Back to Journals » International Journal of General Medicine » Volume 16
Prognostic and Immune Infiltration Analysis of Transaldolase 1 (TALDO1) in Hepatocellular Carcinoma
Received 6 July 2023
Accepted for publication 10 November 2023
Published 8 December 2023 Volume 2023:16 Pages 5779—5788
DOI https://doi.org/10.2147/IJGM.S425490
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Kedan Cen, Caide Lu
Department of Hepatopancreatobiliary Surgery, The Affiliated Lihuili Hospital, Ningbo University, Ningbo, Zhejiang, People’s Republic of China
Correspondence: Caide Lu, Department of Hepatopancreatobiliary Surgery, The Affiliated Lihuili Hospital, Ningbo University, 1111 Jiangnan Road, Ningbo, 315040, Zhejiang, People’s Republic of China, Email [email protected]
Background: Transaldolase 1 (TALDO1) deficiency was associated with hepatocellular carcinoma (HCC), but the association between TALDO1 and prognosis in HCC remains unclear.
Material and Methods: RNA-seq and clinical data of HCC from The Cancer Genome Atlas (TCGA) database and Gene Expression Omnibus (GEO) database were analyzed. CCK8 and EdU assays were utilized to examine the effect of TALDO1 on HCC proliferation. Transwell assay was used to explore the effect of TALDO1 on HCC migration. Western Blot was applied to detected the expression levels of pathway-related proteins.
Results: TALDO1 mRNA level was higher in HCC tissues than in control normal tissues in TCGA and GEO databases (P< 0.001, P< 0.001). Expression of TALDO1 mRNA was associated with histologic grade (P=0.021). Multivariate regression analysis revealed that high TALDO1 mRNA expression was an independent risk factor for prognosis (P< 0.001). High expression of TALDO1 had poor overall survival (OS) (P=0.046). Additionally, Nomogram prognostic model of TALDO1 and clinicopathological parameters could predict the prognostic OS of HCC patients. Functional enrichment and immune infiltration analysis revealed that TALDO1 negatively regulates immune response. Furthermore, knockdown TALDO1 expression suppressed the proliferation and migration ability of HCC cells. Mechanistically, TALDO1 leaded to activation of the mitogen-activated protein kinase (MAPK) pathway and enhancement of epithelial-mesenchymal transition (EMT).
Conclusion: Our findings demonstrated that TALDO1 could serve as a promising novel biomarker for HCC patients, which might modulate the immune microenvironment resulting in a poor prognosis.
Keywords: transaldolase 1, prognosis, immune infiltration, proliferation, migration
Introduction
Liver cancer is one of the most fatal malignant tumors worldwide. In the global cancer epidemic research in 2020, liver-related malignant tumors ranked third in all cancer mortality.1 Hepatocellular carcinoma (HCC) is the most common primary liver cancer.2 Surgical resection, liver transplantation, and radiofrequency ablation are effective treatments for HCC,3 but many patients have overall survival (OS) shortly after treatment. Therefore, developing new prognostic biomarkers and treatment methods for HCC is significant.
The pentose phosphate pathway (PPP) is critical for protecting against oxidative stress and cell apoptosis, and its dysregulation has been implicated in the occurrence and development of numerous tumors.4,5 Transaldolase 1 (TALDO1), a key enzyme in the PPP, is responsible for generating ribo-5-phosphate (R5P) for nucleic acid synthesis and producing nicotinamide adenine dinucleotide phosphate (NADPH) for lipid biosynthesis.6 TALDO1 deficiency is closely related to hepatocarcinogenesis.7 Furthermore, TALDO1 abnormal expression has been associated with the prognosis of several types of cancer.8–10 However, there are few published research papers on the prognostic value of TALDO1 in HCC.
