Back to Journals » Infection and Drug Resistance » Volume 13
Prevalence of HIV-1 Integrase Strand Transfer Inhibitor Resistance in Treatment-Naïve Voluntary Counselling and Testing Clients by Population Sequencing and Illumina Next-Generation Sequencing in Taiwan
Authors Tsai HC , Chen IT, Tsai KW , Lee SSJ , Chen YS
Received 12 August 2020
Accepted for publication 1 December 2020
Published 17 December 2020 Volume 2020:13 Pages 4519—4529
DOI https://doi.org/10.2147/IDR.S273704
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Hung-Chin Tsai,1– 4 I-Tzu Chen,1 Kuo-Wang Tsai,5 Susan Shin-Jung Lee,1,2 Yao-Shen Chen1,2
1Division of Infectious Diseases, Department of Medicine, Kaohsiung Veterans General Hospital, Kaohsiung, Taiwan; 2Faculty of Medicine, School of Medicine, National Yang-Ming University, Taipei, Taiwan; 3Department of Parasitology, Kaohsiung Medical University, Kaohsiung, Taiwan; 4Institute of Biomedical Sciences, National Sun Yat-Sen University, Kaohsiung, Taiwan; 5Department of Research, Taipei Tzu Chi Hospital, Buddhist Tzu Chi Medical Foundation, New Taipei, Taiwan
Correspondence: Hung-Chin Tsai
Division of Infectious Diseases, Department of Medicine, Kaohsiung Veterans General Hospital, #386 Ta-Chung 1st Road, Kaohsiung 813, Taiwan
Tel +886 7 3422121 ext. 2029
Fax +886 7 346 8292
Email [email protected]
Purpose: Integrase strand transfer inhibitors (INSTIs) are used as first-line therapy for HIV-1-infected patients. Next-generation sequencing (NGS) can detect low-frequency mutants; however, the clinical value of NGS to detect resistance variants is unknown. This study aimed to evaluate the prevalence of INSTI resistance in southern Taiwan and determine the clinical implications of using NGS to detect integrase region low-level resistant variants.
Patients and Methods: This retrospective cohort study included antiretroviral therapy-naïve HIV-1-infected individuals at Kaohsiung Veterans General Hospital, Taiwan, from 2013 to 2017. Drug-resistance mutations were determined, and an in-house polymerase chain reaction was used for genotyping INSTI resistance. NGS was used to assess INSTI resistance (≧1%), and the results were compared with those from population sequencing. Drug resistance-associated mutations were defined according to the 2019 IAS-USA HIV drug resistance-associated mutations list, and accessory mutations by a Stanford HIVdb score ≥ 10 to at least one INSTI.
Results: A total of 224 patients were included. Subtype B HIV-1 strains were found in 96% of the individuals and subtype CRF01_AE in 4%. The prevalence rates for nucleoside reverse transcriptase inhibitors, non-nucleoside reverse transcriptase inhibitors, protease inhibitors and INSTI resistance were 4%, 5.8%, 0.4% and 0.9%, respectively. The most common INSTI resistance-associated mutations were G163K (0.4%) and E138A (0.4%). Of the 38 patients diagnosed in 2017 who had both NGS and population sequencing data, none had INSTI resistance-associated mutations by population sequencing; however, NGS detected four more INSTI resistance-associated mutations with low frequencies (G163R 3.25%, S153F 3.21%, S153Y 1.36% and Y143H 2.06%). Two patients with S153F and S153Y low frequencies mutations started INSTI-based highly active antiretroviral therapy, and none had virological failure by week 48.
Conclusion: Our findings showed a low rate of HIV drug resistance to INSTIs (0.9%) in treatment-naïve patients. NGS detected more INSTI resistance-associated mutations at a low frequency. Low-level drug resistance-associated mutations to INSTIs identified by NGS did not have an impact on the treatment response to INSTI-based first-line therapy.
