Back to Journals » Breast Cancer: Targets and Therapy » Volume 14
Predictive and Prognostic Value of TRIM58 Protein Expression in Patients with Breast Cancer Receiving Neoadjuvant Chemotherapy
Authors Zheng YZ, Li JY, Ning LW , Xie N
Received 23 August 2022
Accepted for publication 30 November 2022
Published 21 December 2022 Volume 2022:14 Pages 475—487
DOI https://doi.org/10.2147/BCTT.S387209
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Pranela Rameshwar
Yi-Zi Zheng,1,* Jia-Ying Li,2,3,* Lv-Wen Ning,3 Ni Xie3
1Department of Thyroid and Breast Surgery, Shenzhen Breast Tumor Research Center for Diagnosis and Treatment, the First Affiliated Hospital of Shenzhen University, Shenzhen Second People’s Hospital, Shenzhen University, Shenzhen, Guangdong, People’s Republic of China; 2Hengyang Medical School, University of South China, Hengyang, Hunan, People’s Republic of China; 3Biobank, First Affiliated Hospital of Shenzhen University, Shenzhen Second People’s Hospital, Shenzhen University, Shenzhen, Guangdong, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ni Xie, Biobank, First Affiliated Hospital of Shenzhen University, Shenzhen Second People’s Hospital, Shenzhen University, 3002 Sungang West Road, Shenzhen, 518035, Guangdong, People’s Republic of China, Tel +86-13501580802, Fax +86-0755-83003435, Email [email protected]
Introduction: Tripartite motif-containing protein (TRIM) family members play crucial roles in carcinogenesis and chemotherapy resistance. In this study, we aimed to determine whether TRIM58 protein expression is related to patient responses to neoadjuvant therapy (NAT) and their survival outcome.
Methods: Immunohistochemistry was performed on female breast cancer samples from biopsies before NAT in Shenzhen Second People’s Hospital. Univariate and multivariate logistic regression tests were used to analyze the association between TRIM58 protein expression and pathological complete response (pCR). The Cox proportional hazards model was used to calculate the adjusted hazard ratio (HR) with a 95% confidence interval (95% CI). The Kaplan–Meier plotter database was used to analyze the prognostic value of TRIM58.
Results: High TRIM58 expression was associated with small tumor size in all the patients (n = 58). Multivariate analysis suggested that low TRIM58 expression was an independent predictive factor for higher pCR (odds ratio = 0.06, 95% CI 0.005– 0.741, P = 0.028). The Kaplan–Meier Plotter dataset suggested that the TRIM58 high-expression group showed a worse 5-year overall survival than the low-expression group (HR = 1.34, 95% CI 1.07– 1.67, P = 0.01). Pathway analysis revealed the potential mechanisms of TRIM58 in chemoresistance.
Discussion: Our study suggests that TRIM58 is a promising biomarker for both neoadjuvant chemosensitivity and long-term clinical outcomes in breast cancer. It may also help to identify candidate responders and determine treatment strategies.
Keywords: chemosensitivity, pathological complete response, biomarker, patient stratification, predictive diagnostics
Introduction
Breast cancer is the most common malignancy in women worldwide. Its incidence is increasing annually, and it has the highest mortality rate among women with malignant tumors.1 Its high clinical heterogeneity is partially explained by its biological heterogeneity, even within well-characterized subtypes.2 Although clinical classification according to hormone receptor (HR) and human epidermal growth factor receptor 2 (HER2) status is the current basis for defining therapeutic strategies, the complexity of breast cancer treatment in both early and advanced disease is further increased by the lack of reliable predictive and prognostic biomarkers. In fact, most new drugs introduced in recent years have been administered according to clinical criteria, and the identification of efficacy markers is a difficult challenge. Neoadjuvant therapy (NAT) is the standard treatment for locally advanced breast cancer to downstage tumors, facilitating surgical treatment or breast-conserving surgery. Pathological complete response (pCR) is the state in which no tumor cells remain, or only ductal carcinoma in situ remains in the surgical specimen after preoperative treatment. Several large-scale clinical trials have confirmed that patients who achieved pCR following NAT presented with a much better prognosis than patients without pCR.3–5 