The tumor microenvironment plays a vital role in cancer occurrence, progression, and treatment.11 The tumor microenvironment is made up of various crucial elements, such as tumor infiltration immune cells, chemokines, and cytokines. In recent years, the field of cancer therapy has been transformed by immune-checkpoint inhibitors (ICIs), which have garnered heightened attention for their potential in HCC treatment.12 Nevertheless, there is a lack of supplementary biomarkers to inform clinical decision-making.
In this study, we analyzed the TALDO1 expression differences between normal liver tissue and liver cancer tissue and explored the relationship between TALDO1 and the prognosis of HCC patients. We also conducted functional enrichment and immunological infiltration analysis in HCC. Then, we explore the effect of the downregulation of TALDO1 in HCC cells. We found that TALDO1 is a potential prognostic biomarker and plays a vital role in the biological behavior of HCC.
Materials and Methods
Data Collection
RNA-seq and clinical data of TCGA-LIHC were downloaded from UCSU Xena, which contained 50 non-tumor liver tissues and 371 liver tissues. RNA-seq data of GSE3637613 was obtained from the GEO database, which had 193 normal liver tissues and 240 liver tissues.
Immune Infiltration Analysis
The tumor immune estimation resource, version 2 (TIMER2; http://timer.cistrome.org/)13 web server’s “Immune-Gene” module was utilized to explore the relationship between TALDO1 expression and immune infiltration in HCC. In terms of CD4+ T cells and regulatory T cells were chosen. The Spearman’s rank correlation test was applied to calculate the p values and partial correlation coefficients. TISIDB (http://cis.hku.hk/TISIDB/)14 was employed to explore the association of TALDO1 with immune inhibitors and immune stimulators in HCC.
Functional Enrichment Analysis
The 150 most relevant positive and negative genes of TALDO1 were submitted to the Database for Annotation, Visualization, and Integrated Discovery (DAVID, v6.8). The official gene symbols were used as identifiers, and Homo sapiens was selected as the species. Subsequently, Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed to obtain enrichment results.
Cell Culture
The human HCC cell line Huh-7 was acquired from the Chinese Academy of Sciences Cell Bank (CASCB, Shanghai, China), and SK-Hep-1 was acquired from Procell Life Science & Technology Co., Ltd. (Procell, Wuhan, China); both cell lines came with short tandem repeat (STR) appraisal certificates. The cells were cultivated in DMEM (Gibco, Rockville, MD, USA) containing 10% fetal bovine serum (Gibco), 100 mg/mL streptomycin and 100 IU/mL penicillin and cultivated in 5% CO2 at 37 °C.
Transfection
The siRNAs were purchased from GenePharma (Jiangsu, China). The interference sequences were as follows: si-NC, sense, 5’-UUCUCCGAACGUGUCACGUTT-3’, antisense, 5’-ACGUGACACGUUCGGAGAATT-3’; si-TALDO1, sense, 5’- GGCAAACACCGACAAGAAATT-3’, antisense, 5’-UUUCUUGUCGGUGUUUGCCTT-3’. For transfection, we used Lipofectamine 2000 (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions. The siRNAs were transfected into cells with Lipofectamine 2000, and the cells were cultured for 48 h.
Real-Time Quantitative PCR (qPCR)
The total RNA of cells was extracted by TransZol Up Plus RNA Kit (TransGen, Beijing, China). TransScript All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (TransGen, Beijing, China) was utilized to reverse transcribe extracted RNA into cDNA. Subsequently, qPCR was conducted with PerfectStart Green qPCR SuperMix (TransGen, Beijing, China) on the LightCycler 480 System (Roche, Germany). Setting up the thermal cycling program on qPCR instrument. Common conditions include: Initial denaturation: Typically at 94°C for 30 seconds. Cycling conditions: Typically 40–45 cycles consisting of denaturation at 94°C for 5 seconds, annealing at 50–60°C for 15 seconds, and extension at 72°C for 10 seconds. The relative expression of TALDO1 was normalized to β-actin by the 2−ΔΔCt method. The primer sequences are listed as follows. TALDO1 forward: 5’-CCCCTCGGTCTTGCTATGTC-3’, TALDO1 reverse: 5’-TACTCGTCGATGGCGTGGAA-3’; β-actin forward: 5’- TGGCACCCAGCACAATGAA-3’, β-actin reverse: 5’-CTAAGTCATAGTCCGCCTAGAAGCA −3’.
