Back to Journals » International Journal of Chronic Obstructive Pulmonary Disease » Volume 18
Prognostic Biomarkers Based on Proteomic Technology in COPD: A Recent Review
Authors Fang H , Liu Y , Yang Q , Han S, Zhang H
Received 28 February 2023
Accepted for publication 25 June 2023
Published 30 June 2023 Volume 2023:18 Pages 1353—1365
DOI https://doi.org/10.2147/COPD.S410387
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Min Zhang
Hanyu Fang,1,* Ying Liu,2,* Qiwen Yang,1 Siyu Han,1 Hongchun Zhang1– 3
1Beijing University of Chinese Medicine, Beijing, 100029, People’s Republic of China; 2The Second Health and Medical Department, China-Japan Friendship Hospital, Beijing, 100029, People’s Republic of China; 3Department of Traditional Chinese Medicine for Pulmonary Diseases, Center of Respiratory Medicine, China-Japan Friendship Hospital, Beijing, 100029, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Hongchun Zhang, Department of Traditional Chinese Medicine for Pulmonary Diseases, Center of Respiratory Medicine, China-Japan Friendship Hospital, Beijing, 100029, People’s Republic of China, Tel +86 13701226664, Email [email protected]
Abstract: Chronic obstructive pulmonary disease (COPD) is a common heterogeneous respiratory disease which is characterized by persistent and incompletely reversible airflow limitation. Due to the heterogeneity and phenotypic complexity of COPD, traditional diagnostic methods provide limited information and pose a great challenge to clinical management. In recent years, with the development of omics technologies, proteomics, metabolomics, transcriptomics, etc., have been widely used in the study of COPD, providing great help to discover new biomarkers and elucidate the complex mechanisms of COPD. In this review, we summarize the prognostic biomarkers of COPD based on proteomic studies in recent years and evaluate their association with COPD prognosis. Finally, we present the prospects and challenges of COPD prognostic-related studies. This review is expected to provide cutting-edge evidence in prognostic evaluation of clinical patients with COPD and to inform future proteomic studies on prognostic biomarkers of COPD.
Keywords: chronic obstructive pulmonary disease, omics, proteome, biomarker, prognosis
Introduction
Chronic obstructive pulmonary disease (COPD) is a common chronic lung disease characterized by persistent airflow limitation and corresponding respiratory symptoms. COPD severely affects the quality of life of patients and has become an important cause of death worldwide, imposing a heavy economic burden on patients, families and society.1,2 The pathogenesis of COPD is complex, and inhalation of harmful particles or gases, such as tobacco smoke, can cause oxidative stress, inflammatory responses and protease/anti-protease imbalance in the airways, etc.3 Although COPD is common, it is often undiagnosed and thus the best time for treatment is missed.4 The current assessment of lung function includes forced expiratory volume in the first second (FEV1), forced vital capacity (FVC) and FEV1/FVC ratio. They are the “gold standards” for COPD diagnosis and the most commonly used indexes for COPD severity assessment, disease progression monitoring, prognosis and treatment response evaluation.5,6 Due to the heterogeneity and phenotypic complexity of COPD, the information provided by spirometry is limited, making it difficult to make an accurate diagnosis, therapeutic effect, and prognostic evaluation at an early stage.7,8 Furthermore, it often exhibits low accuracy in cases of predicting disease worsening and possible biasing due to the operator, patient, and conditions.9 In addition to spirometry, quantitative computed tomography (QCT) is increasingly being used in the evaluation of COPD. QCT can accurately describe the pathology in COPD by quantifying the patterns of emphysema, bronchial wall thickness and gas trapping.10 QCT is advantageous in the early diagnosis of COPD, as it can identify lung injury in patients with early COPD even when spirometry is normal.11 Furthermore, QCT can be used to assess the progression of lung function12,13 and the risk of acute exacerbations14,15 in COPD. There are various QCT-based indicators, including cross-sectional area (CSA),16 parametric response map (PRM),17 the ratio of the mean lung density at end expiration to end inspiration (E/I),18 total airway count (TAC),19 etc. However, there is no universal protocol for the analysis of QCT for COPD diagnosis and prognostic assessment, and these indicators still need further clinical validation. Moreover, the complexity of quantitative data analysis and the potential radiation exposure of QCT further limit the application of these image-based markers.
With the development of omics technology in recent years, proteomics is widely used in the study of COPD, providing an effective tool for discovering biomarkers and elucidating the complex mechanism of COPD. This makes the study of biomarkers based on proteomic technology a noteworthy area for understanding COPD. Biomarkers are classified as screening, diagnostic, or prognostic markers based on their ability to predict, diagnose, or assess disease status, respectively.20,21 A typical COPD patient with a decades-long smoking history does not manifest signs of disease until the latter stages of life, so the search for new prognostic biomarkers (especially those superior to spirometry in evaluating prognosis) is imperative and urgent. Promising prognostic biomarkers require strong clinical utility and discrimination, as well as ease of collection, which allow them to play an important role in clinical practice.
In this review, we focused on prognostic biomarkers in COPD based on proteomic technology and aimed to provide an update on the status of relevant studies. We summarized the progress of recent studies using proteomic technologies to screen and validate prognostic biomarkers in specimens from different sources of COPD and assessed the association of these biomarkers and COPD prognosis. The search for prognostic biomarkers in COPD first requires a clear definition of COPD prognosis—which can be evaluated in terms of the following:22 (1) The rate of decline in lung function; (2) The risk of acute exacerbations; (3) Mortality.
Proteomics of COPD
Wilkins and Williams first introduced the concept of proteome.23 It is defined as “all proteins expressed in the genome of a cell/tissue” and can be used to study the composition, classification, expression levels, post-translational modifications, protein-nucleic acid interactions, protein–metabolite interactions, etc. at macro and micro levels. Proteomics not only provides qualitative or quantitative protein detection and comparison of differences but also allows dynamic analysis of protein localization and modification, as well as the interactions between specific proteins and other proteins, nucleic acids, genes, etc., thus revealing the biological functions of specific proteins and their relevance to the occurrence and development of diseases.24,25 In addition, proteomics has the potential to reveal some disease mechanisms that cannot be identified at the genetic level.26 Meanwhile, most types of specimens in proteomic studies can be easily collected in a less invasive and reproducible way. Because of these advantages, proteomic approaches have been used in many chronic lung diseases, including COPD, idiopathic pulmonary fibrosis (IPF),27 pulmonary sarcoidosis,28 asthma,29 etc.
