Back to Journals » Journal of Inflammation Research » Volume 15
Probiotic Combination CBLEB Alleviates Streptococcus pneumoniae Infection Through Immune Regulation in Immunocompromised Rats
Authors Lv L, Peng L , Shi D, Shao L, Jiang H, Yan R
Received 5 November 2021
Accepted for publication 28 January 2022
Published 15 February 2022 Volume 2022:15 Pages 987—1004
DOI https://doi.org/10.2147/JIR.S348047
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Longxian Lv,1,* Ling Peng,2,* Ding Shi,1 Li Shao,3,* Huiyong Jiang,1 Ren Yan1
1State Key Laboratory for the Diagnosis and Treatment of Infectious Diseases, National Clinical Research Center for Infectious Diseases, Collaborative Innovation Center for Diagnosis and Treatment of Infectious Diseases, The First Affiliated Hospital, College of Medicine, Zhejiang University, Hangzhou, Zhejiang, People’s Republic of China; 2Department of Respiratory Disease, Zhejiang Provincial People’s Hospital, Affiliated People’s Hospital, Hangzhou Medical College, Hangzhou, Zhejiang, People’s Republic of China; 3Institute of Translational Medicine, Affiliated Hospital of Hangzhou Normal University, Hangzhou, Zhejiang, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Huiyong Jiang; Ren Yan, The First Affiliated Hospital, College of Medicine, Zhejiang University, No. 79 Qingchun Road, Hangzhou, Zhejiang, 310003, People’s Republic Of China, Tel/Fax +86-571-87236453, Email [email protected]; [email protected]
Background: Streptococcus pneumoniae (SP) is the most common cause of bacterial pneumonia, especially for people with immature or compromised immune systems. In addition to vaccination and antibiotics, immune regulation through microbial intervention has emerged in recent anti-SP infection research. This study investigated the therapeutic effect of a combination of live Bifidobacterium, Lactobacillus, Enterococcus, and Bacillus (CBLEB), a widely used probiotic drug, on SP infection in rats.
Methods: An immunocompromised SP-infection rat model was established by intraperitoneal injection of cyclophosphamide and nasal administration of SP strain ATCC49619. Samples from SP-infected, SP-infected and CBLEB-treated, and healthy rats were collected to determine blood indicators, serum cytokines, gut microbiota, faecal and serum metabolomes, lung- and colon-gene transcriptions, and histopathological features.
Results: CBLEB treatment alleviated weight loss, inflammation, organ damage, increase in basophil percentage, red cell distribution width, and RANTES levels and decrease in total protein and albumin levels of immunocompromised SP-infection rats. Furthermore, CBLEB treatment alleviated dysbiosis in gut microbiota, including altered microbial composition and the aberrant abundance of opportunistic pathogenic bacterial taxa such as Eggerthellaceae, and disorders in gut and serum metabolism, including altered metabolomic profiles and differentially enriched metabolites such as 2,4-di-tert-butylphenol in faeces and L-tyrosine in serum. The transcriptome analysis results indicated that the underlying mechanism by which CBLEB fights SP infection is mainly attributed to its regulation of immune-related pathways such as TLR and NLR signalling in the lungs and infection-, inflammation- or metabolism-related pathways such as TCR signalling in the colon.
Conclusion: The present study shows a potential value of CBLEB in the treatment of SP infection.
Keywords: Streptococcus pneumonia infection, probiotics, microbiota, metabolism, transcriptome
Introduction
Pneumonia is the single greatest infectious cause of death worldwide and is associated with a tremendous economic burden.1 According to the World Health Organization’s report, pneumonia accounted for 15% of all deaths among children under 5 years old, killing 808,694 children in 2017.2 Streptococcus pneumoniae (SP) is one of the major human pathogens and the most common cause of bacterial pneumonia, septicaemia and meningitis.3,4 However, unlike some major bacterial respiratory pathogens, such as Mycobacterium tuberculosis, SP is also a ubiquitous human commensal of the upper respiratory tract, specifically the nasopharynx.5 Colonization with different pneumococcal strains occurs in children up to 2 years of age, with the prevalence of nasopharyngeal carriage has been estimated at 27–65% among infants.6 Because the SP strains causing invasive infections are surrounded by a polysaccharide capsule layer that inhibits innate and adaptive immune responses to infection,7 such asymptomatic colonization in people with an immature or compromised immune system can progress to mild disease (ie, sinusitis and otitis media) and occasionally to pneumonia, sepsis, etc.8
Vaccination and the use of antibiotics are the most conventional methods of prevention and treatment of SP infection. However, antibody-based immunity induced by protein-polysaccharide conjugate pneumococcal vaccines (PCVs) is highly serotype specific, protecting against only certain serotype strains whose capsular polysaccharide types are included in the vaccines,9 and PCVs are less effective at preventing pneumonia than septicaemia and meningitis.10,11 The excessive use of antibiotics has led to a serious situation of high-level antimicrobial resistance worldwide,12 and the antibiotics themselves can also have an impact on health (ie, disrupting the normal human microbiota, leading to secondary infections), especially for immunocompromised individuals, elderly individuals, and children under 2 years of age. Thus, new strategies to fight against pneumococci, such as targeting enhancement of the immunity of high-risk groups, are also required.
The human microbiota is important for the development and maintenance of immunity, and it also helps the host resist pathogen invasion. Therefore, appropriate microbiota regulation is considered a potential solution to prevent and treat SP infection. In recent years, oral and nasal administration of some lactic acid bacteria, such as Lacticaseibacillus rhamnosus CRL150513 and Lacticaseibacillus casei CRL431,14 has been reported to improve T cell-mediated immunity against SP infection. The probiotic drug combination of live Bifidobacterium infantis CGMCC0460.1, Lactobacillus acidophilus CGMCC0460.2, Enterococcus faecalis CGMCC0460.3 and Bacillus cereus CGMCC0460.4 (CBLEB), is widely used in China. It has been reported that taking CBLEB has a therapeutic effect on immunodeficiency.15 The present study aimed to investigate the effect of CBLEB against pneumococcal infection in immunocompromised rats and the underlying mechanism.
Materials and Methods
Microorganisms
The probiotic drug CBLEB was purchased from Hangzhou Yuanda Biopharmaceutical Co., Ltd. The probiotic powders were dissolved in sterile saline to prepare a CBLEB solution (B. infantis, 4.7×109 colony-forming units (CFU)/kg; L. acidophilus, 4.7×109 CFU/kg; E. faecalis, 4.7×108 CFU/kg; and B. cereus, 4.7×107 CFU/kg). S. pneumoniae ATCC49619 (serotype 19) was purchased from the American Type Culture Collection (ATCC). The pathogen was incubated at 37 °C in an atmosphere of 5% CO2 and harvested by centrifugation at 3600 ×g for 10 min at 4 °C. Then, the concentration of the pathogen was adjusted to the dose used in the nasal administration.