Western Blotting
The total proteins in the human HCC cells were extracted with RIPA lysis buffer (Solarbio, Beijing, China) containing 1% PMSF (Solarbio), subjected to 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, and then transferred from the gel to a PVDF membrane (Bio-Rad Laboratories, Hercules, CA, USA). The PVDF membrane was blocked with 5% skim milk for 1 h at room temperature and incubated with primary antibodies at 4 °C overnight. After washing with Tris-buffered saline with 0.1% Tween-20 (TBST), the membrane was incubated with the secondary antibody at room temperature for 2 h. Finally, the protein bands were visualized using ECL luminous liquid (Advansta, San Jose, CA, USA) and quantified via ImageJ software (National Institutes of Health, USA). Antibodies against TALDO1, E-cadherin, ERK1/2, p-ERK1/2 and secondary antibodies were obtained from ABclonal Technology (Wuhan, China). Antibody against β-actin was obtained from Cell Signaling Technology (Danvers, MA, USA). The size marker used was the Blue Plus V Protein Marker from TransGen (Beijing, China).
CCK-8 Assay
SK-Hep-1 or Huh7 cells transfected with si-NC and si-TALDO1 were placed in 96-well plates with 2000 cells in each well. CCK-8 solution (10 μL) was added to each well and incubated for 2 h at 37 °C. The optical density value at 450 nm was detected with a microplate reader (Thermo Fisher Scientific, Waltham, MA, USA) and recorded at 0, 24, 48, and 72 h to determine cell viability.
5-Ethynyl-2′-Deoxyuridine (EdU) Assay
Cell proliferation was evaluated through the EdU cell proliferation kit (Invitrogen, USA). Following the manufacturer’s specifications, cells were incubated with 1× EdU working solution for 2 h. After removing the culture medium, the cells were fixed by 4% paraformaldehyde (Solarbio, China) and permeabilized with 0.5% Triton Χ-100. Subsequently, the cells were incubated at room temperature for 20 min, stained with DAPI for 5 min, and observed and photographed with a fluorescence-inverted microscope (Leica, Germany).
Migration Assays
Migration assays were plated in 24-well transwell plates (8 μm pore size, Corning, NY, USA). Then, 50,000 HCC cells in serum-free DMEM were plated in the upper chamber. Then, 500 µL of complete culture medium containing 20% FBS was added to the lower chamber. After 24 h, the cells that migrated to the lower surface of the membrane were fixed with 4% paraformaldehyde (Solarbio, China) and stained with crystal violet. The cells were photographed under an optical microscope.
Statistical Analysis
Statistical analyses were conducted by SPSS 21.0 or Prism 8. A t or U-test was used to analyze the experimental results between groups. All statistical data are expressed as the mean ± SD (mean ± SD) unless otherwise specified, with a P value of < 0.05 (two-tailed) was considered to indicate statistical significance.
Results
Upregulated TALDO1 Expression is Associated with Poor Prognosis in HCC
We explored the expression of TALDO1 mRNA between normal and tumor tissue in HCC (Figure 1A). The results showed that the level of TALDO1 mRNA is increased in tumor tissues. Also, we validated the upregulation of TALDO1 expression in HCC using the GSE36376 databases. Patients with varying expression levels of TALDO1 showed distinct clinical and pathological characteristics patterns (Figure 1B). Increases in TALDO1, tumor stage, T stage, histologic grade, weight, age and gender showed asymmetric distributions in the TCGA database. TALDO1 was notably enriched in higher histologic grade HCC (P=0.021, Figure 1C). We conducted Cox proportional hazard model and Kaplan-Meier analyses to explore the correlation between TALDO1 and prognosis in HCC (Figure 1D). Univariate and multivariate Cox regression analysis showed that T stage and TALDO1 were independent prognostic factors. Kaplan-Meier analysis demonstrated that patients with high expression of TALDO1 exhibited poor overall survival (OS) (P = 0.046, Figure 1E) in the TCGA database. Therefore, TALDO1 can be a predictive marker for overall survival in HCC patients.