However, not all patients achieve pCR after NAT, and its efficacy is often affected by an individual’s sensitivity to the drug and endogenous resistance. The NeoSphere study demonstrated that for HER2-positive breast cancer, the combination of chemotherapy with trastuzumab and pertuzumab resulted in a higher pCR rate of 45.8% when compared to that with chemotherapy with trastuzumab alone.6 This indicates that half of the patients failed to achieve pCR. Patients with other molecular subtypes of breast cancer have disappointing therapeutic outcomes.7 Molecular typing is also associated with NACT efficacy in breast cancer. However, in an era of individualized treatment, these alone are far from enough. Therefore, in the context of Predictive Preventive Personalised Medicine,8 it is necessary to distinguish between patients who would benefit from NAT and those who would not, suggesting the need to explore more effective biomarkers and disease monitoring tools to estimate individual chemosensitivity and long-term survival outcomes.9,10
Almost all oncoproteins and tumor suppressors are regulated by post-translational modifications, including the ubiquitin-proteasome system.11 Therefore, the identification and targeting of E3 ligases involved in the regulation of oncoproteins and tumor suppressor proteins is a current focus of cancer research.11–14 Tripartite motif-containing (TRIM) proteins, characterized by RING-finger domains or B-boxed and associated coiled-coil regions, possess E3 ubiquitin ligase activity in diverse processes and diseases, including cancers.11,15,16 Aberrant gene methylation of TRIM58, a member of the TRIM protein family, is involved in carcinogenesis.17 Previous studies reported that TRIM58 served as tumor suppressor and conferred better survival in gastric cancer, thyroid cancer, colorectal cancer and renal cell carcinoma.18–21 Our previous study identified that TRIM58 was highly expressed in drug-resistant breast-cancer cell strains and inactivated p53/p21 to promote chemoresistance by ubiquitination of DEAD-Box Helicase 3 X-Linked (DDX3) in breast cancer.22 However, it remains unclear whether TRIM58 expression levels influence the individual clinical benefits of NAT. We aimed to investigate the predictive and prognostic value of TRIM58 in neoadjuvant settings for breast cancer. We propose that TRIM58 overexpression may be involved in the reduction of chemosensitivity in breast cancer, leading to poor survival. Underlying mechanisms were also explored by pathway analysis.
Materials and Methods
Patients
A total of 58 female patients with primary breast cancer were enrolled in the study. The inclusion criteria were as follows: I. Patients with NAT indications who underwent NAT at the Shenzhen Second People’s Hospital between January 2018 and December 2020. II. No previous treatment before NAT. III. They were 18–80 years of age and had only one primary tumor. The exclusion criterion was the lack of a pathological tissue section. The therapeutic regimen decisions were based on the Chinese Anti-Cancer Association guidelines for the diagnosis and treatment of breast cancer. Taxanes (paclitaxel or docetaxel) and anthracycline (combined or sequential) were prescribed for 4–8 cycles as the NAT regimen for all patients. HER2+ patients were treated with trastuzumab concomitantly, at a loading dose of 4 mg/kg (or 8 mg/kg every 3 weeks), followed by a maintenance dose of 2 mg/kg (or 6 mg/kg every 3 weeks) on day 1, weekly (or every 3 weeks). Patients underwent surgery sequentially after receiving NAT. The outcomes of patients were retrospectively analyzed. All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the Helsinki declaration.
Pathology and Immunohistochemistry
Data on age, menstrual status, tumor size, lymph node (LN) metastases, estrogen receptor (ER), progesterone receptor (PR), HER2, Ki-67, and histological grade were obtained from the hospital information system. All tissues were histologically diagnosed with invasive breast cancer by the pathology department of Shenzhen Second People’s Hospital. Paraffin wax containing the patient’s tissue specimen was cut into slices and subjected to hematoxylin and eosin (H&E) staining. Immunohistochemical (IHC) staining was used to assess ER, PR, HER2, Ki-67, and TRIM58 expression. The tissue slides were examined by two pathologists from the First Affiliated Hospital of Shenzhen University.