Animal Experimental Design and Sample Collection
Specific pathogen-free male Sprague-Dawley rats (6–8 wks old, 200–300 g) were randomly divided into three groups, the healthy control (HC), SP infection (SP) and probiotic treatment (SP+CBLEB) groups, and each group consisted of ten rats. All animals were supplied with balanced rodent food and water and maintained with a controlled light/dark cycle. The entire experiment lasted for 30 days.
The immunocompromised rat model representing a group with high risk of SP infection was established by intraperitoneal injection of cyclophosphamide (40 mg/kg), and the rats were subsequently infected with SP. Briefly, the rats in the SP and SP+CBLEB groups received cyclophosphamide on days 12, 13, 14, and 21 and nasal administration of SP (1.0 × 108 CFU per rat) on day 22, while the rats in the HC group received the same dose of saline. Additionally, the rats in the SP+CBLEB group were gavaged with CBLEB solution once daily for 28 days, while the rats in the HC and SP infection groups were administered normal saline. On day 30, rats were anaesthetized with 400 mg/kg chloral hydrate by intraperitoneal injection. Spleen and lung samples were fixed in 10% paraformaldehyde for 24 h, embedded in paraffin, cut into 3-μm sections and stained with haematoxylin and eosin (H&E). Blood and faecal samples were collected and immediately used or stored at −80 °C until use. All experiments were approved by the Institutional Animal Care and Research Advisory Committee at Zhejiang University.
Quantitative Detection of SP Invasion
SP invasive infection was verified by using real-time PCR (RT-PCR), as previously described.16 Briefly, DNA was extracted from lung samples with DNeasy Blood & Tissue Kits (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions. The burden of SP was estimated from the number of ply gene copies per gram of lung sample. Quantitative PCR was performed with a ViiA7 RT-PCR system (Applied Biosystems, Waltham, Massachusetts, USA) and using SYBR Premix Ex TaqTM (RR420A; TaKaRa, Tokyo, Japan). The reaction mixture (10 μL) contained 5 μL of TB Green, 0.4 μL (1 μM) of each primer (forward: TGCAGAGCGTCCTTTGGTCTAT; reverse: CTCTTACTCGTGGTTTCCAACTTGA), 0.2 μL of ROX reference dye, 2 μL of DNA template, and 2 μL of PCR-grade water.16 The reaction was hot-started at 95 °C for 30s, followed by 40 cycles of 95 °C for 5 s, 57 °C for 30s and 72 °C for 30s, and the subsequent melt curve was assessed from 60 °C to 95 °C at 0.2 °C/read. Plasmids harbouring the PCR-amplified ply gene were used as standards to produce a calibration curve.
Histology
Spleen index = spleen weight/body weight × 100.
Thymus index = thymus weight/body weight × 100.
Spleen and lung sections were scanned with a section scanner after H&E staining, and the images were used to evaluate the degree of tissue damage, inflammation, and necrosis. Pulmonary histological damage was quantified using the American Thoracic Society 2010 Lung Injury Scoring System.17 The spleen pathological severity score was graded from 0 to 3 points as described previously.18
Haematological Tests
Routine blood indexes, such as white blood cell, neutrophil, lymphocyte, monocyte, eosinophil and basophil counts, were measured with an XN-2000 automatic haematology analyser (Sysmex, Tokyo, Japan). Liver and kidney function indicators, including total protein, albumin, globulin, alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (ALP), total bilirubin, direct bilirubin, indirect bilirubin, g-glutamyltransferase (GGT), creatinine, urea nitrogen and uric acid levels, were measured with an automatic biochemical analyser (Hitachi 7600–210; Tokyo, Japan).
Cytokine Assay
Serum cytokine concentrations were measured by using a Bio-Plex Pro™ rat cytokine 23-plex assay kit (Bio-Rad Laboratories, Hercules, USA), including interleukin-1α (IL-1α), IL-1b, IL-2, IL-4, IL-5, IL-6, IL-7, IL-10, IL-12 (p70), IL-13, IL-17A, IL-18, granulocyte colony-stimulating factor (G-CSF), granulocyte-macrophage colony-stimulating factor (GM-CSF), macrophage colony-stimulating factor (M-CSF), monocyte chemoattractant protein 1 (MCP-1), macrophage inflammatory protein 1a (MIP-1α), MIP-3α, tumour necrosis factor-a (TNF-α), interferon-g (IFN-γ), vascular endothelial growth factor (VEGF), growth-regulated a protein (GRO/KC), and regulated upon activation normal T cell expressed and secreted protein (RANTES).
Gut Microbiota Analysis
DNA was extracted from faecal samples using a QIAamp Fast DNA Stool Mini Kit (Qiagen, Valencia, CA, USA). The V3-V4 region of the 16S rDNA gene was amplified using the primers 338F (ACTCCTACGGGAGGCAGCAG) and 806R (GGACTACHVGGGTWTCTAAT). PCR amplification, library preparation, and DNA sequencing were conducted as described previously on the Illumina MiSeq platform (Illumina, San Diego, CA, USA).5 The raw reads were trimmed, filtered and then merged using FLASH v1.2.11. Quality control was conducted using FastQC v0.11.8. Quantitative Insights Into Microbial Ecology (QIIME) v1.9.1 software was used for the operational taxonomic unit (OTU) assignment, clustering, identification against the Greengenes database and the NCBI 16S Microbial database, and subsequent statistical analysis of microbial diversity and differential enrichment.
Metabolomic Analysis
Serum (200 μL) and faecal (20 mg) samples were pretreated before gas chromatography-mass spectrometry (GC-MS) analysis as described previously.19 Briefly, samples were added to 800 μL of prechilled methanol before homogenization, centrifugation and filtration. The supernatant was subsequently dried with nitrogen, methoxymated and trimethylsilylated with 20 μL of heptadecanoic acid (1 μg/μL) as an internal standard. The prepared samples were then assayed with an Agilent 7890A-5975C GC-MS system (Agilent, CA, USA) using an Agilent J&W Hp-5 MS column. Raw GC-MS data were processed using Agilent Qualitative Analysis software (vB.07.00). Metabolite identification was based on the results of a programmed comparison with the National Institute of Standards and Technology (NIST) 17 mass spectral library using a matching score higher than 80 as the threshold. The identified compounds were then manually screened to remove the derivatization reagent to obtain the final result. Data were normalized to the internal standard prior to multivariate analysis. Orthogonal partial least squares discriminant analysis (OPLS-DA) was performed to visualize metabolic differences between groups using Umetrics SIMCA software v14.1.