The proteomic approach has unique advantages over spirometry and QCT in the assessment of COPD. As mentioned above, spirometry has limitations in assessing COPD in the early stage. Biomarkers of inflammation may play key roles both in earlier diagnosis and evaluation of COPD prognosis.30 Proteomics provides an effective tool for elucidating the overall changes in complex inflammatory diseases such as COPD.31 Therefore, proteomic studies are expected to find biomarkers that are superior to spirometry in the early stage. The use of QCT for the evaluation of COPD is still in the investigation stage and is limited by issues such as complicated data analysis and potential radiation exposure, and it is not a routine test for COPD in current clinical practice.32 In contrast, proteomic specimens are, in most cases, easy to collect repeatedly and less invasive, without causing too much damage to the subjects, which means that relevant clinical studies are easier to conduct.
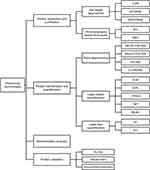
Proteomic technologies can be divided into four parts according to the research steps (Figure 1), and previous reviews have described in detail the specific applications of these technologies.33–36
- Protein separation and purification: Gel-based approaches and chromatography are represented. The former includes two-dimensional gel electrophoresis (2-DE), two-dimensional differential gel electrophoresis (2D-DIGE), and sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), which are commonly used for the separation of complex protein specimens. The latter includes ion-exchange chromatography (IEC), size-exclusion chromatography (SEC), etc., which are common protein purification techniques. COPD proteomic studies are mainly supported by gel-based approaches.37
- Protein identification and quantification: Mass spectrometry (MS) is the main method for protein identification and is also the core technology and important support for protein analysis.38,39 It includes surface-enhanced laser desorption/ionization-time-of-flight mass spectrometry (SELDI-TOF-MS), matrix-assisted laser desorption/ionization-time-of-flight mass spectrometry (MALDI-TOF-MS), electrospray ionization mass spectrometry (ESI-MS), liquid chromatography tandem-mass spectrometry (LC-MS/MS), etc. The quantitative techniques in proteomics are based on the known protein types and the signal intensity given by mass spectrometry to quantify their expression and abundance. The common quantification techniques include label-based quantification and label-free quantification, including isotope-coded affinity tags (ICAT), isotope-coded protein labels (ICPL), isobaric tag for relative and absolute quantitation (iTRAQ), tandem mass tag (TMT), stable isotope labeling by amino acids in cell culture (SILAC), spectral counting (SC), extracted ion chromatograms (XIC), etc. Label-based quantification has been used more often than label-free quantification in COPD proteomic studies. The reason is that, in iTRAQ, for example, the multiplexing potential of isotopic labeling increases the statistical relevance and accuracy of results compared to label-free mass spectrometry techniques.40
- Bioinformatics analysis: Bioinformatics is an interdisciplinary field that uses computer science and mathematical modeling to capture and interpret large amounts of biological and medical data. Bioinformatics is essential for data management in modern biology and medicine. UniProt,41 GenBank42 and MaxQB43 are representative protein bioinformatics databases. The biological processes, cellular components, molecular functions, signaling pathways and interactions associated with the candidate proteins can be further analyzed and evaluated.
- Protein validation: Candidate proteins screened by bioinformatics techniques are used to validate the reliability of the samples. This technique includes enzyme-linked immunosorbent assay (ELISA), Westernblot, immunohistochemistry, etc., which are widely used nowadays.
In recent years, more studies on COPD proteomics have been reported, among which there are some prognostic biomarkers with potential applications. They can make up for the shortcomings of existing clinical tests and traditional pathological observation methods. Protein-targeted clinical tests can reduce or avoid the harm to patients from invasive examinations such as bronchoscopy and allow dynamic observation of the network regulation of signaling pathways associated with the disease and the onset and progression of the disease.
The detection and identification of proteins in different fluids/tissues is currently an area of increasing interest to understand whether they are tools for monitoring disease progression. Proteomic studies in COPD can screen for biomarkers associated with the prognosis of the disease from various types of specimens such as serum/plasma, sputum, lung tissue, etc., from patients for qualitative or quantitative analysis and dynamically reflect the heterogeneous foci of COPD and changes in the type/quantity of pathogenic proteins in different disease stages.44 The following reviews the research progress of the prognostic biomarkers of COPD screened and validated using proteomic techniques in recent years, respectively, according to the source of the specimens, and summarized in Table 1.
 |
Table 1 COPD-Associated Proteomic Prognostic Biomarkers |
Serum/Plasma
Serum/plasma is often used to test people with different diseases and dominates clinical proteomic studies. Protein detection and analysis in blood specimens is most used in COPD proteomic studies. This is because peripheral blood specimens are rich in protein and have the advantage of being collected in a less invasive and reproducible way. Lung cells release some of their contents into the blood at the time of damage or death, which makes serum/plasma one of the most important sources of information for COPD proteomics.26
Pratte et al45 confirmed that soluble receptor for advanced glycation end products (sRAGE) is a prognostic biomarker for COPD by analyzing plasma sRAGE levels in four independent cohorts and found that lower plasma sRAGE levels associated with reduced forced expiratory volume in the first second (FEV1) and exacerbation of emphysema. Plasma sRAGE is a predictor of emphysema progression, and it is also the most promising blood COPD biomarker.65,66
In a cross-sectional study and a prospective longitudinal study, Bozinovski et al46 found that serum amyloid A (SAA) was upregulated in acute exacerbations compared to stable COPD. Also in patients with acute exacerbations, SAA levels were significantly higher in patients with respiratory failure than in other hospitalized patients. Therefore, SAA may become a novel biomarker for acute exacerbations of COPD, ie, a higher SAA level is associated with a poorer prognosis.