Establishing Nomogram for Predicting Prognosis of OS for HCC Patients
To predict the prognostic OS of HCC patients, we developed a Nomogram prognostic model using clinicopathological parameters and TALDO1. In a Nomogram, each predictor is assigned a specific numerical value or score based on its relative importance in predicting the OS. The total score is calculated by summing the scores of all the predictors for an individual patient. (Figure 2A). In addition, the calibration curves demonstrated a strong agreement between the observed and predicted OS without deviation from the reference line (Figure 2B), and the AUC of the 1-year, 3-year, and 5-year ROC curves of the line graph prognostic model were 0.74, 0.72, and 0.70, respectively (Figure 2C). These findings validate the reliability and accuracy of our Nomogram prognostic model in effectively estimating the OS of HCC patients.
Functional Enrichment and Immune Infiltration Analysis of TALDO1
To explore the biological function of TALDO1, we chose the most related gene by Pearson correlation in the TCGA databases (Figure 3A and B). In positive genes with TALDO1, Biological processes most associated with TALDO1 were mRNA splicing via spliceosome, mRNA processing and positive regulation of transcription, which was DNA-templated. Cellular components were nucleoplasm, nucleus and ribonucleoprotein complex. Molecular functions were RNA binding, protein binding and mRNA binding. Signaling pathways were spliceosome and ubiquitin-mediated proteolysis. In positive genes with TALDO1, Biological processes most related to TALDO1 were acute phase response, complement activation and positive regulation of immune response. Cellular components were the mitochondrial matrix, extracellular space and extracellular region. Molecular functions were identical protein binding, complement binding, and component C3b binding. Signaling pathways were complement and coagulation cascades and PPAR signaling pathways. These results suggested that TALDO1 plays an essential role in the biology and immunity of HCC.
To further explore the relationship between TALDO1 and the immune microenvironment, we analyzed the relationship between TALDO1 expression and various tumor-infiltration immune cells (Figure 3C and D). TALDO1 was significantly negatively associated with the infiltration level of CD4+ T cells (Rho=−0.158, P=0.003). However, TALDO1 had a significant positive correlation with the infiltration level of regulatory T cells (Rho=0.134, P=0.012). Then, we explored the relationship between TALDO1 expression and various immune signatures using the TISIDB database (Figure 3E and F). Our findings revealed a correlation between TALDO1 expression and immune inhibitor as well as immune stimulator in HCC, including CD276 (Rho=−0.165, P=0.001), CXCL12 (Rho=−0.243, P<0.001), IL 6R (Rho=−0.221, P<0.001), TNFSF15 (Rho=−0.317, P<0.001), LGALS9 (Rho=0.162, P=0.002), LAG3 (Rho=0.141, P=0.007), CTLA4 (Rho=0.14, P=0.007). Therefore, the evidence revealed that TALDO1 may act as a negative immunoregulatory factor to regulate immune infiltration in HCC.
Knockdown TALDO1 Inhibited the Proliferation and Migration of HCC Through ERK/EMT Pathway
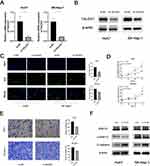
Based on the clinical evidence suggesting a potential involvement of TALDO1 in HCC progression, we conducted gene silencing experiments to explore its role in Huh7 and SK-Hep-1 cell lines. The efficacy of TALDO1-siRNA was validated using both qPCR and Western Blot (Figure 4A and B). Then, we assessed the viability and proliferation of HCC cells through CCK8 and EdU assays. The results demonstrated that the knockdown of TALDO1 led to a reduction in the proliferation of HCC (Figure 4C and D). Additionally, there was a consistent decrease in the migratory ability of HCC cells as observed in the transwell assay (Figure 4E).