Keywords: HIV, integrase strand transfer inhibitor, drug resistance, next-generation sequencing
Introduction
Although modern new antiretroviral agents have reduced the prevalence of HIV drug resistance,1 the widespread use of antiretroviral therapy still has an impact on the emergence of drug resistance. Such drug resistance can compromise the clinical treatment outcomes in both treatment-naïve patients and also in those who have received treatment.2,3
The prevalence rate of transmitted drug resistance (TDR) in Taiwan, where routine genotypic drug-resistance testing is not available, is about 10% for nucleoside reverse transcriptase inhibitors (NRTIs), non-nucleoside reverse transcriptase inhibitors (NNRTIs) and protease inhibitors (PIs).4,5 Treatment with the first integrase strand transfer inhibitor (INSTI), raltegravir, combined with two NRTIs was introduced for HIV-1-infected treatment-naïve patients in 2012. Abacavir/dolutegravir/lamivudine (Triumeq) was introduced as the first INSTI-based single-tablet regimen in June 2016, and elvitegravir/cobicistat/emtricitabine/tenofovir alafenamide (Genvoya) as the second INSTI-based single-tablet regimen in 2017. The reported rates of TDR-associated mutations to INSTIs in Taiwan are low, at 1.2% in northern Taiwan from 2006 to 20156 and zero in southern Taiwan from 2013 to 2016.7 As HIV INSTI resistance is low in HIV-1-infected treatment-naïve individuals, routine Sanger genotypic drug-resistance testing is not recommended. However, with the increasing use of INSTIs as first-line treatment, monitoring TDR to INSTIs has become increasingly important. Next-generation sequencing (NGS) is a powerful tool due to its ability to detect lower frequency mutants.8 However, the clinical value of NGS to detect resistance variants has yet to be elucidated. Therefore, the aim of this study was to monitor the prevalence of INSTI resistance in southern Taiwan and to discuss the clinical implications of using NGS for low-level resistant variants.
Materials and Methods
Ethics Statement
This study was approved by the Institutional Review Board of Kaohsiung Veterans General Hospital in Taiwan (VGHKS15-CT5-10). The study subjects were signed the written informed consent and all of the participants had their records used in this study. This study complied with the Declaration of Helsinki.
Study Design and Participants
We conducted this retrospective cohort study on the prevalence of HIV-1 TDR in HIV-1-infected voluntary counselling and testing (VCT) clients in southern Taiwan from 2013 to 2017. Detailed descriptions of the VCT procedures and government policies for HIV care have been well described previously.9 Standard follow-up of the HIV-infected patients consisted of out-patient visits every 3 months, with tests for CD4+ T cell count, viral load, hematology, and biochemistry every 3 months in the first year of an HIV diagnosis, and every 6 months thereafter.
The Taiwan Centers for Disease Control (CDC) limits the prescription of combination antiretroviral therapy (cART) to anti-retroviral-naive HIV-1-infected patients who received their first cART after 1 June 2012 due to financial constraints. First-line cARTs are limited to zidovudine/lamivudine (ZDT/3TC) with efavirenz (EFV) or nevirapine (NVP). The reimbursement criteria were changed in June 2016 so that all treatment-naïve HIV-1-infected patients can receive a single-tablet regimen (STR). Efavirenz/emtricitabine/tenofovir disoproxil fumarate (Atripla) was first introduced to Taiwan in 2010, followed by emtricitabine/rilpivirine/tenofovir disoproxil fumarate (Complera) and abacavir/dolutegravir/lamivudine (Triumeq) in June 2016, elvitegravir/cobicistat/emtricitabine/tenofovir alafenamide (Genvoya) in September 2018, rilpivirine/emtricitabine/tenofovir alafenamide (Odefsey) and bictegravir/emtricitabine/tenofovir alafenamide (Biktarvy) in October 2019 as the recommended first-line therapy. Atripla, Complera and Triumeq were the only three STRs available when this study was conducted.