Transcriptomic Analysis
Total RNA was extracted from lung and colon samples using an RNeasy Plus Mini Kit (Qiagen, Valencia, CA, USA). RNA quantification, library preparation and sequencing were conducted as described previously.15 Prior to alignment, raw reads were trimmed using Trimmomatic to generate clean data. The paired-end clean reads were then aligned to a reference genome (Rnor_6.0, GCA_000001895.4) using HISTA2 v2.0.5. Each gene’s expression value (fragments per kilobase of transcripts per million mapped fragments, FPKM) was calculated using FeatureCounts v1.5.0-p3. The R package “DESeq2” was used to analyse differential expression between two groups, and “ClusterProfiler” package was used to determine the gene annotation, functional classification, and pathway enrichment. The significant transcriptome results were verified by RT-PCR. The mRNA of representative genes was reverse transcribed and then measured in triplicate. Gene transcription was transformed into relative expression based on internal control, the housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The primer sequences are provided in Supplemental Table S1.
Statistics
The Shapiro–Wilk test was used to determine whether the data were normally distributed. The Wilcoxon test was used to compare any two data sets that were not normally distributed; otherwise, one-way ANOVA followed by the Student-Newman-Keuls method was used. Differentially abundant metabolites were selected according to the variable importance in the projection (VIP) values obtained from the OPLS-DA model and the P-values from the Wilcoxon test or one-way ANOVA. Spearman’s rank test was used to analyse correlations between every two significant factors. P-values of the hypergeometric test were used to determine significance in gene set enrichment analysis. P-values were adjusted for the false discovery rate using the Benjamini-Hochberg method where necessary (*Padj < 0.05; **Padj < 0.01; ***Padj < 0.001).
Results
CBLEB Treatment Alleviates Weight Loss, Inflammation and Organ Damage in Immunocompromised SP-Infection Rats
The number of SP in the lung of rats was quantified. The SP burden in the lung samples of the SP and SP+CBLEB groups increased significantly, indicating that SP invasive infection occurred (Supplemental Figure S1). The changes in body weight of rats in each group were measured during the study (Figure 1A). In the First 11 days (before cyclophosphamide injection), the weight of the rats in each group increased, and there were no significant differences among these groups. From 12 to 21 days, compared with the rats in the HC group, the rats in the SP group (injected with cyclophosphamide, gavaged with normal saline) had significant weight loss, and this weight loss trend slowed in the SP+CBLEB group (injected with cyclophosphamide, gavaged with CBLEB). On the 22nd day, the body weight of rats in all groups decreased whether SP or saline was nasally administered. This outcome may be because anaesthesia administration affected the normal diet of the rats. Among the groups of rats, the weight loss of rats in the SP group was the most significant, reflecting the detrimental effects of SP infection on health.
Administration of CBLEB partially alleviated abnormality of blood indicator levels and immune dysfunction in immunocompromised SP-infection rats. First, CBLEB administration significantly alleviated the elevation in blood basophil percentage and red cell distribution width (RDW) and serum RANTES levels, as well as the decline in blood total protein (TP) and albumin (ALB) levels. Second, in the SP+CBLEB group, the decrease in blood indirect bilirubin (IB) levels and increase in basophil count and uric acid levels in the SP group became insignificant when compared with those in the HC group (Figure 1C).
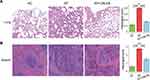
CBLEB administration also partially alleviated organ damage in immunocompromised SP-infection rats. First, the spleen index was significantly increased, while the thymus index was decreased in both the SP and SP+CBLEB groups compared with those in the HC group (Figure 1B). CBLEB administration also tended to alleviate this increase in the spleen index (P = 0.065 in SP vs SP + CBLEB). Second, as shown in the photomicrographs of H&E-stained tissue sections, CBLEB administration significantly ameliorated pulmonary pathological changes, such as inflammatory cell infiltration, alveolar cavity collapse, alveolar wall and interstitial thickening, and fibrinoid exudation in the bronchial cavity in immunocompromised SP-infection rats (Figure 2A), as well as changes in the spleen, including enlarged red pulp area, decreased white pulp area, histiocyte proliferation, hyperaemia, haemosiderin deposition, and increased multinucleated giant cell (MGC) numbers (Figure 2B). Furthermore, CBLEB administration significantly lowered the histological scores in the lung and spleen (Figure 2A and B).
CBLEB Treatment Rescues Dysbiosis of the Gut Microbiota in Immunocompromised SP-Infection Rats
We obtained a total of 2,632,886 reads from 30 faecal samples from the three groups by 16S rDNA amplicon sequencing. Alpha diversity analyses showed that no significant differences in the Shannon and Chao1 indexes were present among the HC, SP, and SP+CBLEB groups, indicating similar community diversity in these groups (Figure 3A).
For beta diversity, a principal coordinates analysis (PCoA) plot based on the unweighted UniFrac distance demonstrated that the overall microbial compositions of the three groups varied (Figure 3B). This result was confirmed by analysis of similarity (ANOSIM) (R = 0.47, P = 0.001).
Then, we further analysed alterations in the relative abundance of bacterial taxa in the three groups. Compared with that of HC rats, the gut microbiota of rats in the SP group was significantly altered (Figure 3C). First, the bacterial taxa from the phylum Firmicutes, such as Streptococcaceae, Fournierella, Monoglobus and Eubacterium ventriosum group, were significantly enriched in the SP group, whereas Peptostreptococcaceae, Jeotgalicoccus, Tuzzerella, Eubacterium, Romboutsia and an unclassified Peptostreptococcaceae genus were significantly depleted. Among them, Streptococcaceae, Peptostreptococcaceae, Romboutsia and the unclassified genus Peptostreptococcaceae were the most predominant taxa whose abundance was altered by SP (relative abundances were more than 1% in at least one group), suggesting their special role in the gut microbiota changes associated with SP infection. Second, the bacterial taxa from the phylum Actinobacteria, such as Eggerthellaceae, Adlercreutzia, Gordonibacter and Coriobacteriaceae bacterium CHKCI002, were significantly enriched in the SP group, whereas the bacterial taxa from the phylum Bacteroidetes, such as Muribaculum and Prevotellaceae UCG-001, were significantly depleted.
Compared with SP infection only, administration of CBLEB significantly alleviated the enrichment of Streptococcaceae, Eggerthellaceae, Monoglobaceae, Coriobacteriales Incertae Sedis, Monoglobus, Adlercreutzia, Fournierella and Eubacterium ventriosum group and the depletion of Eubacteriaceae, Eubacterium and Jeotgalicoccus in immunocompromised rats (Figure 3C). At the same time, compared with those in the HC group, the relative abundances of Peptostreptococcaceae and Romboutsia in the SP+CBLEB group did not change significantly, which indirectly implied the regulatory effect of CBLEB on the gut microbiota.
CBLEB Treatment Alleviates Disorder of Gut Metabolism in Immunocompromised SP-Infection Rats
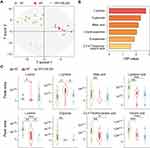
To investigate the regulatory effect of CBLEB on the gut metabolism of SP-infected rats, we analysed faecal samples using GC-MS. In total, we identified 81 metabolites from the three groups. A OPLS-DA plot showed that the metabolic profiles of rats in the HC, SP, and SP+CBLEB groups were clearly separated from each other (Figure 4A), indicating significant differences in their faecal metabolomic profiles. The VIP values of 12 metabolites (arachidonic acid, D-ribose, D-fructose, pantothenic acid, heptanoic acid, L-methionine, 1-linolenoylglycerol, L-sorbose, 5-hydroxyhexanoic acid, m-cresol, 2-propenoic acid, and phosphoric acid) were greater than 1.5, suggesting their important contribution to distinguishing the metabolic profiles of the three groups in this OPLS-DA model (Figure 4B).