ER, PR, Ki-67, HER2, and TRIM58 analyses were performed on paraffin-embedded tumor samples from biopsies. ER, PR, HER2, and Ki-67 were detected using rabbit monoclonal antibodies (MXB Biotechnologies, Fuzhou, CHN). TRIM58 was detected using a rabbit anti-TRIM58 polyclonal antibody (ab254768; Abcam, Cambridge, UK; 1:200). Tumors were considered ER- or PR-positive when ≥ 1% of tumor nuclei were positive for these hormone receptors, according to the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) guidelines.23 HER2 assessments were conducted according to ASCO/CAP recommendations (2013).24 Ki-67 levels were recorded as continuous values. A sample was defined as having a high proliferation index if its Ki-67 index was ≥ 30%. TRIM58 evaluation was conducted according to the following criteria; the percentage of positively stained cancer cells was graded on a four-point scale:25 0 (< 5% positive cells), 1 (6–25% positive cells), 2 (25–50% positive cells), 3 (50–75% positive cells), or 4 (> 75% positive cells); and staining intensity of breast cancer cells was subjectively assessed according to a four-point scoring system: 1 (−), 2 (+), 3 (++), or 4 (+++). The final TRIM58 results were scored by multiplying the staining intensity by the percentage of positively stained tumor cells, dividing TRIM58 expression into low (≤ 6) and high (> 6) staining. The pathological complete response (pCR) was defined as the absence of invasive breast cancer cells in breast and nodes (ypT0/Tis ypN0).
Pathway Analysis
TRIM58 expression profiles, including all available 1075 primary breast cancers measured by read counts, were downloaded from The Cancer Genome Atlas (TCGA) database. Quantile normalization and 10-based log transformations were performed on the raw read counts of expression data. Differentially expressed genes (DEGs) were calculated between the low- and high-TRIM58 groups using a two-tailed Student’s t-test. Gene expression of specific genes was determined to be significantly altered if the absolute log fold change (|logFC|) was > 1 and the Benjamini–Hochberg false discovery rate–corrected P-value was < 0.05. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Gene Ontology (GO) category enrichment analyses were performed on the top 2000 DEGs using g:Profiler (https://biit.cs.ut.ee/gprofiler/). Protein‒protein interaction (PPI) networks were constructed using STRING database (https://string-db.org/).
Statistical Analysis
The correlations between TRIM58 expression and other clinicopathological characteristics were tested using the chi-squared or Fisher’s exact tests. The TRIM58 expression value as a continuous variable was further compared between subgroups using the Mann–Whitney U-test.
The endpoint of this study was pCR, defined as the absence of invasive cancer in the breast and no residual cancer cells in LN samples obtained at the time of surgery. Univariate and multivariate logistic regression tests were used to analyze the associations between different TRIM58 expression levels and pCR outcomes using calculated hazard ratio (HR).
Receiver operating characteristic (ROC) curves were generated to identify whether TRIM58 expression combined with important clinicopathological characteristics or American Joint Committee on Cancer (AJCC) stages (8th edition) could better predict patients’ responses to therapy. The areas under the curve (AUCs) were compared using a z-test.
The Kaplan–Meier Plotter website (http://kmplot.com/analysis/) was used to analyze the prognostic value of TRIM58 expression in overall survival (OS). Furthermore, the patients were stratified according to the hormone receptor (HorR) (positive versus negative), HER2 (positive versus negative), and pathological status. The survival curve was derived using the Kaplan–Meier method, and the Log rank test was used to compare survival rates. All statistical analyses were performed using GraphPad 8.0 (GraphPad Software LLC, California, USA) or R language version 3.5.1 (www.r-project.org). Statistical significance was set at P < 0.05.
Results
Baseline Characteristics
Fifty-eight patients were enrolled in this study, 25 (43.1%) were classified into the high TRIM58 expression group and 33 (58%) into the low TRIM58 expression group. Associations between baseline clinicopathological characteristics and TRIM58 expression were investigated as categorical variables. The results showed no significant associations, except for a marginal significance between high TRIM58 expression and smaller tumor size (P = 0.059; Table 1). To further determine the association between TRIM58 expression and other clinicopathological features, TRIM58 expression values, as a continuous variable, were compared between different subgroups. Higher TRIM58 expression was associated with smaller tumor size (P = 0.02; Figure 1A). Additionally, luminal A-like and triple-negative patients exhibited higher TRIM58 expression with marginal significance, compared with that of luminal B-like patients (P = 0.02; Figure 1B). However, no significant differences in ER status (Figure 1C), PR status (Figure 1D), Ki-67 index (Figure 1E), or LN status (Figure 1F) were observed between the groups.