Population Sequencing for Genotypic Drug-Resistance Testing (GRT)
Resistance testing for PR/RT (pol gene) was performed by using ViroSeq HIV-1 Genotyping System version v2.8 (Celera, Alameda, CA, USA). INSTI (pol gene) resistance was determined by in-house population sequencing.10,11 Resistance-associated mutations (RAMs) were defined by the presence of at least one mutation included in the 2019 drug-resistance mutation list of the International AIDS Society-USA consensus guidelines.12 INSTI RAMs in the Stanford database with a Stanford HIVdb score ≧10 were also studied.6 Antiretroviral resistance to PR/RT and INSTI was interpreted using the HIVdb program of the Stanford University HIV Drug-Resistance Database (http://hivdb.stanford.edu; date last accessed, 1 Jan 2019). Those patients classified as having low-level resistance, intermediate resistance and high-level resistance were defined as having drug resistance.13
Illumina NGS for HIV INSTI Resistance
HIV pol amplicons obtained for sanger sequencing were also deep sequenced for the INSTI region using a MiSeq instrument (Illumina, USA). DNA libraries were generated for the pol PCR products using FastTaqPlus Master Mix (Topgen Biotech, Taiwan) with two sets of HIV pol primer pairs (Table 1) and a TopPrep Index Kit for Illumina (Topgen Biotech, Taiwan), according to the manufacturer’s instructions. For all libraries, DNA Clean Beads (Topgen Biotech, Taiwan) were used to purify the amplicon product, and the purified amplicon length was checked using a DNA-2500 Kit on a MultiNA-MCE202 system (Shimadzu, Japan). Prior to validation and sequencing, all libraries were quantified using Qubit Fluorometer (Thermo Fisher Scientific, USA) and Qubit dsDNA HS Assay kits (Thermo Fisher Scientific, USA). Multiplexed runs including 38 viral sample libraries were performed using a 500-cycle MiSeq Reagent Kit Nano v2 (Illumina, USA), achieving a median coverage for drug-resistance mutations sites after filtering reads for size and quality of 7647x.
 |
Table 1 Primer List for Next-Generation Sequencing Studies |
Bioinformatic Analysis
Drug-resistance mutation frequency was analyzed by using HyDRA by the National Microbiology Laboratory of the Public Health Agency of Canada from FASTQ files of each patient.14 Bowtie215 was used for reference mapping. Amino acid mutations were queried against a merged drug-resistance mutation database including the International AIDS Society-USA12 and the Stanford HIVDR Database.13 A minimum threshold of 1% was used to define the presence of drug-resistance-associated mutations.
Phylogenetic Analysis
Phylogeny from 38 patients underwent NGS study for INSTI resistance was inferred from the integrase region sequences derived from sanger sequencing. The sequences were then aligned using the Clustal W program in the MEGA (Molecular Evolutionary Genetics Analysis) analytical package (version 5.0; MEGA, Tempe, AZ, USA), with minor manual modifications. Phylogenetic trees were constructed using the neighbor-joining method on the Kimura two-parameter distance matrix in MEGA software, with the branching pattern being calculated from 1000 bootstrap replicates. Bootstrap values >700 of 1000 replicates were considered to be significant. Reference sequences were retrieved from the Los Alamos HIV database (http://www.hiv.lanl.gov), and included sequences of various subtypes across Asia.
Statistical Analysis
The Mann–Whitney U-test was used to compare the median values of continuous variables between groups (resistance and wild virus), and Fisher’s exact test was used to compare categorical variables between two groups. A two-sided p<0.05 was considered to be statistically significant. Statistical calculations were performed using SPSS version 12.0 (SPSS Inc., Chicago, IL).
Results
A total of 224 patients from our free VCT program were enrolled for GRT from 2013 to 2017. All of the 224 patients were male, with a medium age (interquartile range, IQR) of 26 (23–31) years, and 97% were men who have sex with men (MSM). The prevalence rates of hepatitis A, B and C infections were 11%, 8% and 2%, respectively. The seroprevalence rates of syphilis, amoebiasis (defined as indirect hemagglutination ≥1:256) and Toxoplasma gondii were 32%, 2% and 7%, respectively. The median CD4 cell count (IQR) was 305 (207–429) cells/µL, and the viral load (IQR) was 4.8 log (4.4–5.1) (Table 2).