Kim et al47 conducted a 2-year follow-up of 12 patients with acute exacerbation of chronic obstructive pulmonary disease (AECOPD) and analyzed paired blood samples collected from each patient at the beginning of treatment for AECOPD and during recovery. The results showed that the levels of 14 proteins were higher in the AECOPD samples than in the recovery samples. After validation by ELISA, alpha-1-antichymotrypsin (SERPINA3) was suggested as a possible biomarker for early identification of AECOPD, ie, higher SERPINA3 levels may be associated with increased risk of AECOPD. Earlier studies have suggested that serum SERPINA3 levels are associated with poor prognosis in COPD, including exacerbation of systemic inflammation and increased 10-year mortality.67
Leung et al48 used multiple reaction monitoring mass spectrometry (MRM-MS) to detect and analyze plasma from multiple cohorts of patients with exacerbation and stable COPD and found that the biomarker scores of a panel of five proteins were significantly higher in the acute exacerbation phase than in the stable phase, so they may be prognostic proteins for determining the risk of AECOPD. Of which, fibronectin is noteworthy. Earlier studies showed the potential of fibronectin to determine COPD prognosis: Fibronectin expression in bronchial vessels was negatively correlated with FEV1 values in COPD patients,68 and the ratio of circulating fibronectin to C-reactive protein (CRP) was independently associated with all-cause mortality in COPD patients.69
Diao et al49 found that thyroxine-binding globulin (THBG) levels were negatively associated with percentage of predicted FEV1 value, and COPD patients with higher baseline THBG levels had a greater risk of acute exacerbation than those with lower THBG levels. Diao’s team50 found in an earlier study that fetuin-B (FETUB) predicted the occurrence of acute exacerbation or frequent acute exacerbation and positively correlated with the percentage of emphysema assessed by CT. THBG and FETUB can assist in the management of stable stage and acute exacerbation in COPD patients as a potential prognostic biomarker.
In addition to common clinical risk factors (environmental exposures, allergy, asthma and infections, etc.), alpha-1 antitrypsin deficiency (AATD), one of the most common autosomal recessive inheritable diseases worldwide, has been associated with an increased risk for developing COPD.70–72 In a multicenter, prospective 3-year study of 49 adults with severe AATD, Beiko et al51 measured 317 proteins from serum. After adjustments were made for age and sex, C-reactive protein (CRP), adipocyte fatty acid-binding protein (AFBP), and tissue plasminogen activator (tPA) were found to be associated with emphysema progression.
BALF
Bronchoalveolar lavage (BAL), performed during flexible bronchoscopy, has gained widespread acceptance that provides important information about immunologic, inflammatory, and infectious processes taking place at the alveolar level.73 Advantages with BAL include its non-invasive nature and ability to readily sample alveolar contents; as a result, this technique has been widely used in patients with interstitial lung disease, lung tumors, infections, etc.74 As clinical testing techniques continue to improve, the analysis of bronchoalveolar lavage fluid (BALF) has expanded from routine cytology testing to proteomics, which means that BAL can play a role in the evaluation of more lung diseases, including COPD.
Lv et al52 found that the lower the lung surfactant protein A (SP-A) and D (SP-D) in BALF, the worse the lung function and the higher the probability of complications and ventilator use. Meanwhile, the results of animal experiments showed that COPD model mice with emphysema and airway wall thickening were more likely to have reduced SP-A and SP-D. This study suggests that SP-A and SP-D are potentially associated with airway remodeling, inflammatory response, and prognosis in COPD. In another recent clinical study, similar results were obtained by Hristova et al.53 SP-A and SP-D levels were positively correlated with FEV1/FVC, while they were negatively correlated with computed tomography small airways disease measures (expiratory to inspiratory mean lung density).
Yang et al54 studied the cellular proteins of BALF from healthy never-smokers versus smokers with normal lung function and quantified 506 differential proteins, enriched in 15 signaling pathways. Among them, proteins in 2 pathways (phagosome and leukocyte transendothelial migration pathways) were significantly correlated with activated cytotoxic T cells and airway obstruction levels in smokers, suggesting that these proteins may play an important role in the molecular events preceding the development of COPD in susceptible smokers and are potential prognostic biomarkers. The results also provide a new vision for the study of the molecular mechanisms of smoking, a well-recognized risk factor for COPD.
Sputum
Sputum provides a wealth of information about the state of the lung and contains various components such as mucus, microbial products of colonized bacteria or viruses, particles from the external environment, inflammatory cells and mediators from the airway lumen or tissues, etc. Particularly, inflammatory cells and mediators can reflect the phenotypic features of chronic respiratory disorders such as COPD, and thus proteomic profiles of sputum can be used to screen for COPD.75 The work of Casado et al76 demonstrated for the first time that distinct differences in protein profiles of sputum samples were related to the phenotype of COPD and cigarette smoking illness severity. In recent years, proteomic analysis of sputum has gained increasing attention in the study of COPD.77
In a recent pilot study, Dasgupta et al55 explored the profile of sputum proteins in asthma, COPD, and chronic bronchitis. Eight proteins (Azurocidin1, Neutrophil defensin 3, Lactotransferrin, Calmodulin 3, Coronin1A, Mucin 5B, Mucin 5AC and BPI fold containing family B1) which could differentiate between exacerbation prone and nonexacerbators were identified. It is also worth noting that total protein concentration was demonstrated for the first time as a useful biomarker for “frequent exacerbators”. These findings will help in the identification of those with exacerbations of COPD. With the limitation of sample size, larger studies are required to validate the results.
BPIFB1 has been identified in the proteome of sputum,76,78 and its multiple protective roles against respiratory infections and inflammation have been well discussed in a previous review.79 The aim of Gao et al56 was to further investigate the relationship between sputum bactericidal/permeability-increasing fold-containing protein B1 (BPIFB1) levels and changes in lung function in patients with smoking and COPD, based on the screening results of previous proteomic studies, the longitudinal changes in lung function in nonsmokers, smokers without COPD and smokers with COPD were followed up for 4 years and longitudinal changes in lung function were compared with baseline BPIFB1 levels in sputum. The results showed that baseline BPIFB1 levels in sputum were higher in COPD patients compared to nonsmokers and smokers without COPD. Besides, baseline BPIFB1 levels in sputum correlated with lung function at baseline and after 4 years of follow-up in all participants. While in COPD patients who smoked, changes in lung function over 4 years were significantly correlated with baseline BPIFB1 levels. This study strongly suggests that higher BPIFB1 levels in sputum are associated with quicker changes in lung function over time, and therefore BPIFB1 is likely to be a prognostic biomarker for COPD.
Mallia-Milanes et al57 examined sputum samples from stable and exacerbated COPD and identified a total of 299 proteins. Analysis showed that elastase activity was significantly increased in exacerbated phase compared to stable phase, suggesting that the risk of exacerbation of COPD may be associated with increased elastase activity.
Lung Tissues
To obtain an accurate diagnosis, invasive methods are required in some cases. Lung tissue samples are typically obtained by transbronchial lung biopsy or open lung biopsy procedures. This invasive approach is mostly limited to research purposes and is not applicable to daily clinical settings.75 In the case of proteomic studies, the main limitation of this approach is the small sample size. However, accurate and valuable information can be obtained by analyzing protein expression in a real biological environment that directly reflects lung pathology.