According to the comparison of each metabolite, the levels of 11 metabolites (2,4-di-tert-butylphenol, D-glucose, D-lyxose, 1-hexadecanol, 1-octadecanol, 1-pentadecanol, D-fructose, glyceric acid, m-cresol, L-alanine, and L-threonine) were higher, while the levels of (Z,Z)-9,12-octadecadienoic acid, 1-monolinolein, and 2-propenoic acid were lower in the SP group than in the HC group. Upon treatment with CBLEB, the changes in 7 of these metabolites (2,4-di-tert-butylphenol, D-lyxose, 1-octadecanol, glyceric acid, m-cresol, (Z,Z)-9,12-octadecadienoic acid, and 2-propenoic acid) became nonsignificant (Figure 4C), reflecting the regulatory effect of CBLEB on gut metabolism disorder.
CBLEB Treatment Alleviates Serum Metabolism Disorder in Immunocompromised SP-Infection Rats
Next, we investigated the regulatory effect of CBLEB on serum metabolism. A total of 60 metabolites were identified from the HC, SP, and SP+CBLEB groups. An OPLS-DA plot showed that the metabolic profiles of the rats in the three groups were also clearly separated from each other (Figure 5A), indicating significant differences in their serum metabolomic profiles. The VIP values of 6 metabolites (L-proline, D-glucose, malic acid, L-hydroxyproline, D-arabinose, and 2,3,4-trihydroxybutyric acid) were greater than 1.5, suggesting their important contribution to distinguishing the metabolic profiles of the three groups in this OPLS-DA model (Figure 5B).
During the comparison of each metabolite among the groups, we found that L-lysine and L-tyrosine were enriched, whereas malic acid, L-glutamic acid, L-proline, D-glucose, 2,3,4-trihydroxybutyric acid, and glyceric acid were depleted in the SP group compared with those in the HC group (Figure 5C). Upon treatment with CBLEB, the enrichment of L-tyrosine was significantly reversed in immunocompromised SP-infection rats, and differential serum levels of L-lysine, malic acid and D-glucose between the SP and HC groups were no longer observed, indicating the regulatory effect of CBLEB on serum metabolism disorder.
CBLEB Treatment Partially Reversed Transcriptional Regulatory Changes in the Lungs and Colons of Immunocompromised SP-Infection Rats
Compared with that in the lung samples of rats in the HC group, the transcription of 597 genes was increased and that of 399 genes was decreased in the lung samples of rats in the SP group (Padj < 0.05). Mapping these genes to the KEGG pathway database revealed 21 upregulated pathways and 6 downregulated pathways. Among them, the upregulation of 11 pathways, which mainly relate to the immune response (Toll-like receptor (TLR) signalling, natural killer (NK) cell-mediated cytotoxicity, NOD-like receptor (NLR) signalling, haematopoietic cell lineage, and chemokine signalling), infection (malaria, measles, influenza A, and Epstein-Barr virus infection), and immune signalling (cytokine-cytokine receptor interaction, viral protein interaction with cytokines and cytokine receptors), was relieved by CBLEB treatment (Figure 6A). To validate these transcriptome results, we conducted RT-PCR analysis of representative genes involve in above immune response pathways, including Tlr2, Tlr9, Cxcl10, Ccl5, Tnf, Il-1b, Il-6, Cd80, Cd48 and Nod2. We found that their expression was similar to that in the transcriptome analysis (Supplemental Figure S2A and 2C). Moreover, when comparing the SP+CBLEB and HC groups, the changes in the upregulated pathways, including homologous recombination, Fanconi anaemia, p53 signalling, transcriptional misregulation in cancer and hepatitis C, and the downregulated pathways, including oxidative phosphorylation, metabolism of xenobiotics by cytochrome P450, and hepatocellular carcinoma, caused by SP infection were no longer significant. These results indicate that the regulation of pathways such as immunity, infection, and metabolism in the lung may be involved in the mechanism of CBLEB activity against SP infection.
In the colon, the transcription of 210 genes was upregulated, while that of 360 genes was downregulated in the SP group compared with the HC group (Padj < 0.05). Mapping these genes to the KEGG database revealed 23 downregulated pathways and 5 upregulated pathways. Among them, the upregulation of 3 pathways, axon guidance, African trypanosomiasis and Staphylococcus aureus infection, as well as the downregulation of 3 pathways, phenylalanine metabolism, renin-angiotensin system and chemical carcinogenesis, were reversed by CBLEB treatment (Figure 6B). To validate these results, we conducted RT-PCR analysis of representative genes involve in above pathways, including Pak2 and Pak4 in axon guidance, Krt14 in Staphylococcus aureus infection, Hbb in African trypanosomiasis, Aldh3a1 in phenylalanine metabolism and chemical carcinogenesis, and Mcpt1 in renin-angiotensin system. We found that their expression was similar to that in the transcriptome analysis (Supplemental Figure S2B and 2D). Moreover, when comparing the SP+CBLEB and HC groups, the upregulation of pathways, including pantothenate and CoA biosynthesis, nonhomologous end-joining, T cell receptor (TCR) signalling, ErbB signalling, cytokine-cytokine receptor interaction, phosphatidylinositol signalling system, cell adhesion molecules, and p53 signalling, and the downregulation of pathways, including arachidonic acid metabolism, caused by SP infection became nonsignificant. These results indicate that these pathways in the colon may be involved in the mechanism of CBLEB activity against SP infection.
The Gut Microbiota and Metabolites Involved in CBLEB Treatment are Closely Correlated with Improved Health
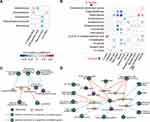
First, we analysed the correlations between the relative abundances of gut microbes and the levels of metabolites involved in CBLEB treatment using Spearman’s rank correlation test. Both the absolute value of the correlation coefficient r > 0.4 and P < 0.01 were used as the screening threshold to identify extremely significant results.20 Adlercreutzia and Gordonibacter were positively correlated with 1-octadecanol and m-cresol, while Peptostreptococcaceae, Eubacterium and Romboutsia were negatively correlated with glyceric acid (Figure 7A). Additionally, Adlercreutzia was also found to be negatively correlated with 2-propenoic acid.