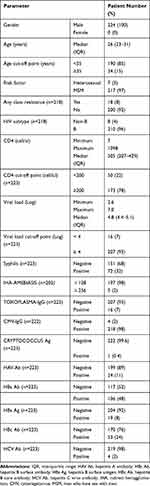 |
Table 2 Demographic Data of the Treatment-Naïve HIV-1-Infected Patients (n=224) |
The success rate for sanger sequencing the PR/RT region was 97% (218/224) and 100% for the INSTI region. Most of the 218 patients (96%, 210/218) had HIV subtype B, and 4% (8/218) had CRF01_AE. A total of 8.9% harbored TDR to the PR/RT and INSTI regions by sanger sequencing. Among them, 4% had resistance to NRTIs, 5.8% to NNRTIs, 0.4% to PIs and 0.9% to INSTIs (Figure 1). The detailed rates of individual antiretroviral drug resistance are summarized in Figure 2 and Table 3. The most common RAMs to NRTIs were M184V and K65R (1.3%), while those for NNRTIs were V179D (4.5%), V106I (2.7%), and K103N (1.3%), and those for PIs were L10I (13.4%), A71T (5.8%), and L10V (4.0%). The most common INSTI RAMs were G163K (0.4%) and E138A (0.4%) (Figure 3). A total of 38 patients enrolled in 2017 also had successfully Illumina NGS data for the INSTI resistance. There were no significant differences in demographic information between the 38 patients enrolled in 2017 (Table 4) and 186 patients enrolled from 2013 to 2016 (data not shown). Among the 38 patients who had both sanger sequencing and Illumina NGS data, 5% (2/38) had NRTI/NNRTI/PI resistance and none had INSTI resistance (one had an L74V substitution) by sanger sequencing however Illumina NGS for INSTI resistance detected four more INSTI resistance (≧1%) associated mutations with low frequency (G163R 3.25%, S153Y 1.36%, S153F 3.21% and Y143H 2.06%) (Table 5).
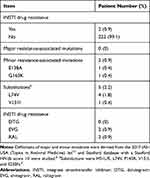 |
Table 3 INSTI Drug Resistance and Resistance-Associated Mutations by Sanger Sequencing Among Treatment-Naïve HIV-1-Infected Patients (n=224) |
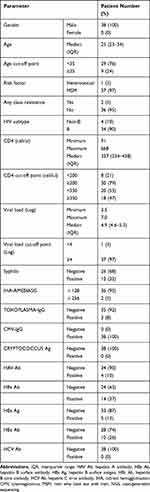 |
Table 4 Demographic Data of the HIV-1-Infected Patients Who Underwent Both Sanger Sequencing and NGS for INSTI Resistance (n=38) |
 |
Table 5 Overall Frequency (%) of INSTI Resistance-Associated Mutations Among HIV-1-Infected Patients by NGS (n=38)a |
Two patients with S153F and S153Y low frequencies mutations started INSTI-based highly active antiretroviral therapy and none had virological failure by week 48. Phylogeny from the 38 patients who received both sanger sequencing and NGS were inferred from the integrase gene sequences. One cluster was detected (sample 19 and 31) (Figure 4). The sample 19 and 31 have the same sanger sequencing in the integrase region by phylogenetic tree analysis. However, they have different mutation percentage and position in the NGS data (sample19: L74M (0.47%); sample31: L74M (0.51%) and 92G (0.49%)).
Discussion
In this study, we identified a low rate of HIV drug resistance to INSTIs (2/224, 0.9%) in treatment-naïve patients by population sequencing. Illumina NGS detected four more INSTI resistance-associated mutations at a low frequency (1.36–3.25%), however, they did not affect INSTI-based antiretroviral therapy. The clinical consequences of low-frequency RAMs detected by NGS are still controversial. Low-frequency NNRTI drug-resistant variants less than 20% level have been strongly associated with virologic failure for patients initiating NNRTI-based therapy.16–18 However, in a study analyzing the impact of TDR mutations on future viral suppression of HIV infection, researchers collected historical samples from 80 recently infected HIV-1 patients diagnosed in Israel between 2000 and 2014. GRT was performed using Illumina NGS and sanger-based population sequencing, and TDR was detected in 31.3% of the patients by NGS and 8.8% by sanger sequencing. All sanger-based population sequencing-detected mutations were also identified by NGS. The minor TDR variants did not become major variants in later samples and did not hinder successful treatment. However, they did not perform INSTI resistance testing in their samples.19 In another study from France, researchers evaluated the prevalence of virological failure to INSTI-based regimens and the impact of baseline minor resistant variants on the virological response to an INSTI-based regimen using ultra-deep sequencing. Their results showed that there was no association between the presence of baseline minor resistant variants and the response to INSTI-based therapies.20 In addition, Dalmat et al compared the variant detection capacity of NGS versus sanger sequencing for resistance genotyping in 144 drug-resistance tests (105 PR/RT and 39 INSTI tests) in 2016, and found that NGS could detect all of the high-frequency drug-resistance mutations (>20% frequency) found by sanger sequencing. Moreover, NGS also detected low-frequency mutations (1% to 20% frequency) associated with higher levels of drug resistance in 30/105 (29%) PR/RT and 4/39 (10%) INSTI tests. In clinical follow-up of 69 patients for a median of 674 days, none of the minor alleles detected by NGS resulted in subsequent virological failure.21 Although our study results are similar to theirs in that baseline INSTI resistance variants did not have an impact on the subsequent treatment response to INSTI-based antiretroviral therapy.
