrated in Figure 5, there were no significant statistical differences in OS among different levels of NLR, PLR, MLR, ELR, SII, PNI, ALI, NEUT, HB, PLT, LY, EO, BASO and MON (P>0.05), while there were statistical differences in OS among different levels of WBC (P<0.05).
PFS Statistical Results of Liver, Kidney, Myocardium, Inflammation, and Coagulation Indexes
From Figure 6, it can be observed that there are no significant statistical differences in OS among different levels of GGT, ALP, PA, ALB, CK-MB, LDH, CRP, PCT, D-D, FIB, and FDP (P>0.05). However, there is a statistically significant difference in OS among different levels of UA (P<0.05).
OS Statistical Results of Tumor Markers and Thyroid Function Groups
From Figure 7, it was evident that there are no significant statistical differences in OS among different levels of CEA, T3, T4, and TSH (P>0.05). However, there were significant statistical differences in OS among different levels of CA125, ProGRP, SCCA, and CYFRA21 (P<0.05).
Cox Multivariate Analysis
In the single-factor analysis influencing PFS and OS times, indicators with P<0.05 were subjected to Cox multivariate analysis. Table 3 demonstrated that male gender, treatment response assessed as PD, and SCCA > 1.6 were independent prognostic risk factors for PFS in LC patients. Table 4 indicates that there were no significant independent risk factors for OS time in LC patients (P>0.05).
 |
Table 3 Cox Multivariate Analysis of PFS Time |
 |
Table 4 Cox Multivariate Analysis of OS Time |
Concordance Assessment of Nomogram Prognostic Model
In this work, a validation set of 120 patients was utilized to validate the PFS Nomogram prognostic model. To assess the predictive value of the Nomogram prognostic model, we conducted inflammation treatment and employed the Harrell consistency index for evaluation. The consistency index for predicting PFS was 0.782 (95% CI: 0.743–0.804). A consistency index greater than 0.7 indicates a high similarity between the predicted results from the Nomogram and the actual results, with differences within an acceptable range.
Discussion
Due to LC being one of the malignancies with the poorest prognosis worldwide and having a high mortality rate, improving the prognosis of LC patients is a crucial topic in clinical research.19 The prerequisite for enhancing the prognosis of LC patients is understanding the risk factors that influence patient outcomes. Wang et al20 confirmed that higher age, advanced clinical TNM staging, increased expression of miRNA-21 and miRNA-155 in NSCLC tissues were independently associated with lower survival rates in NSCLC patients. Huang et al21 proposed that the pretreatment lymphocyte percentage is a reliable indicator for predicting cancer prognosis and an accurate response marker for lung adenocarcinoma and advanced cancer. Thus, factors influencing the prognosis of LC patients may include clinical baseline conditions and laboratory indicators. However, there is currently no comprehensive analysis study linking these factors with LC. Based on this study, general clinical data, blood laboratory indicators related to liver and kidney function, cardiac markers, inflammation, and tumor markers were collected and analyzed for 322 patients.
The progression of LC may impact other organs such as the liver and kidneys. Moreover, during the treatment process, drug metabolism and toxicity are associated with the liver and kidneys,22 prompting the collection of liver and kidney indicators. As LC is a systemic disease, it may also trigger a systemic inflammatory response23. Studies suggest that the occurrence of thyroid dysfunction may be higher in LC patients,24 leading to the collection of thyroid function indicators. Additionally, research proposes that LC patients may exhibit abnormal coagulation enzyme activity, possibly related to the tumor itself or changes in the anticoagulation mechanism. Abnormal coagulation enzyme activity is correlated with poor patient prognosis.25 PFS and OS are crucial indicators commonly used to assess the prognosis of cancer patients, closely linked to survival evaluation. In this study, the analysis examined the associations between PFS, OS, clinical factors, and laboratory indicators, evaluating the risk factors influencing PFS and OS. The results revealed that male gender, treatment response assessed as PD, and SCCA > 1.6 are independent prognostic risk factors for PFS in LC patients. The poorer prognosis in male patients may be associated with differences in sex hormones, immune