Ohlmerier et al58 identified lung tissue from nonsmokers, smokers, and patients with mild to severe COPD. Cathepsin D (CTSD), dihydropyrimidinase-related protein 2 (DPYSL2), transglutaminase 2 (TGM2), and tripeptidyl-peptidase 1 (TPP1) were validated as COPD-related specific proteins, where TGM2 levels correlated with lung function in COPD patients and were independent of smoking. It indicates that TGM2 is a potential prognostic biomarker in COPD, but further studies in larger cohorts are still needed, along with the study of the mechanism of the TGM2 pathway, to accurately assess the prognostic value of TGM2 in COPD progression.
Sun et al59 examined changes in the lung proteome of COPD patients and found that 23 proteins were differentially expressed in the lung tissue of frequent exacerbators compared to infrequent exacerbators. Three significantly regulated proteins [major histocompatibility complex, class II, DQ alpha 1 (HLA-DQA1), polymeric immunoglobulin receptor (pIgR) and biglycan] were more expressed in those with frequent exacerbations than in those with infrequent exacerbations. Sun’s team took full account of the heterogeneity of COPD and innovatively grouped the included patients according to the frequency of acute exacerbations. This study provides new insights into the mechanism of frequent COPD exacerbations, while the above differentially expressed proteins can be considered as potential prognostic biomarkers associated with the risk of acute exacerbations.
Liu et al60 compared the protein expression profiles of lung tissue from COPD patients requiring lung transplantation and healthy individuals and found that platelet glycoprotein VI (GP6), platelet factor 4 (PF4) and thrombospondin-1 (THBS1), which are associated with platelet activation and wound healing, were significantly downregulated in COPD patients, suggesting that COPD patients are more prone to impaired hemostasis, which may impede the repair process of lung tissue. This study identified some of the key proteins involved in the pathogenesis of COPD, and although the sample size was small, it still provides clues for the future search of prognostic biomarkers for COPD. The relationship between the expression levels of the above key proteins and changes in lung function in COPD patients deserves further investigation.
Skerrett-Byrne et al61 induced a mouse COPD model by cigarettes and mapped changes in the lung tissue proteome over 12 weeks. Lung tissue levels of heterogeneous nuclear ribonucleoproteins C1/C2 (HNRNPC), Musashi homologue 2 (MSI2) and S100 calcium-binding protein A1 (S100A1) were significantly higher in cigarette-exposed mice than in control healthy mice, as well as total lung volume (TLC) and alveolar diameter. The above three proteins were correlated with the severity of COPD, and their value as prognostic biomarkers can be further evaluated by observing the changes in their expression in lung tissues of COPD patients with different severity.
To investigate the role played by anterior gradient 3 (AGR3) in the lung tissue of COPD patients, Ye et al62 collected lung tissue from current-smoking patients, patients with infrequent COPD exacerbations and frequent COPD exacerbations. The result suggested that lower AGR3 expression in the lung tissue might promote viral and bacterial infection and induce immune inflammation to increase susceptibility to COPD exacerbation. This allows AGR3 to be a possible biomarker for evaluating the risk of acute exacerbation.
ELF
Epithelial-lining fluid (ELF) is a thin fluid layer produced by epithelial cells lining the airway mucosa and alveolar surfaces, containing high concentrations of biologically active molecules that play an important role in forming a barrier against the external contaminants such as air pollutants, toxic components of cigarette smoke, and allergens. As the first barrier between the lung and the external environment, ELF is an attractive target for proteomic studies of COPD80 and contains little DNA or RNA but high levels of proteins.81
The Franciosi team has made an important contribution to the application of ELF in COPD proteomic study. Initially, they used bronchoscopic microprobe (BMP) to sample the ELF of healthy individuals and COPD patients.82 In contrast to other methods used for the collection of biofluids from the lung, the advantage of this technique is that ELF is not diluted, contains a high concentration of biomolecules, and without contamination from oropharyngeal bacteria or saliva. Further study showed the feasibility of performing LC-MS/MS and quantitative proteomics in ELF samples from COPD patients.80 In their latest work,63 they collected baseline ELF from subjects as well as ELF 24 hours after smoking 3 cigarettes and analyzed epithelial lining fluid from healthy controls and COPD patients using iTRAQ. They found that after acute smoke exposure, the expression of peroxiredoxin I, s100 calcium-binding protein A9 (S100A9), s100 calcium-binding protein A8 (S100A8) and aldehyde dehydrogenase 3a1 (ALDH3A1) decreased in healthy controls, while peroxiredoxin I, serpin family b member 3 (SerpinB3) and ALDH3A1 expression increased in the COPD patient group. The authors concluded that the bronchial tree of COPD patients has dramatically altered after many years of smoking and is therefore capable of upregulating these major protective proteins for at least 24 hours after smoke inhalation. Regular monitoring of changes in these proteins in the smoking population could be able to reflect the prognosis at the proteomic level, which remains to be confirmed by prospective studies.
EBC
Exhaled breath condensate (EBC), a non-invasive method of collecting cooled coagulated material from exhaled breath, has great potential as a safe and clinically meaningful measurement of biomarkers of COPD.75,83 However, in recent years, EBC-related proteomic studies in COPD have been mostly devoted to exploring diagnostic biomarkers84–86 and lacked attention on prognostic biomarkers. Given that EBC is safe to sample, easy to perform and reproducible, EBC-related COPD proteomic studies are worth further investigation, especially prospective studies that help to find prognostic markers.
Urine
Urine has gained increasing interest as an attractive resource for biomarker studies with non-invasive and continuous collection. In addition to the urinary system, biomarkers in urine can be a sensitive indicator of respiratory diseases. Urine proteomic studies have identified biomarkers for several respiratory diseases, such as lung cancer87, pulmonary fibrosis,88 ventilator-induced lung injury,89 and acute respiratory distress syndrome (ARDS).90 Therefore, urine is a promising resource for studying biomarkers of lung diseases. Qin et al64 compared the dynamic changes in urine proteomics as well as lung function in cigarette smoke-induced COPD rats and normal rats over an 8-week period and found trefoil factor 2 (TFF2) down-regulated significantly in COPD rats at week 8, when obvious airflow obstruction was detected, compared to normal rats. TFF2 was verified as a potential biomarker for prognostic assessment of cigarette smoke-induced COPD. However, this was a preliminary study and variables such as medication, surgery, patient habits, and renal function that may affect urine samples were not considered. Most of the current urinary proteomic studies are focused on kidney diseases,91 and clinical studies of urinary proteomics related to COPD are lacking.