The main strength of this study is that our study population was homogenous who predominantly had subtype B, and all of them were enrolled from our VCT program without selection. The phylogenetic tree study from in those patients only found a very small cluster., indicating that the minor resistance variants were not transmission those newly diagnosed VCT clients. However, there are also several limitations to this study. The 1% cut-off point was chosen because it is well known that low-level variants with more than 1% for NNRTI TDR have clinical significance if a patient initiates an NNRTI-based regimen.16–18 In addition, our patients were enrolled from a VCT program, and the majority was MSM. Therefore, the results of this study may not be expanded to heterosexual populations and intravenous drug users. Second, the number of HIV-1-infected patients who underwent NGS was relatively small compared to those of other studies. Third, the prevalence of drug RAMs would be different depending on the choice of interpretation tool, for example, the French National Agency for AIDS Research (ANRS) HIV drug-resistance interpretation algorithm, HIVdb drug-resistance interpretation algorithm, IAS-USA HIV drug-resistance mutations list, Los Alamos National Laboratories HIV Sequence database, Rega Institute Drug-Resistance Interpretation Algorithm or WHO 2009 list of mutations. Finally, the calculated prevalence of INSTI resistance-associated mutations was based on specific data from 1 year (2017). However, there were no significant differences in the demographic information between the patients who underwent NGS or not, and the phylogenetic tree study in those patients did not identify any large major clusters. Nevertheless, the possibility of underestimating the prevalence of minor resistance-associated variants by NGS still cannot be totally excluded given the small number of patients who underwent NGS studies.
Conclusion
In conclusion, there was a low rate of HIV drug resistance to INSTIs (0.9%) in treatment-naïve patients in this study. Illumina NGS detected more low-frequency INSTI resistance-associated mutations. Low-level drug resistance-associated mutations to INSTIs as detected by NGS did not have an impact on the treatment response to INSTI-based first-line therapy. However, large study is needed to verify this finding.
Acknowledgments
The authors thank the laboratory technical support from Miss Pei-Yun Chou, Department of Infectious Diseases, Kaohsiung Veterans General Hospital, Taiwan.
Funding
This study was funded by the Medical Foundation in Memory of Dr. Deh-Lin Cheng (MF-DLC100053S5C2Y1) and by the Veterans General Hospitals and University System of Taiwan Joint Research Program Grant (VGHUST106-G3-1-1).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Davy-Mendez T, Eron JJ, Brunet L, Zakharova O, Dennis AM, Napravnik S. New antiretroviral agent use affects prevalence of HIV drug resistance in clinical care populations. AIDS. 2018;32(17):2593–2603. doi:10.1097/QAD.0000000000001990
2. Wittkop L, Günthard HF, de Wolf F, et al. Effect of transmitted drug resistance on virological and immunological response to initial combination antiretroviral therapy for HIV (EuroCoord-CHAIN joint project): a European multicohort study. Lancet Infect Dis. 2011;11(5):363–371. doi:10.1016/S1473-3099(11)70032-9
3. Rupérez M, Pou C, Maculuve S, et al. Determinants of virological failure and anti-retroviral drug resistance in Mozambique. J Antimicrob Chemother. 2015;70(9):2639–2647. doi:10.1093/jac/dkv143
4. Weng YW, Chen IT, Tsai HC, et al. Trend of HIV transmitted drug resistance before and after implementation of HAART regimen restriction in the treatment of HIV-1 infected patients in southern Taiwan. BMC Infect Dis. 2019;19(1):741. doi:10.1186/s12879-019-4389-1