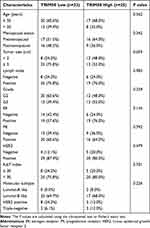 |
Table 1 Associations Between TRIM58 Expression Level and Baseline Clinicopathological Characteristics in Breast Cancer |
The median age of the patients in this study was 45.5 years-old (range 26–73 years). Moreover, 56.9% of the patients were premenopausal, 34.5% exhibited tumor sizes < 3 cm, 62.1% had PR+ tumors, 34.5% had ER+ tumors, 84.5% had HER2+ tumors, and 77.6% exhibited tumors with Ki-67 > 30%. Histological grade III tumors were present in 44.8% of patients. Fine-needle aspiration revealed that 75.9% of the patients were positive for cancer in their LNs before chemotherapy (Figure 2).
TRIM58 Expression and pCR
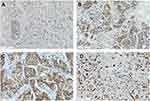
Representative tissue staining for TRIM58 is shown in Figure 3. Patients with high TRIM58 expression achieved a lower pCR rate (20.0%) than patients with low TRIM58 expression [48.5%; Odds ratio (OR) = 0.266, 95% confidence interval (CI) 0.08–0.877; P = 0.03; Table 2, Figure 2]. Multivariate logistic regression analysis suggested that TRIM58 expression was an independent predictive factor for pCR (OR = 0.061, 95% CI 0.005–0.741; P = 0.028; Table 2). LN status was also an independent predictive factor for pCR (OR = 0.013, 95% CI 0.001–0.206; P = 0.002; Table 2). No significant association was observed between pCR and any of the other potential factors, included age, menopausal status, tumor size, ER status, PR status, HER2 status, Ki67 index, and histological grade.
 |
Table 2 Univariate and Multivariate Analyses for the Predictive Factors of Pathological Complete Response |
Furthermore, the accuracy of different models to evaluate the predictive value of TRIM58 for pCR was compared using ROC curves. The largest AUC value was 0.885 when TRIM58 expression was combined with clinicopathological variables. The AUC values of the considered clinicopathological variables, TRIM58 expression combined with AJCC stage, and AJCC stage alone were 0.844, 0.778, and 0.719, respectively (P = 0.0308; Figure 4).
TRIM58 Expression and Survival
Kaplan–Meier method estimates of survival according to TRIM58 expression levels using RNA-seq data from the Kaplan–Meier Plotter database for all available breast cancer patients were performed to ensure sufficient follow-up time to perform survival analysis.26 The TRIM58 overexpression group showed significantly worse 5-year OS compared with the low-expression group (n = 2976; log-rank P = 0.01; HR = 1.34; 95% CI 1.07–1.67; Figure 5A). Additionally, in the groups of HorR+, HER2+, or HER2− patients, the TRIM58 high-expression group always exhibited worse 5-year OS compared to the TRIM58 low-expression groups (HorR+, n = 2575; log-rank P = 0.035; HR = 1.33; 95% CI 1.02–1.72; Figure 5B; HER2+, n = 379; log-rank P = 0.021; HR = 2.06; 95% CI 1.1–3.87; Figure 5D; HER2−, n = 2496; log-rank P = 0.046; HR = 1.28; 95% CI 1.0–1.64; Figure 5E). However, in HorR− patients there were no significant differences observed in OS between the two groups (n = 185; log-rank P = 0.12; HR = 1.79; 95% CI 0.84–3.78; Figure 5C).
GO, KEGG, and PPI Network Analyses
The potential mechanism of action of TRIM58 in breast cancer by downloading RNA-seq data from TCGA and analyzing the DEGs between the TRIM58 high and low expression groups using GO and KEGG enrichment analyses.27,28 The enriched GO terms included cellular component (CC), molecular function (MF), and biological process (BP) (Figure 6A), which involved a large number of oxidoreductase, hydrolase, and peptidase activities. We identified the top KEGG pathways, among which drug metabolism (related to cytochrome P450) highlighted its potential effects on chemotherapy resistance (Figure 6B). To further explore the potential role of TRIM58, PPIs related to TRIM58 were also identified using STRING (Figure 6C).29
Discussion
This study aimed to identify TRIM58 expression as an independent negative predictor of pCR in patients with breast cancer. In addition, the association between TRIM58 expression and baseline tumor features was analyzed. Moreover, the prognostic value of TRIM58 for OS was revealed in HorR+, HorR−, HER2+, HER2−, and all patients with breast cancer. As far as we know, this is the first study to identify TRIM58 as a predictive biomarker of the efficacy of neoadjuvant chemotherapy for breast cancer. Additionally, we first demonstrated the associations between high TRIM58 expression and smaller tumor size. Moreover, we revealed, for the first time, the prognostic value of TRIM58 for long-term outcomes in breast cancer.