Conclusions and Prospects
Over the past decade, many advances have been made in the use of proteomics to study prognostic biomarkers in COPD. Proteomic studies in COPD are characterized by a rich source of samples, and most of the samples collected are non-invasive and easily reproducible. In the future, the field of proteomics will further benefit from technological advances in sample preparation, mass spectrometry, bioinformatics, etc.
Today, lung function indicators such as FEV1, FVC and FEV1/FVC, although not perfect, they are still the most used indicators for COPD diagnosis and prognosis. The above-mentioned prognostic biomarkers screened by proteomic techniques are highly expected by researchers, but unfortunately none of them are currently used in the clinic, which is attributed to 5 obstacles. (1) Many of these studies are cross-sectional in nature, and as observational studies, they can only suggest correlation rather than causation between exposure factors and outcome, regardless of their results, so predictive studies are needed to further validate the value of these prognostic biomarkers. (2) Most proteomic studies have attempted to find one or several disease-specific biomarkers. Given the heterogeneity and phenotypic complexity of COPD, it is unlikely that only one or a few proteins are responsible for the progression of the disease. (3) Some studies use cellular or animal models to mimic the disease in humans, but it is difficult to assess how much of these experimental results can be successfully translated to real-world patient situations.61,64 (4) Differences in study populations, different sample sources, different research platforms or methods used by laboratories, and subject bias all contribute to limited reproducibility of results. (5) The workflow of proteomics requires a high level of expertise,33,34 which limits its widespread use by general clinicians and primary hospitals. This may also contribute to the limited sample size of proteomic studies.
For the above problems, we propose some solutions that may be helpful. Firstly, screening the most promising biomarkers from the existing studies is an important future task. Many of these studies are cross-sectional, which means they are usually easy to conduct and are useful for establishing preliminary evidence in a further study.92 The existing studies identified a large number of potential biomarkers, and the screening works made further validation feasible. Secondly, conducting predictive studies to validate the value of these screened biomarkers in predicting COPD prognosis, comparing to FEV1/FVC and QCT, can help to transform them from the laboratory to the clinical use. It also requires close collaboration between scientists, clinicians and regulatory authorities. Thirdly, with the development of statistical and bioinformatics tools, multi-omics studies are the current trend.31,93 Even more recently, deep learning methods have been combined with multi-omics studies to improve the accuracy of disease prediction.94 These tools can be used to systematically integrate metabolomics, proteomics, and transcriptomics data sets, and improve the opportunity to discover repeatable and reliable biomarkers. Also, multi-omic studies can avoid the limitations of finding a single biomarker as mentioned above.
Despite all these obstacles, combined with the current research results, we still believe that in the era of precision medicine and personalized medicine, proteomic technology has great potential in evaluating COPD prognosis. It is evident from recent studies that sample sources for COPD prognostic biomarker studies are abundant, but sample sources are mostly concentrated in blood, sputum, and lung tissue, and less frequently from BALF, ELF and urine. EBCs are easy to collect and non-invasive; however, none of the recent COPD prognostic biomarker studies have EBC as a sample source. Future screening of COPD prognostic biomarkers is worth starting with EBC.
Funding
This study is supported by the National Administration of Traditional Chinese Medicine (QI Huang Project) of China (2019-QTL-003).
Disclosure
The authors declare that there is no conflict of interest.
References
1. López-Campos JL, Tan W, Soriano JB. Global burden of COPD. Respirology. 2016;21(1):14–23. doi:10.1111/resp.12660
2. Naghavi M, Abajobir AA, Abbafati C, et al. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980–2016: a systematic analysis for the global burden of disease study 2016. Lancet. 2017;390(10100):1151–1210. doi:10.1016/S0140-6736(17)32152-9
3. Lange P, Ahmed E, Lahmar ZM, Martinez FJ, Bourdin A. Natural history and mechanisms of COPD. Respirology. 2021;26(4):298–321. doi:10.1111/resp.14007
4. Gershon AS, Thiruchelvam D, Chapman KR, et al. Health services burden of undiagnosed and overdiagnosed COPD. Chest. 2018;153(6):1336–1346. doi:10.1016/j.chest.2018.01.038
5. Balasubramanian A, MacIntyre NR, Henderson RJ, et al. Diffusing capacity of carbon monoxide in assessment of COPD. Chest. 2019;156(6):1111–1119. doi:10.1016/j.chest.2019.06.035
6. Boutou AK, Shrikrishna D, Tanner RJ, et al. Lung function indices for predicting mortality in COPD. Eur Respir J. 2013;42(3):616. doi:10.1183/09031936.00146012
7. Csikesz N, Gartman E. New developments in the assessment of COPD: early diagnosis is key. Int J Chronic Obstruct. 2014;9(1):277–286. doi:10.2147/COPD.S46198
8. Lowe KE, Regan EA, Anzueto A, Austin E, Austin JHM, Beaty TH. COPDGene 2019: redefining the diagnosis of chronic obstructive pulmonary disease. Am J Physiol Lung Cell. 2019;5(6):384–399. doi:10.15326/jcopdf.6.5.2019.0149
9. Dong T, Santos S, Yang Z, Yang S, Kirkhus NE. Sputum and salivary protein biomarkers and point-of-care biosensors for the management of COPD. Analyst. 2020;145(5):1583–1604. doi:10.1039/C9AN01704F
10. Occhipinti M, Paoletti M, Bartholmai BJ, et al. Spirometric assessment of emphysema presence and severity as measured by quantitative CT and CT-based radiomics in COPD. Resp Res. 2019;20(1):101. doi:10.1186/s12931-019-1049-3