response, and other factors. Studies indicate that androgens can promote the growth and differentiation of tumor cells, potentially making LC tumors more prone to proliferation and spreading in males, thereby increasing invasiveness. Additionally, gender differences in the immune system result in relatively weaker anti-cancer immune responses in males, affecting the survival period of LC, possibly related to the inhibitory effects of androgens on the immune system.26,27 A treatment response assessed as PD suggests that patients evaluated as having disease progression after treatment have a higher risk of progression-free survival compared to other assessment results (such as stability and partial remission). This aligns with the general understanding in clinical practice, typically associated with an adverse prognosis.28 SCCA is a biomarker associated with SCC and is commonly used to assess specific types of cancer. Studies indicate that elevated SCCA levels are associated with the malignancy, invasiveness, or other adverse biological characteristics of tumors. Increased SCCA is considered a marker of poor prognosis in LC patients, linked to disease progression, recurrence, or shortened survival.29
A Nomogram prognostic model, also known as a nomograph or a nomogram, can comprehensively analyze multiple quantitative and qualitative variables and predict the occurrence of a specific event (such as PFS and OS). It visually evaluates survival risk through graphical representation.30 The Nomogram prognostic model is based on the results of Cox regression analysis.31 According to the study findings, we further combined the complete dataset, including variables such as gender, treatment response (PD or other), SCCA levels, and PFS information for each patient. We fitted a Cox proportional hazards model, ensuring the continuity of each variable, and then extracted the model’s results to create a Nomogram, illustrating the impact of each variable. After establishing the Nomogram prognostic model, we evaluated its performance using the Harrell concordance index (C-index) on a validation set of 120 patients.32 The Harrell C-index is a metric used to assess the predictive performance of survival analysis models, such as the Cox proportional hazards model.33 It measures the accuracy of the model in ordering patient survival times, indicating whether the model correctly predicts longer survival times for patients with longer actual survival times among adjacent patients. The Harrell C-index ranges from 0 to 1, where 1 indicates perfect prediction of survival time order, 0.5 indicates predictions equivalent to random chance, and values below 0.5 indicate predictions opposite to actual outcomes. Upon evaluation, the consistency index for predicting PFS using the Nomogram model was 0.782, indicating good consistency. However, it is essential to note that the Harrell C-index has certain limitations, such as low sensitivity to model changes, the neglect of time dependence, and susceptibility to sample size influences.34,35 Therefore, further exploration will be conducted to validate more effective evaluation methods in future studies.
Conclusion
In summary, by analyzing the clinical and hematological laboratory parameters affecting LC prognosis, it was concluded that being male, having a treatment response assessed as PD, and SCCA > 1.6 are independent risk factors affecting the survival of LC patients. The PFS Nomogram model demonstrated a good fit. However, this result is contrary to the conclusions of other studies or normal logic, which may be because this is retrospective and unicentric, resulting in certain limitations in the observation of prognostic indicators, which causes errors in the conclusions. Therefore, a large-sample prospective validation study is required in the future. This study also provides a research basis for the nomogram model in the clinical application of prognostic evaluation in LC.
Acknowledgment
Jie Cui and Jinpeng Liu should be regard as co-corresponding authors for this study. We want to especially acknowledge all the participants in this study.
Funding
The work was supported by Xi’an Science and Technology Plan Project (Grant NO.21YXYJ0128), Xi’an International Medical Center Hospital-level topic (Grant NO:2021QN002).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Inoue A, Johnson TF, Walkoff LA, et al. Lung cancer screening using clinical photon-counting detector computed tomography and energy-integrating-detector computed tomography: a prospective patient study. J Comput Assist Tomogr. 2022;47(2):229–235. PMID: 36573321. doi:10.1097/RCT.0000000000001419