EMT (epithelial-mesenchymal transition) was associated with cancer metastasis.15 In addition, ERK MAP kinase signaling pathway could regulate cell proliferation and migration.16 Therefore, we detected the potential effects of knockdown TALDO1 on ERK/EMT pathway. Western blot data revealed that downregulation of TALDO1 in HCC cells inhibited the phosphorylation ratio of ERK1/2 (Figure 4F). Besides, knockdown TALDO1 increased the expression of the epithelial biomarker E-cadherin in SK-Hep-1 and Huh7 cells. Above all, our results suggested that TALDO1 knockdown could suppress the proliferation and migration in HCC by regulating the ERK/EMT pathway.
Discussion
HCC is characterized by high mortality and a poor prognosis,17 mainly due to the absence of specific biomarkers and recognizable clinical symptoms in the early stages. The accumulating evidence reported there is an abnormal expression of TALDO1 in various tumors.8,18 In addition, TALDO1 is closely associated with HCC. In this study, we analyzed the TALDO1 expression in HCC patients and accessed its correlation with overall survival. In the clinical analysis, TALDO1 expression upregulated significantly in HCC tissues than in normal tissues. High expression of TALDO1 indicated poor prognosis. In addition, TALDO1 might regulate the immune response by immunological infiltration cells. In vitro validation studies showed that knockdown TALDO1 inhibited the proliferation and migration in HCC cells.
Verhoeven et al19 identified the initial TALDO1 deficiency, characterized primarily by cirrhosis and hepatosplenomegaly in terms of clinical manifestations. Since then, researchers have also uncovered that TALDO1 deficiency predisposes to cirrhosis and HCC in mice and humans.7,9,20 It demonstrated that TALDO1 plays a crucial role in liver carcinogenesis and the development of liver cancer. However, the relationship between the expression of TALDO1 and HCC prognosis remains unknown. In this study, we comprehensively analyzed TALDO1 expression in liver cancer using RNA-seq data from the TCGA and GEO databases. And we found that the expression level of TALDO1 mRNA in liver cancer tissues was higher than in normal tissues. The expression of TALDO1 was significantly up-regulated in rat liver pre-neoplastic foci by cDNA microarray assays.21 Furthermore, the abnormal expression of TALDO1 was found in various cancers. In upper tract urothelial carcinoma,8 the expression of TALDO1 in tumor tissues was also significantly higher than in adjacent normal tissues. In addition, the specific activity of TALDO1 in microvesicles in breast cancer cells was higher than in non-cancerous breast cells.18 It suggested that the abnormal expression of TALDO1 is connected with hepatocarcinogenesis. Next, we explored the correlation of TALDO1 with clinical characteristics in HCC patients through the TCGA database. HCC patients with high expression of TALDO1 were associated with higher histological grades. In both mice and human liver cancer, the protein level of TALDO1 in the serum and tissues of metastatic cases was significantly higher than that of non-metastatic cases.22 And high TALDO1 expression is associated with a poor response to HER2 inhibition in a cohort of breast cancer patient.23 These findings showed that TALDO1 abnormal expression has correlated to poor clinical characteristics. In both upper urothelial carcinoma8 and luminal breast cancer,24 high expression of TALDO1 was significantly associated with a low survival rate of patients. According to survival analysis, the overall survival time of HCC patients with high expression of TALDO1 was shorter than that of patients with low expression of TALDO1. Our results were similar to earlier findings, in which increased expression of TALDO1 has poor overall survival. As a result, high expression of TALDO1 in HCC tissues could predict poor prognosis in patients. TALDO1 could be used as a prognostic biomarker in HCC.