Second, some gut microbes and metabolites may affect host health and diseases. Therefore, we analysed the correlations between gut microbes/metabolites and CBLEB treatment-improved serum indicators. We found that gut bacterial taxa, including Eggerthellaceae, Adlercreutzia, Gordonibacter, Streptococcaceae, Fournierella and Monoglobus, as well as gut metabolites, including 1-octadecanol, D-lyxose, glyceric acid and m-cresol, were negatively correlated with at least one of the following markers: serum malic acid, TP, ALB, and IB. Additionally, gut Streptococcaceae abundance was negatively correlated with serum basophil count and percentage and RDW; gut Adlercreutzia and 1-octadecanol were positively correlated with serum RDW and RANTES; and gut Jeotgalicoccus was positively correlated with serum TP and ALB and negatively correlated with uric acid and RANTES (Figure 7B).
The Gut Microbiota and Metabolites are Closely Correlated with the Pathways Involved in CBLEB Treatment
To reveal how gut microbes or metabolites affect the gut-lung axis during CBLEB treatment, we mapped the differentially expressed genes in the lung and colon that were significantly correlated with gut microbes and/or metabolites involved in CBLEB treatment to the KEGG pathway database.
In the colon, differential genes negatively correlated with Eggerthellaceae were significantly enriched in the TCR signalling pathway; those negatively correlated with Adlercreutzia or 2.4-di-tert-butylphenol were significantly enriched in ribosome biogenesis in eukaryotes; and those negatively correlated with (Z,Z)-9,12-octadecadienoic acid were significantly enriched in leukocyte transendothelial migration. Furthermore, the differential colon-expressed genes positively correlated with Eubacterium were enriched in NK cell-mediated cytotoxicity, and those positively correlated with Adlercreutzia were enriched in axon guidance (Figure 7C).
In the lung, differential genes negatively correlated with Eubacterium were significantly enriched in the measles, inflammatory bowel disease, cell adhesion molecules and TNF signalling pathways, those negatively correlated with Romboutsia were significantly enriched in Fanconi anaemia and malaria, those negatively correlated with Eggerthellaceae were significantly enriched in Huntington’s disease, those negatively correlated with Adlercreutzia were significantly enriched in fatty acid metabolism, and those negatively correlated with 2.4-di-tert-butylphenol were significantly enriched in fatty acid degradation. Moreover, the differential lung-expressed genes positively correlated with Fournierella, Streptococcaceae, or Eggerthellaceae were significantly enriched in NK cell-mediated cytotoxicity; those positively correlated with Adlercreutzia, Fournierella, Eggerthellaceae, 2.4-di-tert-butylphenol or glyceric acid were significantly enriched in malaria; those positively correlated with Fournierella or glyceric acid were significantly enriched in NLR signalling; those positively correlated with Adlercreutzia were significantly enriched in cytokine-cytokine receptor interaction and haematopoietic cell lineage; those positively correlated with Fournierella were significantly enriched in tuberculosis; and those positively correlated with 1-octadecanol were significantly enriched in mTOR signalling (Figure 7D).
Discussion
S. pneumoniae is the leading cause of bacterial pneumonia, the single greatest infectious cause of death worldwide.3 Microbiota regulation by probiotic administration has been reported to have great application potential in the preventing and treating SP infections. For example, heat-killed Lactiplantibacillus pentosus b240 protects mice from SP infection by augmenting the innate immune response;21 dietary supplementation with L. casei CRL43122 and L. rhamnosus CRL150523 is effective in modulating SP infection-induced coagulation activation, the inflammatory process, and lung damage in malnourished mice; and oral administration of a fermented milk drink containing L. rhamnosus GG (LGG), Bifidobacterium sp. B420, L. acidophilus 145, and Streptococcus thermophiles reduces the SP burden in the upper respiratory tract.24 However, there have been no reports on the use of any probiotic drug in the clinical prevention and treatment of SP infection. The probiotic drug CBLEB is popular in China as a gut microbiota regulator and is used by nearly 10 million people every year. A previous study reported that taking CBLEB has a therapeutic effect on immunodeficiency.15 In this study, we found that CBLEB can prevent weight loss in immunocompromised rats with SP infection, reduce lung inflammation and injury, and alleviate gut microbiota dysbiosis and gut and serum metabolism disorders. Its mechanism is related to the regulation of pathways involved in immunity, infection, and metabolism.
The gut microbiota plays an important role in host defence against pneumococcal pneumonia. The gastrointestinal tract is the primary interaction site between the host immune system and microbes, and it is well known that the gut microbiota is critical in establishing immune balance.25,26 In a mouse model, the use of broad-spectrum antibiotics after SP infection causes the depletion of the gut microbiota, increases bacterial loads in the lungs and blood, augments inflammation, accelerates organ failure and even results in death. Faecal microbiota transplantation to gut microbiota-depleted mice leads to normalization of pulmonary bacterial loads and TNF-α and IL-10 levels after SP infection.27 We found that CBLEB administration alleviated the enrichment of Streptococcaceae and the depletion of Eggerthellaceae, Adlercreutzia, Eubacterium, Fournierella, and Romboutsia in the gut of immunocompromised SP-infection rats. Some of these microbes have been reported to have important physiological or pathological functions. For example, members of the genus Eubacterium have been recognized as beneficial microbes since they produce butyrate and contribute to the homeostasis of bile acid and cholesterol, and alterations in Eubacterium spp. have been linked with various human diseases.28 Correspondingly, we found that the colon-expressed genes that positively correlated with Eubacterium were significantly enriched in the NK cell-mediated cytotoxicity pathway, while the lung-expressed genes that negatively correlated with this bacterial taxon were enriched in the measles, inflammatory bowel disease, cell adhesion molecule and TNF signalling pathways. These findings suggest that it is possible to infer the physiological and pathological functions of gut microbes through their correlated signalling pathways in the host. For example, we also found that colon-expressed genes positively correlated with Fournierella were enriched in the NK cell-mediated cytotoxicity and NLR signalling pathways; and the lung-expressed genes positively correlated with Adlercreutzia were enriched in the malaria, cytokine-cytokine receptor interaction and haematopoietic cell lineage pathways. These correlations imply the important roles of Fournierella and Adlercreutzia in the regulation of SP infection.
Some components and metabolites of gut microbes, such as lipopolysaccharides and short-chain fatty acids, which can be transported via the circulatory system, are important means of communication between the gut microbiota and the lungs.29 However, there are no reports on faecal metabolome alterations during SP infection. We found that CBLEB alleviates the faecal enrichment of 1-octadecanol, 2-propenoic acid, 2.4-di-tert-butylphenol, D-lyxose, glyceric acid and m-cresol, as well as the depletion of (Z,Z)-9,12-octadecadienoic acid in immunocompromised SP-infection rats. Among these metabolites, 1-octadecanol, glyceric acid and (Z,Z)-9,12-octadecadienoic acid are commonly found in animals, plants and microbes and can also be used in cosmetics, medicines or food additives;30–32 D-lyxose, a rare pentose found in bacterial glycolipids, is not usually utilized by microbes and is a precursor to antitumour and immunostimulatory α-galactosylceramide agents;33 and 2-propenoic acid, 2.4-di-tert-butylphenol and m-cresol are compounds with low toxicity and antioxidant functions.34–36 Nevertheless, the effects of these compounds on the lung and gut are not fully understood. In this work, we found that colon-expressed genes that negatively correlated with 2.4-di-tert-butylphenol were enriched in the ribosome biogenesis in eukaryotes pathway, as well as those negatively correlated with (Z,Z)-9,12-octadecadienoic acid were enriched in leukocyte transendothelial migration. Additionally, the differential lung-expressed genes that negatively correlated with 2.4-di-tert-butylphenol were enriched in fatty acid degradation, while those positively correlated genes were enriched in the malaria pathway; the differential lung-expressed genes positively correlated with 1-octadecanol were enriched in mTOR signalling, and those positively correlated with glyceric acid were enriched in the malaria and NLR signalling pathways. These findings elucidate how CBLEB-regulated gut metabolites function in the prevention and treatment of SP infection.