11. Regan EA, Lynch DA, Curran-Everett D, et al. Clinical and radiologic disease in smokers with normal spirometry. JAMA Intern Med. 2015;175(9):1539–1549. doi:10.1001/jamainternmed.2015.2735
12. Firdaus AAMH, Pim ADJ, Jan-Willem JL, et al. Airway wall thickness associated with forced expiratory volume in 1 second decline and development of airflow limitation. Eur Respir J. 2015;45(3):644. doi:10.1183/09031936.00020714
13. Koo HJ, Lee SM, Seo JB, et al. Prediction of pulmonary function in patients with chronic obstructive pulmonary disease: correlation with quantitative CT parameters. Korean J Radiol. 2019;20(4):683–692. doi:10.3348/kjr.2018.0391
14. McAllister DA, Ahmed FS, Austin JHM, et al. Emphysema predicts hospitalisation and incident airflow obstruction among older smokers: a prospective cohort study. PLoS One. 2014;9(4):e93221. doi:10.1371/journal.pone.0093221
15. Chaudhary MFA, Hoffman EA, Guo J, et al. Predicting severe chronic obstructive pulmonary disease exacerbations using quantitative CT: a retrospective model development and external validation study. Lancet Digit Health. 2023;5(2):e83–e92. doi:10.1016/S2589-7500(22)00232-1
16. Matsuoka S, Washko GR, Dransfield MT, et al. Quantitative CT measurement of cross-sectional area of small pulmonary vessel in COPD: correlations with emphysema and airflow limitation. Acad Radiol. 2010;17(1):93–99. doi:10.1016/j.acra.2009.07.022
17. Galbán CJ, Han MK, Boes JL, et al. Computed tomography–based biomarker provides unique signature for diagnosis of COPD phenotypes and disease progression. Nat Med. 2012;18(11):1711–1715. doi:10.1038/nm.2971
18. Bodduluri S, Reinhardt JM, Hoffman EA, et al. Signs of gas trapping in normal lung density regions in smokers. Am J Resp Crit Care. 2017;196(11):1404–1410. doi:10.1164/rccm.201705-0855OC
19. Kirby M, Tanabe N, Vasilescu DM, et al. Computed tomography total airway count is associated with the number of micro–computed tomography terminal bronchioles. Am J Resp Crit Care. 2019;201(5):613–615. doi:10.1164/rccm.201910-1948LE
20. Biomarkers Definitions Working Group. Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Ther. 2001;69(3):89–95. doi:10.1067/mcp.2001.113989
21. Fleming TR, Powers JH. Biomarkers and surrogate endpoints in clinical trials. Stat Med. 2012;31(25):2973–2984. doi:10.1002/sim.5403
22. Shaw JG, Vaughan A, Dent AG, et al. Biomarkers of progression of chronic obstructive pulmonary disease (COPD). J Thorac Dis. 2014;6(11):1532–1547. doi:10.3978/j.issn.2072-1439.2014.11.33
23. Kahn P. From genome to proteome: looking at a cell’s proteins. Science. 1995;270(5235):369–370. doi:10.1126/science.270.5235.369
24. Haines DS, Lee JE, Beauparlant SL, et al. Protein interaction profiling of the p97 adaptor UBXD1 points to a role for the complex in modulating ERGIC-53 trafficking. Mol Cell Proteom. 2012;11(6). doi:10.1074/mcp.M111.016444
25. Marko-Varga G, Omenn GS, Paik Y, Hancock WS. A first step toward completion of a genome-wide characterization of the human proteome. J Proteome Res. 2013;12(1):1–5. doi:10.1021/pr301183a
26. Cagnone M, Salvini R, Bardoni A, Fumagalli M, Iadarola P, Viglio S. Searching for biomarkers of chronic obstructive pulmonary disease using proteomics: the current state. Electrophoresis. 2019;40(1):151–164. doi:10.1002/elps.201800305
27. Foster MW, Morrison LD, Todd JL, et al. Quantitative proteomics of bronchoalveolar lavage fluid in idiopathic pulmonary fibrosis. J Proteome Res. 2015;14(2):1238–1249. doi:10.1021/pr501149m
28. Landi C, Bargagli E, Carleo A, et al. A functional proteomics approach to the comprehension of sarcoidosis. J Proteomics. 2015;128:375–387. doi:10.1016/j.jprot.2015.08.012
29. Takahashi K, Pavlidis S, Ng Kee Kwong F, et al. Sputum proteomics and airway cell transcripts of current and ex-smokers with severe asthma in U-BIOPRED: an exploratory analysis. Eur Respir J. 2018;51(5):1702173. doi:10.1183/13993003.02173-2017
30. Paone G, Leone V, Conti V, et al. Blood and sputum biomarkers in COPD and asthma: a review. Eur Rev Med Pharmacol Sci. 2016;4(20):698–708.
31. Tang Y, Chen Z, Fang Z, Zhao J, Zhou Y, Tang C. Multi-omics study on biomarker and pathway discovery of chronic obstructive pulmonary disease. J Breath Res. 2021;15(4):044001. doi:10.1088/1752-7163/ac15ea
32. Halpin DMG, Criner GJ, Papi A, et al. Global initiative for the diagnosis, management, and prevention of chronic obstructive lung disease. the 2020 GOLD science committee report on COVID-19 and chronic obstructive pulmonary disease. Am J Resp Crit Care. 2021;203(1):24–36. doi:10.1164/rccm.202009-3533SO
33. Reisdorph NA, Reisdorph R, Bowler R, Broccardo C. Proteomics methods and applications for the practicing clinician. Ann Allergy Asthma Immunol. 2009;102(6):523–529. doi:10.1016/S1081-1206(10)60128-7
34. Aslam B, Basit M, Nisar MA, Khurshid M, Rasool MH. Proteomics: technologies and their applications. J Chromatogr Sci. 2017;55(2):182–196. doi:10.1093/chromsci/bmw167
35. Chen C, Hou J, Tanner JJ, Cheng J. Bioinformatics methods for mass spectrometry-based proteomics data analysis. Int J Mol Sci. 2020;21(8):2873.
36. Rozanova S, Barkovits K, Nikolov M, Schmidt C, Urlaub H, Marcus K. Quantitative mass spectrometry-based proteomics: an overview. In: Marcus K, Eisenacher M, Sitek B, editors. Quantitative Methods in Proteomics. New York, NY: Springer US; 2021:85–116.
37. Rossi R, De Palma A, Benazzi L, Riccio AM, Canonica GW, Mauri P. Biomarker discovery in asthma and COPD by proteomic approaches. Proteomics. 2014;8(11–12):901–915. doi:10.1002/prca.201300108
38. Wilhelm M, Schlegl J, Hahne H, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509(7502):582–587. doi:10.1038/nature13319
39. Noor Z, Ahn SB, Baker MS, Ranganathan S, Mohamedali A. Mass spectrometry–based protein identification in proteomics—a review. Brief Bioinform. 2021;22(2):1620–1638. doi:10.1093/bib/bbz163
40. Núñez EV, Domont GB, Nogueira FCS. iTRAQ-based shotgun proteomics approach for relative protein quantification. In: Guest PC, editor. Multiplex Biomarker Techniques: Methods and Applications. New York, NY: Springer; 2017:267–274.