According to the functional enrichment analysis, TALDO1 was positively related to common tumor biological functions. Additionally, acute-phase response, as part of the innate immune system, can activate the complement factor and coagulation cascades. TALDO1 was negatively linked to the acute phase and regulation of immune response. TALDO1 is involved in autoimmune diseases, such as multiple sclerosis25 and systemic lupus erythematosus.26 This suggests that TALDO1 might regulate the immune microenvironment in HCC. So, we further explore the relationship between immunological infiltration cells and TALDO1 in HCC. The CD4+ cell was adversely related to TALDO1 expression. However, T cell regulation was positively associated with TALDO1 expression. Besides, TALDO1 expression was related to some immune stimulators and immune inhibitors. Therefore, we hypothesize that TALDO1 can promote HCC by regulating immune cells and immune factors, but it needs more experiments to verify.
We further explored the impact of TALDO1 knockdown in human liver cancer cells. We knocked down the expression of TALDO1 by transfection of siRNAs into Huh7 and SK-Hep-1. Our findings showed that knockdown TALDO1 inhibited liver cancer cells’ proliferation and migration ability. In addition, knockdown TALDO1 inhibited HCC cells metastasis might be linked to ERK/EMT pathway. Therefore, TALDO1 might be a potential tumor promoter in vitro, just like the previous bioinformatics analysis results.
Nevertheless, there are still limitations in the study. First, the study lacks clinical data validation from our research center. Second, functional enrichment and immune infiltration analysis need more experimental assays to prove. Third, further validation of the role of TALDO1 in HCC are still needs to be confirmed in animal models.
This study identified that high expression of TALDO1 has a poorer prognosis than low expression of TALDO1. And TALDO1 is also associated with immune infiltration. Knockdown of TALDO1 can significantly inhibit the proliferation and migration of HCC cells.
Conclusion
Our study reveals the high expression of TALDO1 in HCC, and it is associated with poor clinical outcomes. We also highlight the potential role of TALDO1 in regulating tumor immunity based on functional enrichment analyses and immune infiltration analysis. Furthermore, the knockdown of TALDO1 inhibited cell proliferation and migration. These findings provide a basis for further experimentation and clinical trials to assess the clinical relevance of TALDO1 in cancer treatment and prognosis prediction.
Ethics Approval
This study was conducted in accordance with the Declaration of Helsinki and approved by Ethics Committee of Lihuili Hospital (No. KY2022SL389-01).
Funding
Our research was funded by the Ningbo Medical and Health Brand Discipline (PPXK2018-03).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33.
2. European Assoc Study L. EASL clinical practice guidelines: management of Hepatocellular carcinoma. J Hepatol. 2018;69(1):182–236. doi:10.1016/j.jhep.2018.03.019
3. Forner A, Llovet JM, Bruix J. Hepatocellular carcinoma. Lancet. 2012;379(9822):1245–1255. doi:10.1016/S0140-6736(11)61347-0
4. Liu C-L, Hsu Y-C, Lee -J-J, et al. Targeting the pentose phosphate pathway increases reactive oxygen species and induces apoptosis in thyroid cancer cells. Mol Cell Endocrinol. 2020;2020:499.
5. Li Q, Qin T, Bi Z, et al. Rac1 activates non-oxidative pentose phosphate pathway to induce chemoresistance of breast cancer. Nat Commun. 2020;11(1):1.
6. Jiang P, Du W, Wu M. Regulation of the pentose phosphate pathway in cancer. Protein Cell. 2014;5(8):592–602. doi:10.1007/s13238-014-0082-8
7. Grammatikopoulos T, Hadzic N, Foskett P, et al. Liver disease and risk of Hepatocellular carcinoma in children with mutations in TALDO1. Hepatol Commun. 2022;6(3):473–479. doi:10.1002/hep4.1824
8. Wu Y-R, Lee Y-C, W-M L, et al. High transaldolase 1 expression predicts poor survival of patients with upper tract urothelial carcinoma. Pathol Int. 2021;71(7):463–470. doi:10.1111/pin.13101
9. Leduc CA, Crouch EE, Wilson A, et al. Novel association of early onset hepatocellular carcinoma with transaldolase deficiency. JIMD Rep. 2014;12:121–127.