We also found that CBLEB alleviated the enrichment of L-tyrosine and L-lysine and the depletion of D-glucose and malic acid in the serum of immunocompromised SP-infection rats. Among these metabolites, the depletion of glucose in serum was observed in both a mouse model and patients challenged with SP infection, which may be due to liver metabolism disorder and excessive glucose consumption by the pathogen.37 In contrast, aberrant tyrosine, lysine and malic acid levels have not been reported in SP infection, which may be mainly due to the scarcity of serum metabolomic research on these patients or models. In serum samples from tuberculosis patients, enrichment of lysine and tyrosine has also been observed, and the reason may be related to liver damage.38 Importantly, malic acid is an intermediate metabolite of the citric acid cycle, and changes in its content reflect the regulation of the tricarboxylic acid (TCA) cycle by CBLEB treatment.39 Therefore, the changes in serum metabolism also indicate the positive regulatory effect of CBLEB on SP infection. Since serum components directly reflect and affect health, the rehabilitation effect of CBLEB on SP infection-related dysbiosis of serum metabolism is one of the important manifestations of its therapeutic role and mechanism.
Our results also showed that the most likely mechanism of CBLEB combatting SP infection is its regulation of immune-related pathways in the host’s lungs. First, CBLEB administration alleviates the upregulation of pathways involved in innate immunity, including TLR signalling, NLR signalling, and NK cell-mediated cytotoxicity. TLR2 recognizes lipoteichoic acid and peptidoglycan and thereby plays a protective role in SP infection; streptolysin can bind to TLR4 to induce an inflammatory response, and NOD2 mainly recognizes the bacterial muramyldipeptide, which can induce the production of inflammatory cytokines such as TNF-α and IL-16 through the NF-κB pathway and can also induce caspase-related cell apoptosis.40 It has been reported that intestinal microbes (Lactobacillus reuteri, E. faecalis, Lactobacillus crispatus and Clostridium orbiscindens) promote resistance to lung infection through NOD2 and GM-CSF.41 NK cells are involved in the control of microbial infections and protect the host directly through the production of cytotoxic effectors. During SP-triggered lung infection, NK cells are one of the major cell types responsible for IFN-γ production.42 Second, the upregulation of pathways involved in both innate and adaptive immunity, including the chemokine signalling and haematopoietic cell lineage pathways, was also alleviated by CBLEB treatment. SP strains causing invasive infections are surrounded by a polysaccharide capsule layer that inhibits innate and adaptive immune responses to infection.7 Chemokines and their signalling pathways play a critical role in regulating innate and adaptive immune responses and are involved in many physiological and pathological processes, such as SP infection.43 The haematopoietic cell lineage bears the hallmark characteristics of both the innate and adaptive immune systems. Haematopoietic tissues accelerate both the release of mature granulocytes and the production of new granulocytes to support and reinforce the ongoing recruitment of neutrophils in tissues infected by pathogens.44 Additionally, CBLEB alleviated the upregulation of pathways related to infection (malaria, measles, influenza A, and Epstein-Barr virus infection) and immune signalling (cytokine-cytokine receptor interaction, viral protein interaction with cytokines and cytokine receptors), which also reflects that the improvement in immune-related pathways is an important mechanism of CBLEB activity.
In the colon, we found that partial restoration of those infection-, inflammation- and metabolism-related pathways is the main mechanism by which CBLEB combats SP infection. First, CBLEB alleviates the upregulation of the axon guidance, arachidonic acid metabolism, African trypanosomiasis and S. aureus infection pathways. The expression of genes related to the axon guidance pathway, the critical step in neural circuit development, has been reported to be upregulated in the gastrointestinal tract of neonatal goats with diarrhoea.45 Arachidonic acid may be released from the phospholipids of the cell membrane during phospholipid hydrolysis and is important for the proper function of the immune system, such as promoting allergies and inflammation.46 Second, CBLEB alleviates the downregulation of some important life activity-related pathways, including phenylalanine metabolism, pantothenate and CoA biosynthesis, renin-angiotensin system, nonhomologous end-joining, ErbB signalling, TCR signalling, phosphatidylinositol signalling, cell adhesion molecules, and cytokine-cytokine receptor interaction. Defects in the phenylalanine metabolism pathway are the cause of three well-known disorders: phenylketonuria, albinism, and alkaptonuria;47 coenzyme A (CoA) is an essential cofactor for cell growth and involved in many metabolic reactions, such as phospholipid synthesis, fatty acid synthesis and degradation, and TCA cycle function;48 the renin-angiotensin system is involved in the regulation of smooth muscle of the blood vessels and bowel, the absorption and secretion of glucose, amino acids, fluids and electrolytes, the permeability of the gut mucosa, and gut inflammation;49 non-homologous end-joining is the predominant DNA repair pathway required to detect, process, and ligate DNA double-stranded breaks throughout the cell cycle, and defects in its function may result in severe combined immunodeficiency;50 disruption of the ErbB signalling network, an essential component for mucosal protection and/or the adaptive response to external injury in the gut, will result in major defects in gut epithelial development and in the reparative response to gut injury;51 TCR activation promotes a number of signalling cascades that ultimately determine cell fate through regulating cytokine production, cell survival, proliferation, and differentiation; various phosphoinositides are involved in many signalling pathways, such as the PI3K-Akt pathway that mediates cell proliferation, survival, and metabolism;52 and cell adhesion molecules play a critical role in many biological processes, such as haemostasis, the immune response, inflammation, embryogenesis, and neuronal tissue development.53 Therefore, the regulation of these pathways in the intestine is an important part of the mechanism by which CBLEB combats SP infection.
In conclusion, CBLEB administration reduces the pathogen load in the lungs, ameliorates weight loss and lung inflammatory injury, and improves systemic health in SP-infected rats. Its mechanism of action is related to the role of CBLEB in remodelling the gut microbiota, improving faecal and serum metabolism, and thereby regulating the host’s immune response.
Data Sharing Statement
The original contributions presented in the study are publicly available. This data can be found in NCBI Sequence Read Archive (SRA) database with BioProject ID PRJNA771216.
Ethical Statement
All experiments were approved by the Institutional Animal Care and Research Advisory Committee at Zhejiang University and followed the Laboratory animal-Guidelines for ethical review of animal welfare (GB/T 35892-2018).