2. Boulate D, Fidelle M, Caramella C, et al. Epidemiological study to assess the prevalence of lung cancer in patients with smoking-associated atherosclerotic cardiovascular diseases: PREVALUNG study protocol. BMJ Open. 2022;12(12):e067191. PMID: 36572501. doi:10.1136/bmjopen-2022-067191
3. Shao F, Ling L, Li C, et al. Establishing a metastasis-related diagnosis and prognosis model for lung adenocarcinoma through CRISPR library and TCGA database. J Cancer Res Clin Oncol. 2022. PMID: 36574046. doi:10.1007/s00432-022-04495-z
4. Yadav DK, Bhadresha K, Rao P, Shaikh S, Rawal RM. Identification of hub genes associated with prognosis of lung cancer via integrated bioinformatics and in vitro approach. J Biomol Struct Dyn. 2022;26:1–15. PMID: 36572419. doi:10.1080/07391102.2022.2160816
5. Devi-Marulkar P, Fastenackels S, Karapentiantz P, et al. Regulatory T cells infiltrate the tumor-induced tertiary lymphoïd structures and are associated with poor clinical outcome in NSCLC. Commun Biol. 2022;5(1):1416. PMID: 36566320; PMCID: PMC9789959. doi:10.1038/s42003-022-04356-y
6. Zhang C, Shang X, Wang H. Untargeted metabolomics and lipidomics identified four subtypes of small cell lung cancer. Metabolomics. 2022;19(1):3. PMID: 36574156. doi:10.1007/s11306-022-01964-x
7. Sathiyapalan A, Febbraro M, Pond GR, Ellis PM. Chemo-immunotherapy in first line Extensive Stage Small Cell Lung Cancer (ES-SCLC): a systematic review and meta-analysis. Curr Oncol. 2022;29(12):9046–9065. PMID: 36547123; PMCID: PMC9776593. doi:10.3390/curroncol29120709
8. Brisebarre A, Ancel J, Ponchel T, et al. Transcriptomic FHITlow/pHER2high signature as a predictive factor of outcome and immunotherapy response in non-small cell lung cancer. Front Immunol. 2022;13:1058531. PMID: 36544755; PMCID: PMC9760670. doi:10.3389/fimmu.2022.1058531
9. Kawamoto N, Tsutani Y, Kamigaichi A, et al. Tumour Location Predicts Occult N1 Nodal metastasis in clinical stage I non-small cell lung cancer. Eur J Cardiothorac Surg. 2022:ezac575. PMID: 36571485. doi:10.1093/ejcts/ezac575
10. Cheng H, Bhushan S, Li N, Xiao Z, Gao K. Preoperative neutrophil-to-lymphocyte ratio and platelet-to-lymphocyte ratio are correlated with tumor-node-metastasis stages in patients with non-small cell lung cancer. J Cancer Res Ther. 2022;18(6):1666–1673. PMID: 36412428. doi:10.4103/jcrt.jcrt_473_21
11. Mao JR, Lan KQ, Liu SL, et al. Can the prognosis of individual patients with nasopharyngeal carcinoma be predicted using a routine blood test at admission? Radiother Oncol. 2022;22:109445. PMID: 36566987. doi:10.1016/j.radonc.2022.109445
12. Zhang J, Xu L, Gao J, et al. Leukocyte CH25H is a potential diagnostic and prognostic marker for lung adenocarcinoma. Sci Rep. 2022;12(1):22201. PMID: 36564433; PMCID: PMC9789102. doi:10.1038/s41598-022-24183-9
13. Yin M, Guan S, Ding X, et al. Construction and validation of a novel web-based nomogram for patients with lung cancer with bone metastasis: a real-world analysis based on the SEER database. Front Oncol. 2022;12:1075217. PMID: 36568214; PMCID: PMC9780685. doi:10.3389/fonc.2022.1075217
14. Liu Z, Zhang R, Xv Y, Wang J, Chen J, Zhou X. A novel nomogram integrated with systemic inflammation markers and traditional prognostic factors for adverse events’ prediction in patients with chronic heart failure in the Southwest of China. J Inflamm Res. 2022;15:6785–6800. PMID: 36573109; PMCID: PMC9789703. doi:10.2147/JIR.S366903
15. Zhang H, Liao M, Guo Q, et al. Predicting N2 lymph node metastasis in presurgical stage I-II non-small cell lung cancer using multiview radiomics and deep learning method. Med Phys. 2022. PMID: 36563341. doi:10.1002/mp.16177
16. Yang Z, Cai Y, Chen Y, et al. A CT-based radiomics nomogram combined with clinic-radiological characteristics for preoperative prediction of the novel IASLC grading of invasive pulmonary adenocarcinoma. Acad Radiol. 2022;23:S1076–S6332. PMID: 36567145. doi:10.1016/j.acra.2022.12.006
17. Isla D, Lozano MD, Paz-Ares L, et al. New update to the guidelines on testing predictive biomarkers in non-small-cell lung cancer: a National Consensus of the Spanish Society of Pathology and the Spanish Society of Medical Oncology. Clin Transl Oncol. 2022. PMID: 36571695. doi:10.1007/s12094-022-03046-9
18. Ma L, Fang X, Zhang A. The hypermethylation of FOXP3 gene as an epigenetic marker for the identification of arsenic poisoning risk. Hum Exp Toxicol. 2022;41:9603271221142819. PMID: 36464704. doi:10.1177/09603271221142819