41. The UC. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43(D1):D204–D212. doi:10.1093/nar/gku989
42. Sayers EW, Beck J, Bolton EE, et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021;49(D1):D10–D17. doi:10.1093/nar/gkaa892
43. Schaab C, Geiger T, Stoehr G, Cox J, Mann M. Analysis of high accuracy, quantitative proteomics data in the MaxQB database. Mol Cell Proteom. 2012;11(3):
44. Fujii K, Nakamura H, Nishimura T. Recent mass spectrometry-based proteomics for biomarker discovery in lung cancer, COPD, and asthma. Expert Rev Proteom. 2017;14(4):373–386. doi:10.1080/14789450.2017.1304215
45. Pratte KA, Curtis JL, Kechris K, et al. Soluble receptor for advanced glycation end products (sRAGE) as a biomarker of COPD. Resp Res. 2021;22(1):127. doi:10.1186/s12931-021-01686-z
46. Bozinovski S, Hutchinson A, Thompson M, et al. Serum amyloid A is a biomarker of acute exacerbations of chronic obstructive pulmonary disease. Am J Resp Crit Care. 2008;177(3):269–278. doi:10.1164/rccm.200705-678OC
47. Kim S, Ahn H, Park J, et al. A proteomics-based analysis of blood biomarkers for the diagnosis of COPD acute exacerbation. Int J Chronic Obstruct. 2021;16:1497–1508. doi:10.2147/COPD.S308305
48. Leung JM, Chen V, Hollander Z, et al. COPD exacerbation biomarkers validated using multiple reaction monitoring mass spectrometry. PLoS One. 2016;11(8):e0161129. doi:10.1371/journal.pone.0161129
49. Diao W, Shen N, Du Y, et al. Identification of thyroxine-binding globulin as a candidate plasma marker of chronic obstructive pulmonary disease. Int J Chronic Obstruct. 2017;12:1549–1564. doi:10.2147/COPD.S137806
50. Diao W, Shen N, Du Y, et al. Fetuin-B (FETUB): a plasma biomarker candidate related to the severity of lung function in COPD. Sci Rep. 2016;6(1):30045. doi:10.1038/srep30045
51. Beiko T, Janech MG, Alekseyenko AV, et al. Serum proteins associated with emphysema progression in severe alpha-1 antitrypsin deficiency. Am J Physiol Lung Cell. 2017;3(4):204–216. doi:10.15326/jcopdf.4.3.2016.0180
52. Lv MY, Qiang LX, Wang BC, et al. Complex evaluation of surfactant protein A and D as biomarkers for the severity of COPD. Int J Chronic Obstruct. 2022;17:1537–1552. doi:10.2147/COPD.S366988
53. Hristova VA, Watson A, Chaerkady R, et al. Multiomics links global surfactant dysregulation with airflow obstruction and emphysema in COPD. Erj Open Res. 2023;9(3):00378–2022. doi:10.1183/23120541.00378-2022
54. Yang M, Kohler M, Heyder T, et al. Long-term smoking alters abundance of over half of the proteome in bronchoalveolar lavage cell in smokers with normal spirometry, with effects on molecular pathways associated with COPD. Resp Res. 2018;19(1):40. doi:10.1186/s12931-017-0695-6
55. Dasgupta A, Chakraborty R, Saha B, et al. Sputum protein biomarkers in airway diseases: a pilot study. Int J Chronic Obstruct. 2021;16:2203–2215. doi:10.2147/COPD.S306035
56. Gao J, Ohlmeier S, Nieminen P, et al. Elevated sputum BPIFB1 levels in smokers with chronic obstructive pulmonary disease: a longitudinal study. Am J Physiol-Lung C. 2015;309(1):L17–L26. doi:10.1152/ajplung.00082.2015
57. Mallia-Milanes B, Dufour A, Philp C, et al. TAILS proteomics reveals dynamic changes in airway proteolysis controlling protease activity and innate immunity during COPD exacerbations. Am J Physiol-Lung C. 2018;315(6):L1003–L1014. doi:10.1152/ajplung.00175.2018
58. Ohlmeier S, Nieminen P, Gao J, et al. Lung tissue proteomics identifies elevated transglutaminase 2 levels in stable chronic obstructive pulmonary disease. Am J Physiol-Lung C. 2016;310(11):L1155–L1165. doi:10.1152/ajplung.00021.2016
59. Sun P, Ye R, Wang C, Bai S, Zhao L. Identification of proteomic signatures associated with COPD frequent exacerbators. Life Sci. 2019;230:1–9. doi:10.1016/j.lfs.2019.05.047
60. Liu Y, Liu H, Li C, Ma C, Ge W. Proteome profiling of lung tissues in Chronic Obstructive Pulmonary Disease (COPD): platelet and macrophage dysfunction contribute to the pathogenesis of COPD. Int J Chronic Obstruct. 2020;15:973–980. doi:10.2147/COPD.S246845
61. Skerrett-Byrne DA, Bromfield EG, Murray HC, et al. Time-resolved proteomic profiling of cigarette smoke-induced experimental chronic obstructive pulmonary disease. Respirology. 2021;26(10):960–973. doi:10.1111/resp.14111
62. Ye R, Wang C, Sun P, Bai S, Zhao L. AGR3 regulates airway epithelial junctions in patients with frequent exacerbations of COPD. Front Pharmacol. 2021;12. doi:10.3389/fphar.2021.669403
63. Franciosi L, Postma DS, van den Berge M, et al. Susceptibility to COPD: differential proteomic profiling after acute smoking. PLoS One. 2014;9(7):e102037. doi:10.1371/journal.pone.0102037
64. Qin W, Huang H, Dai Y, Han W, Gao Y. Proteome analysis of urinary biomarkers in a cigarette smoke-induced COPD rat model. Resp Res. 2022;23(1):156. doi:10.1186/s12931-022-02070-1
65. Coxson HO, Dirksen A, Edwards LD, et al. The presence and progression of emphysema in COPD as determined by CT scanning and biomarker expression: a prospective analysis from the ECLIPSE study. Lancet Respir Med. 2013;1(2):129–136. doi:10.1016/S2213-2600(13)70006-7
66. Regan EA, Hersh CP, Castaldi PJ, et al. Omics and the search for blood biomarkers in chronic obstructive pulmonary disease. Insights from COPDGene. Am J Resp Cell Mol. 2019;61(2):143–149. doi:10.1165/rcmb.2018-0245PS
67. Takei N, Suzuki M, Makita H, et al. Serum alpha-1 antitrypsin levels and the clinical course of chronic obstructive pulmonary disease. Int J Chronic Obstruct. 2019;14:2885–2893. doi:10.2147/COPD.S225365