Acknowledgments
This work is supported by the National Key Research and Development Program of China (No. 2018YFC2000500); the National Natural Science Foundation of China (Nos. 81570512, 81790631 and 81790634); the Natural Science Foundation of Zhejiang Province in China (Nos. LQ19H030007, LGF19H030009 and LQ20H030010).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Welte T, Torres A, Nathwani D. Clinical and economic burden of community-acquired pneumonia among adults in Europe. Thorax. 2012;67(1):71–79. doi:10.1136/thx.2009.129502
2. WHO fact sheet on pneumonia provides key facts and information on causes, presenting features, economic costs, treatment, prevention and WHO response. Available from: https://www.who.int/news-room/fact-sheets/detail/pneumonia.
3. O’Brien KL, Wolfson LJ, Watt JP, et al. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374(9693):893–902.
4. Wang Y, Jiang B, Guo Y, et al. Cross-protective mucosal immunity mediated by memory Th17 cells against Streptococcus pneumoniae lung infection. Mucosal Immunol. 2017;10(1):250–259. doi:10.1038/mi.2016.41
5. Yan R, Wang K, Wang Q, et al. Probiotic Lactobacillus casei Shirota prevents acute liver injury by reshaping the gut microbiota to alleviate excessive inflammation and metabolic disorders. Microb Biotechnol. 2021;15(1):247–261. doi:10.1111/1751-7915.13750
6. Hussain M, Melegaro A, Pebody RG, et al. A longitudinal household study of Streptococcus pneumoniae nasopharyngeal carriage in a UK setting. Epidemiol Infect. 2005;133(5):891–898. doi:10.1017/S0950268805004012
7. Hyams C, Camberlein E, Cohen JM, Bax K, Brown JS. The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect Immun. 2010;78(2):704–715. doi:10.1128/IAI.00881-09
8. Ivanov S, Paget C, Trottein F. Role of non-conventional T lymphocytes in respiratory infections: the case of the pneumococcus. PLoS Pathog. 2014;10(10):e1004300. doi:10.1371/journal.ppat.1004300
9. Pilishvili T, Lexau C, Farley MM, et al. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J Infect Dis. 2010;201(1):32–41. doi:10.1086/648593
10. Bonten MJ, Huijts SM, Bolkenbaas M, et al. Polysaccharide conjugate vaccine against pneumococcal pneumonia in adults. N Engl J Med. 2015;372(12):1114–1125. doi:10.1056/NEJMoa1408544
11. Jose RJ, Brown JS. Adult pneumococcal vaccination: advances, impact, and unmet needs. Curr Opin Pulm Med. 2017;23(3):225–230. doi:10.1097/MCP.0000000000000369
12. Yahiaoui RY, Heijer CDD, Bijnen EMV, et al. Prevalence and antibiotic resistance of commensal Streptococcus pneumoniae in nine European countries. Future Microbiol. 2016;11(6):737–744. doi:10.2217/fmb-2015-0011
13. Barbieri N, Herrera M, Salva S, Villena J, Alvarez S. Lactobacillus rhamnosus CRL1505 nasal administration improves recovery of T-cell mediated immunity against pneumococcal infection in malnourished mice. Benef Microbes. 2017;8(3):393–405. doi:10.3920/BM2016.0152
14. Vintini EO, Medina MS. Host immunity in the protective response to nasal immunization with a pneumococcal antigen associated to live and heat-killed Lactobacillus casei. BMC Immunol. 2011;12:46. doi:10.1186/1471-2172-12-46
15. Lv L, Mu D, Du Y, Yan R, Jiang H. Mechanism of the Immunomodulatory Effect of the Combination of Live Bifidobacterium, Lactobacillus, Enterococcus, and Bacillus on Immunocompromised Rats. Front Immunol. 2021;12:694344. doi:10.3389/fimmu.2021.694344
16. Tzanakaki G, Tsopanomichalou M, Kesanopoulos K, et al. Simultaneous single-tube PCR assay for the detection of Neisseria meningitidis, Haemophilus influenzae type b and Streptococcus pneumoniae. Clin Microbiol Infect. 2005;11(5):386–390. doi:10.1111/j.1469-0691.2005.01109.x
17. Matute-Bello G, Downey G, Moore BB, et al. An official American Thoracic Society workshop report: features and measurements of experimental acute lung injury in animals. Am J Respir Cell Mol Biol. 2011;44(5):725–738. doi:10.1165/rcmb.2009-0210ST
18. Xiping Z, Ruiping Z, Binyan Y, et al. Protecting effects of a large dose of dexamethasone on spleen injury of rats with severe acute pancreatitis. J Gastroenterol Hepatol. 2010;25(2):302–308. doi:10.1111/j.1440-1746.2009.05999.x
19. Lv L, Jiang H, Chen Y, et al. The faecal metabolome in COVID-19 patients is altered and associated with clinical features and gut microbes. Anal Chim Acta. 2021;1152:338267. doi:10.1016/j.aca.2021.338267
20. Lv L, Jiang H, Chen X, et al. The Salivary Microbiota of Patients With Primary Biliary Cholangitis Is Distinctive and Pathogenic. Front Immunol. 2021;12:2950. doi:10.3389/fimmu.2021.713647
21. Tanaka A, Seki M, Yamahira S, et al. Lactobacillus pentosus strain b240 suppresses pneumonia induced by Streptococcus pneumoniae in mice. Lett Appl Microbiol. 2011;53(1):35–43. doi:10.1111/j.1472-765X.2011.03079.x
22. Zelaya H, Laino J, Villena J, Marranzino G, Alvarez S, Aguero G. Lactobacillus casei CRL431 modulates hemostatic activation induced by protein malnourishment and pneumococcal respiratory infection. Appl Microbiol Biotechnol. 2020;104(24):10669–10683. doi:10.1007/s00253-020-10957-6
23. Zelaya H, Laino J, Villena J, Alvarez S, Aguero G. Lactobacillus rhamnosus CRL1505 beneficially modulates the immuno-coagulative response after pneumococcal infection in immunocompromised malnourished mice. Can J Microbiol. 2013;59(10):684–693. doi:10.1139/cjm-2013-0361
24. Gluck U, Gebbers JO. Ingested probiotics reduce nasal colonization with pathogenic bacteria (Staphylococcus aureus, Streptococcus pneumoniae, and beta-hemolytic streptococci). Am J Clin Nutr. 2003;77(2):517–520. doi:10.1093/ajcn/77.2.517
25. Schluter J, Peled JU, Taylor BP, et al. The gut microbiota is associated with immune cell dynamics in humans. Nature. 2020;588(7837):303–307. doi:10.1038/s41586-020-2971-8
26. Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9(5):313–323. doi:10.1038/nri2515
27. Schuijt TJ, Lankelma JM, Scicluna BP, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. 2016;65(4):575–583. doi:10.1136/gutjnl-2015-309728
28. Mukherjee A, Lordan C, Ross RP, Cotter PD. Gut microbes from the phylogenetically diverse genus Eubacterium and their various contributions to gut health. Gut Microbes. 2020;12(1):1802866. doi:10.1080/19490976.2020.1802866
29. Lv L, Gu S, Jiang H, et al. Gut mycobiota alterations in patients with COVID-19 and H1N1 infections and their associations with clinical features. Commun Biol. 2021;4(1):480. doi:10.1038/s42003-021-02036-x
30. Jin Z, Wu K, Hou J, Yu K, Shen Y, Guo S. A PTX/nitinol stent combination with temperature-responsive phase-change 1-hexadecanol for magnetocaloric drug delivery: magnetocaloric drug release and esophagus tissue penetration. Biomaterials. 2018;153:49–58. doi:10.1016/j.biomaterials.2017.10.040
31. Fox KJ, Prather KLJ. Production of D-Glyceric acid from D-Galacturonate in Escherichia coli. J Ind Microbiol Biotechnol. 2020;47(12):1075–1081. doi:10.1007/s10295-020-02323-2
32. Su H, Liu R, Chang M, Huang J, Wang X. Dietary linoleic acid intake and blood inflammatory markers: a systematic review and meta-analysis of randomized controlled trials. Food Funct. 2017;8(9):3091–3103. doi:10.1039/C7FO00433H
33. Huang J, Chen Z, Zhang W, Zhang T, Mu W. D-lyxose isomerase and its application for functional sugar production. Appl Microbiol Biotechnol. 2018;102(5):2051–2062. doi:10.1007/s00253-018-8746-6
34. Tong W, Xu Y, Xian M, et al. Biosynthetic pathway for acrylic acid from glycerol in recombinant Escherichia coli. Appl Microbiol Biotechnol. 2016;100(11):4901–4907. doi:10.1007/s00253-015-7272-z
35. Mishra R, Kushveer JS, Khan MIK, et al. 2,4-Di-Tert-Butylphenol Isolated From an Endophytic Fungus, Daldinia eschscholtzii, Reduces Virulence and Quorum Sensing in Pseudomonas aeruginosa. Front Microbiol. 2020;11:1668. doi:10.3389/fmicb.2020.01668
36. Api AM, Belsito D, Biserta S, et al. RIFM fragrance ingredient safety assessment, m-cresol, CAS Registry Number 108-39-4. Food Chem Toxicol. 2021;149(Suppl 1):112043. doi:10.1016/j.fct.2021.112043
37. Musie E, Moore CC, Martin EN, Scheld WM. Toll-Like Receptor 4 Stimulation before or after Streptococcus pneumonia e Induced Sepsis Improves Survival and Is Dependent on T-Cells. PLoS One. 2014;9(1):e86015. doi:10.1371/journal.pone.0086015
38. Zhou A, Ni J, Xu Z, et al. Application of (1)h NMR spectroscopy-based metabolomics to sera of tuberculosis patients. J Proteome Res. 2013;12(10):4642–4649. doi:10.1021/pr4007359
39. Choi I, Son H, Baek JH. Tricarboxylic Acid (TCA) Cycle Intermediates: regulators of Immune Responses. Life. 2021;11(1):69. doi:10.3390/life11010069
40. Harder J, Franchi L, Munoz-Planillo R, Park JH, Reimer T, Nunez G. Activation of the Nlrp3 inflammasome by Streptococcus pyogenes requires streptolysin O and NF-kappa B activation but proceeds independently of TLR signaling and P2X7 receptor. J Immunol. 2009;183(9):5823–5829. doi:10.4049/jimmunol.0900444
41. Brown RL, Sequeira RP, Clarke TB. The microbiota protects against respiratory infection via GM-CSF signaling. Nat Commun. 2017;8(1):1512. doi:10.1038/s41467-017-01803-x
42. Baranek T, Morello E, Valayer A, et al. FHL2 Regulates Natural Killer Cell Development and Activation during Streptococcus pneumoniae Infection. Front Immunol. 2017;8:123. doi:10.3389/fimmu.2017.00123
43. Mohan T, Zhu W, Wang Y, Wang BZ. Applications of chemokines as adjuvants for vaccine immunotherapy. Immunobiology. 2018;223(6–7):477–485. doi:10.1016/j.imbio.2017.12.001
44. Raasch CE, Zhang P, Siggins RW, LaMotte LR, Nelson S, Bagby GJ. Acute alcohol intoxication impairs the hematopoietic precursor cell response to pneumococcal pneumonia. Alcohol Clin Exp Res. 2010;34(12):2035–2043. doi:10.1111/j.1530-0277.2010.01291.x
45. Cheng Y, Yang C, Tan Z, He Z. Changes of Intestinal Oxidative Stress, Inflammation, and Gene Expression in Neonatal Diarrhoea Kids. Front Veterinary Sci. 2021;8:598691. doi:10.3389/fvets.2021.598691
46. Hanna VS, Hafez EAA. Synopsis of arachidonic acid metabolism: a review. J Adv Res. 2018;11:23–32. doi:10.1016/j.jare.2018.03.005
47. Vernon HJ, Manoli I. Milestones in treatments for inborn errors of metabolism: reflections on Where chemistry and Medicine meet. Am J Med Genet A. 2021;185(11):3350–3358. doi:10.1002/ajmg.a.62385
48. Butman HS, Kotze TJ, Dowd CS, Strauss E. Vitamin in the Crosshairs: targeting Pantothenate and Coenzyme A Biosynthesis for New Antituberculosis Agents. Front Cell Infect Microbiol. 2020;10:605662. doi:10.3389/fcimb.2020.605662
49. Garg M, Angus PW, Burrell LM, Herath C, Gibson PR, Lubel JS. Review article: the pathophysiological roles of the renin-angiotensin system in the gastrointestinal tract. Aliment Pharmacol Ther. 2012;35(4):414–428. doi:10.1111/j.1365-2036.2011.04971.x
50. Zhao B, Rothenberg E, Ramsden DA, Lieber MR. The molecular basis and disease relevance of non-homologous DNA end joining. Nat Rev Mol Cell Biol. 2020;21(12):765–781. doi:10.1038/s41580-020-00297-8
51. Yusta B, Holland D, Koehler JA, et al. ErbB signaling is required for the proliferative actions of GLP-2 in the murine gut. Gastroenterology. 2009;137(3):986–996. doi:10.1053/j.gastro.2009.05.057
52. Nguyen T, Deenick EK, Tangye SG. Phosphatidylinositol 3-kinase signaling and immune regulation: insights into disease pathogenesis and clinical implications. Expert Rev Clin Immunol. 2021;1–10.
53. Harjunpaa H, Llort Asens M, Guenther C, Fagerholm SC. Cell Adhesion Molecules and Their Roles and Regulation in the Immune and Tumor Microenvironment. Front Immunol. 2019;10:1078. doi:10.3389/fimmu.2019.01078
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.