5. Liu WC, Chang LH, Wu PY, et al. Seroincidence of HIV and prevalence of transmitted drug resistance of HIV-1 strains among persons seeking voluntary counselling and testing in Taiwan. J Int AIDS Soc. 2014;17(Suppl 4):19758. doi:10.7448/IAS.17.4.19758
6. Chang SY, Lin PH, Cheng CL, et al. Prevalence of Integrase Strand Transfer Inhibitors (INSTI) resistance mutations in taiwan. Sci Rep. 2016;6(1):35779. doi:10.1038/srep35779
7. Tsai HC, Chen IT, Wu KS, et al. HIV-1 integrase strand-transfer inhibitor resistance in southern Taiwan. Oncotarget. 2018;9(38):24927–24935. doi:10.18632/oncotarget.24837
8. Noguera-Julian M, Edgil D, Harrigan PR, Sandstrom P, Godfrey C, Paredes R. Next-generation human immunodeficiency virus sequencing for patient management and drug resistance surveillance. J Infect Dis. 2017;216(suppl_9):S829–S833. doi:10.1093/infdis/jix397
9. Tsai HC, Chou PY, Wann SR, Lee SS, Chen YS. Chemokine co-receptor usage in HIV-1-infected treatment-naïve voluntary counselling and testing clients in Southern Taiwan. BMJ Open. 2015;5(4):e007334. doi:10.1136/bmjopen-2014-007334
10. Canducci F, Barda B, Ceresola E, et al. Evolution patterns of raltegravir-resistant mutations after integrase inhibitor interruption. Clin Microbiol Infect. 2011;17(6):928–934. doi:10.1111/j.1469-0691.2010.03375.x
11. Canducci F, Ceresola ER, Boeri E, et al. Cross-resistance profile of the novel integrase inhibitor Dolutegravir (S/GSK1349572) using clonal viral variants selected in patients failing raltegravir. J Infect Dis. 2011;204(11):1811–1815. doi:10.1093/infdis/jir636
12. Wensing AM, Calvez V, Ceccherini-Silberstein F, et al. update of the drug resistance mutations in HIV-1. Top Antivir Med. 2019;2019(27):111–121.
13. Stanford University HIV Drug Resistance Datbase. Available from: https://hivdb.stanford.edu. Version 8.7,
14. Ji H, Liang B, Knox C, et al. Development of a data processing portal in support of tagged pooled pyrosequencing-based HIV drug resistance analysis.
15. Langmead B, Salzberg S. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359. doi:10.1038/nmeth.1923
16. Simen BB, Simons JF, Hullsiek KH, et al. Low abundance drug resistant viral variants in chronically HIV-infected antiretroviral-naive patients significantly impact treatment. J Infect Dis. 2009;199(5):693–701. doi:10.1086/596736
17. Johnson JA, Li J-F, Wei X, et al. Minority HIV-1 drug resistance mutations are present in antiretroviral treatment-naive populations and associate with reduced treatment efficacy. PLoS Med. 2008;5(7):e158. doi:10.1371/journal.pmed.0050158
18. Li JZ, Paredes R, Ribaudo H, et al. Minority HIV-1 drug resistance mutations and the risk of NNRTI-based antiretroviral treatment failure: a systematic review and pooled analysis. JAMA. 2011;305(13):1327–1335. doi:10.1001/jama.2011.375
19. Moscona R, Ram D, Wax M, et al. Comparison between next-generation and Sanger-based sequencing for the detection of transmitted drug-resistance mutations among recently infected HIV-1 patients in Israel, 2000–2014. J Int AIDS Soc. 2017;20(1):21846. doi:10.7448/IAS.20.1.21846
20. Nguyen T, Fofana DB, Lê MP, et al. Prevalence and clinical impact of minority resistant variants in patients failing an integrase inhibitor-based regimen by ultra-deep sequencing. J Antimicrob Chemother. 2018;73(9):2485–2492. doi:10.1093/jac/dky198
21. Dalmat RR, Makhsous N, Pepper GG, et al. Limited marginal utility of deep sequencing for HIV drug resistance testing in the age of integrase inhibitors. J Clin Microbiol. 2018;56(12):e01443–18. doi:10.1128/JCM.01443-18
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