10. Basta PV, Bensen JT, Tse C-K, et al. Genetic variation in transaldolase 1 and risk of squamous cell carcinoma of the head and neck. Cancer Detect Prev. 2008;32(3):200–208. doi:10.1016/j.cdp.2008.08.008
11. Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi:10.1038/s41591-018-0014-x
12. Donne R, Lujambio A. The liver cancer immune microenvironment: therapeutic implications for hepatocellular carcinoma. Hepatology. 2022. doi:10.1002/hep.32740
13. Li T, Fu J, Zeng Z, et al. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi:10.1093/nar/gkaa407
14. Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi:10.1093/bioinformatics/btz210
15. Fontana R, Mestre-Farrera A, Yang J, et al. Update on epithelial-mesenchymal plasticity in cancer progression. Mech Dis. 2023;19:1.
16. Guan X. Cancer metastases: challenges and opportunities. Acta Pharm Sin B. 2015;5(5):402–418. doi:10.1016/j.apsb.2015.07.005
17. Villanueva A, Longo DL. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–1462. doi:10.1056/NEJMra1713263
18. Risha Y, Susevski V, Huttmann N, et al. Breast cancer-derived microvesicles are the source of functional metabolic enzymes as potential targets for cancer therapy. Biomedicines. 2021;9(2):107. doi:10.3390/biomedicines9020107
19. Verhoeven NM, Huck JHJ, Roos B, et al. Transaldolase deficiency: liver cirrhosis associated with a new inborn error in the pentose phosphate pathway. Am J Hum Genet. 2001;68(5):1086–1092. doi:10.1086/320108
20. Hanczko R, Fernandez DR, Doherty E, et al. Prevention of hepatocarcinogenesis and increased susceptibility to acetaminophen-induced liver failure in transaldolase-deficient mice by N-acetylcysteine. J Clin Invest. 2009;119(6):1546–1557. doi:10.1172/JCI35722
21. Suzuki S, Asamoto M, Tsujimura K, Shirai T. Specific differences in gene expression profile revealed by cDNA microarray analysis of glutathione S-transferase placental form (GST-P) immunohistochemically positive rat liver foci and surrounding tissue. Carcinogenesis. 2004;25(3):439–443. doi:10.1093/carcin/bgh030
22. Wang C, Guo K, Gao D, et al. Identification of transaldolase as a novel serum biomarker for hepatocellular carcinoma metastasis using xenografted mouse model and clinic samples. Cancer Lett. 2011;313(2):154–166. doi:10.1016/j.canlet.2011.08.031
23. Ding Y, Gong C, Huang D, et al. Synthetic lethality between HER2 and transaldolase in intrinsically resistant HER2-positive breast cancers. Nat Commun. 2018;9. doi:10.1038/s41467-018-06651-x
24. Alfarsi LH, El Ansari R, Craze ML, et al. SLC1A5 co-expression with TALDO1 associates with endocrine therapy failure in estrogen receptor-positive breast cancer. Breast Cancer Res Treat. 2021;189(2):317–331. doi:10.1007/s10549-021-06298-1
25. Colombo E, Banki K, Tatum AH, et al. Comparative analysis of antibody and cell-mediated autoimmunity to transaldolase and myelin basic protein in patients with multiple sclerosis. J Clin Invest. 1997;99(6):1238–1250. doi:10.1172/JCI119281
26. Al-Mayouf SM, AlTassan RS, AlOwain MA. Systemic lupus erythematosus in a girl with PTEN variant and transaldolase deficiency: a novel phenotype. Clin Rheumatol. 2020;39(11):3511–3515. doi:10.1007/s10067-020-05205-1
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.