19. Zhao T, Zhao X, Qian K, Shi K, Gu Y, Zhang Y. Radiotherapy prognosis-associated gene GCNT3 promotes the proliferation, migration and invasion of lung adenocarcinoma cells. Heliyon. 2022;8(12):e12100. PMID: 36578381; PMCID: PMC9791338. doi:10.1016/j.heliyon.2022.e12100
20. Wang X, Zhang Y, Zhi X. Correlation between microRNA expression, clinicopathological characteristics, and prognosis in patients with non-small cell lung cancer: a retrospective study. Thorac Cancer. 2017;8(5):511–516. PMID: 28727276; PMCID: PMC5582486. doi:10.1111/1759-7714.12480
21. Huang H, Li L, Luo W, et al. Lymphocyte percentage as a valuable predictor of prognosis in lung cancer. J Cell Mol Med. 2022;26(7):1918–1931. PMID: 35122390; PMCID: PMC8980931. doi:10.1111/jcmm.17214
22. Ma B, Niu F, Qu X, et al. A tetrameric protein scaffold as a nano-carrier of antitumor peptides for cancer therapy. Biomaterials. 2019;204:1–12. PMID: 30861422; PMCID: PMC6441627. doi:10.1016/j.biomaterials.2019.03.004
23. Tomita M, Ayabe T, Maeda R, Nakamura K. The prognostic values of a novel preoperative inflammation-based score in Japanese patients with non-small cell lung cancer. World J Oncol. 2019;10(4–5):176–180. PMID: 31636791; PMCID: PMC6785273. doi:10.14740/wjon1222
24. Ichimura T, Ichikura D, Hinata M, Hida N, Baba T. Thyroid dysfunction with atezolizumab plus bevacizumab after lenvatinib in hepatocellular carcinoma: a case series. SAGE Open Med Case Rep. 2023;11:205. PMID: 37009547; PMCID: PMC10064459. doi:10.1177/2050313X231164488
25. Hojbjerg JA, Bentsen KK, Vinholt PJ, Hansen O, Jeppesen SS, Hvas AM. Increased in vivo thrombin generation in patients with localized non-small cell lung cancer unfit for surgery. Clin Appl Thromb Hemost. 2023;29:10760296231152897. PMID: 36802980; PMCID: PMC9941591. doi:10.1177/10760296231152897
26. Chen Z, Cai A, Zheng H, et al. Carbidopa suppresses prostate cancer via aryl hydrocarbon receptor-mediated ubiquitination and degradation of androgen receptor. Oncogenesis. 2020;9(5):49. PMID: 32404918; PMCID: PMC7220950. doi:10.1038/s41389-020-0236-x
27. Macek Jilkova Z, Aspord C, Decaens T. Predictive factors for response to PD-1/PD-L1 checkpoint inhibition in the field of hepatocellular carcinoma: current status and challenges. Cancers. 2019;11(10):1554. PMID: 31615069; PMCID: PMC6826488. doi:10.3390/cancers11101554
28. Morabito F, Adornetto C, Monti P, et al. Genes selection using deep learning and explainable artificial intelligence for chronic lymphocytic leukemia predicting the need and time to therapy. Front Oncol. 2023;13:1198992. PMID: 37719021; PMCID: PMC10501728. doi:10.3389/fonc.2023.1198992
29. Hou F, Hou Y, Sun XD, et al. Establishment of a prognostic risk prediction modelfor non-small cell lung cancer patients with brainmetastases: a retrospective study. PeerJ. 2023;11:e15678. PMID: 37456882; PMCID: PMC10349557. doi:10.7717/peerj.15678
30. Jin G, Hu W, Zeng L, et al. Development and verification of a nomogram for predicting short-term mortality in elderly ischemic stroke populations. Sci Rep. 2023;13(1):12580. PMID: 37537270. doi:10.1038/s41598-023-39781-4
31. Deng J, Fei Y, Huang B. Nomogram for predicting the recurrence rate in selective radiofrequency thermocoagulation of the trigeminal nerve based on regression via least absolute shrinkage and selection operator. Pain Physician. 2023;26(4):E341–E352. PMID: 37535781.
32. Clift AK, Dodwell D, Lord S, et al. Development and internal-external validation of statistical and machine learning models for breast cancer prognostication: cohort study. BMJ. 2023;381:e073800. PMID: 37164379; PMCID: PMC10170264. doi:10.1136/bmj-2022-073800
33. Chen L, Zhang Y, Yu C, et al.; China Kadoorie Biobank Collaborative Group. Modeling biological age using blood biomarkers and physical measurements in Chinese adults. EBioMedicine. 2023;89:104458. PMID: 36758480; PMCID: PMC9941058. doi:10.1016/j.ebiom.2023.104458
34. Lee S, Choi Y, Seo MK, et al. Magnetic resonance imaging-based radiomics for the prediction of progression-free survival in patients with nasopharyngeal carcinoma: a systematic review and meta-analysis. Cancers. 2022;14(3):653. PMID: 35158921; PMCID: PMC8833585. doi:10.3390/cancers14030653
35. Liu L, Wang Z, Zhao L, Chen X, He S. External validation of non-invasive diabetes score in a 15-year prospective study. Am J Med Sci. 2022;364(5):624–630. PMID: 35640678. doi:10.1016/j.amjms.2022.05.023
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