68. Kranenburg AR, Willems-Widyastuti A, Mooi WJ, et al. Enhanced bronchial expression of extracellular matrix proteins in chronic obstructive pulmonary disease. Am J Clin Pathol. 2006;126(5):725–735. doi:10.1309/JC477FAEL1YKV54W
69. Man SFP, Xing L, Connett JE, et al. Circulating fibronectin to C-reactive protein ratio and mortality: a biomarker in COPD? Eur Respir J. 2008;32(6):1451. doi:10.1183/09031936.00153207
70. Zuo L, Pannell BK, Zhou T, Chuang C. Historical role of alpha-1-antitrypsin deficiency in respiratory and hepatic complications. Gene. 2016;589(2):118–122. doi:10.1016/j.gene.2016.01.004
71. Joanna C. Targeted screening programmes in COPD: how to identify individuals with α1-antitrypsin deficiency. Eur Respir Rev. 2015;24(135):40. doi:10.1183/09059180.00010614
72. Joan BS, Sarah JL, Rupert J, et al. Trends of testing for and diagnosis of α1-antitrypsin deficiency in the UK: more testing is needed. Eur Respir J. 2018;52(1):1800360. doi:10.1183/13993003.00360-2018
73. Meyer KC, Raghu G, Baughman RP, et al. An official American thoracic society clinical practice guideline: the clinical utility of bronchoalveolar lavage cellular analysis in interstitial lung disease. Am J Resp Crit Care. 2012;185(9):1004–1014. doi:10.1164/rccm.201202-0320ST
74. Hogea S, Tudorache E, Pescaru C, Marc M, Oancea C. Bronchoalveolar lavage: role in the evaluation of pulmonary interstitial disease. Expert Rev Resp Med. 2020;14(11):1117–1130. doi:10.1080/17476348.2020.1806063
75. Pelaia G, Terracciano R, Vatrella A, et al. Application of proteomics and peptidomics to COPD. Biomed Res Int. 2014;2014:764581. doi:10.1155/2014/764581
76. Casado B, Iadarola P, Pannell LK, et al. Protein expression in sputum of smokers and chronic obstructive pulmonary disease patients: a pilot study by CapLC-ESI-Q-TOF. J Proteome Res. 2007;6(12):4615–4623. doi:10.1021/pr070440q
77. Amato MD, Iadarola P, Viglio S. Proteomic analysis of human sputum for the diagnosis of lung disorders: where are we today? Int J Mol Sci. 2022;23(10):5692.
78. Ohlmeier S, Mazur W, Linja-aho A, et al. Sputum proteomics identifies elevated PIGR levels in smokers and mild-to-moderate COPD. J Proteome Res. 2012;11(2):599–608. doi:10.1021/pr2006395
79. Britto CJ, Cohn L. Bactericidal/permeability-increasing protein fold–containing family member A1 in airway host protection and respiratory disease. Am J Resp Cell Mol. 2014;52(5):525–534. doi:10.1165/rcmb.2014-0297RT
80. Franciosi L, Govorukhina N, Fusetti F, et al. Proteomic analysis of human epithelial lining fluid by microfluidics-based nanoLC-MS/MS: a feasibility study. Electrophoresis. 2013;34(18):2683–2694. doi:10.1002/elps.201300020
81. Bowler RP, Ellison MC, Reisdorph N. Proteomics in pulmonary medicine. Chest. 2006;130(2):567–574. doi:10.1378/chest.130.2.567
82. Franciosi L, Govorukhina N, Ten Hacken N, Postma D, Bischoff R. Proteomics of epithelial lining fluid obtained by bronchoscopic microprobe sampling. In: Toms SA, Weil RJ, editors. Nanoproteomics: Methods and Protocols. Totowa, NJ: Humana Press; 2011:17–28.
83. Casado B, Luisetti M, Iadarola P. Advances in proteomic techniques for biomarker discovery in COPD. Expert Rev Clin Immunol. 2011;7(1):111–123. doi:10.1586/eci.10.75
84. Sun C, Zhou T, Xie G, et al. Proteomics of exhaled breath condensate in stable COPD and non-COPD controls using tandem mass tags (TMTs) quantitative mass spectrometry: a pilot study. J Proteomics. 2019;206:103392. doi:10.1016/j.jprot.2019.103392
85. López-Sánchez LM, Jurado-Gámez B, Feu-Collado N, et al. Exhaled breath condensate biomarkers for the early diagnosis of lung cancer using proteomics. Am J Physiol-Lung C. 2017;313(4):L664–L676. doi:10.1152/ajplung.00119.2017
86. Fumagalli M, Ferrari F, Luisetti M, et al. Profiling the proteome of exhaled breath condensate in healthy smokers and COPD patients by LC-MS/MS. Int J Mol Sci. 2012:13894–13910. doi:10.3390/ijms131113894
87. Zhou M, Kong Y, Wang X, et al. LC-MS/MS-based quantitative proteomics analysis of different stages of non-small-cell lung cancer. Biomed Res Int. 2021;2021:5561569. doi:10.1155/2021/5561569
88. Wu J, Li X, Zhao M, Huang H, Sun W, Gao Y. Early detection of urinary proteome biomarkers for effective early treatment of pulmonary fibrosis in a rat model. Proteomics. 2017;11(11–12):1700103. doi:10.1002/prca.201700103
89. Qin W, Zhang X, Chen L, et al. Differential urine proteome analysis of a ventilator-induced lung injury rat model by label-free quantitative and parallel reaction monitoring proteomics. Sci Rep. 2021;11(1):21446. doi:10.1038/s41598-021-01007-w
90. Batra R, Uni R, Akchurin OM, et al. Urine-based multi-omic comparative analysis of COVID-19 and bacterial sepsis-induced ARDS. Mol Med. 2023;29(1):13. doi:10.1186/s10020-023-00609-6
91. Wu Q, Fenton RA. Urinary proteomics for kidney dysfunction: insights and trends. Expert Rev Proteom. 2021;18(6):437–452. doi:10.1080/14789450.2021.1950535
92. Wang X, Cheng Z. Cross-sectional studies: strengths, weaknesses, and recommendations. Chest. 2020;158(1):S65–S71. doi:10.1016/j.chest.2020.03.012
93. Liang W, Yang Y, Gong S, et al. Airway dysbiosis accelerates lung function decline in chronic obstructive pulmonary disease. Cell Host Microbe. 2023;31(6):1054–1070.e9. doi:10.1016/j.chom.2023.04.018
94. Zhuang Y, Xing F, Ghosh D, et al. Deep learning on graphs for multi-omics classification of COPD. PLoS One. 2023;18(4):e0284563. doi:10.1371/journal.pone.0284563
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.