It was observed that TRIM58 expression was negatively related with tumor size. And it seemed plausible that TRIM58 expression appeared to be associated with tumor malignancy. A commonly accepted theory states that as a tumor grows, the cells within the tumor will acquire the capability to spread to, survive and proliferate within the regional LNs and other distant sites.30,31 Meanwhile, within any LN grouping, the growth of tumor size worsens the prognosis.32 However. Biologic characteristics might actually be more aggressive in cases of small tumors with LN metastases. There is evidence, including from our previous study, that a very small tumor size is indicative of a biologically aggressive disease in patients with extensive LN involvement.33,34 In contrast, tumors that fail to metastasize to regional LNs, even at a late stage (eg, T3N0 tumors), may reflect a more biologically indolent phenotype.35 Therefore, tumor size does not directly indicate the degree of tumor malignancy, and the severity of small-sized breast cancer needs to be reconsidered. It is therefore explainable that high expression of TRIM58, while indicating lower pCR rate and worse prognosis, is also associated with smaller tumors.
In parallel, our clinical data revealed for the first time that TRIM58 is an independent predictive factor for pCR, with its high expression contributing to a significantly lower pCR rate. So far, few clinical studies have focused on the prediction value of TRIM58 for neoadjuvant chemotherapy in breast cancer. However, several basic studies concerning TRIM58 and other proteins of the TRIM family theoretically support our findings. Previous studies have shown that TRIM family proteins are closely related to the degradation of tumor suppressor proteins, leading to chemoresistance.11 High TRIM protein expression mediates the ubiquitination of downstream substrates, leading to changes in signaling pathways, and eventually chemoresistance.36 For example, TRIM47 is significantly upregulated in breast cancer tissues and forms ternary complexes with protein kinases C-ε and D3, leading to breast cancer cell proliferation and endocrine drug resistance.37 In non-TNBCs, TRIM27 enhanced cancer cell drug resistance through p21 ubiquitination.38 TRIM3 promotes cancer cell resistance to tamoxifen in ER+ breast cancer via the TRIM3/UBC9/ESR1 axis.39 Most importantly, TRIM58 was found to inactivate p53/p21, promoting chemoresistance by the ubiquitination of DDX3 in breast cancer.22 Consistent with the above studies, TRIM58 was shown to be associated with the regulation of a large number of enzyme activities in GO analysis, such as endopeptidase, peptidase and hydrolase. On the other hand, KEGG pathway analyses suggested that drug metabolism (related to cytochrome P450) may have potential effects on chemotherapy resistance. The cytochrome P450 enzyme (CYP) is a crucial metabolic oxidase, and is essential for the activation or inactivation of anticancer drugs, including cyclophosphamide, paclitaxel, doxorubicin, docetaxel, cisplatin and 5-fluorouracil.40 The genetic polymorphisms of CYP enzymes in breast tumors have an effect on the drug treatment outcomes of patients with breast cancer. To be specific, the CYP1B1 4326G allele is associated with a decreased response rate, reduced progression-free survival and shorter overall survival in patients with breast cancer treated with taxanes.41 CYP2B6 serves a pivotal role in the efficacy of doxorubicin and cyclophosphamide.42 Thus, the potential pathway between TRIM58 and the the cytochrome P450 enzyme might be crucial in the development of resistance to chemotherapeutic drugs, leading to the unsatisfactory therapeutic effect in some patients with breast cancer. Since cytochrome P450 plays a pivotal role in the generation of drug resistance and the susceptibility of tumors,43 it is plausible that future studies investigating cytochrome P450 may shed light on the main mechanism of TRIM58 leading to drug resistance in breast cancer cells. GO analysis showed that many oxidoreductase, hydrolase, and peptidase activities were associated with TRIM58, which also suggested that TRIM58 may regulate drug resistance in tumor cells by drug metabolism mechanisms. This suggests the potential of TRIM58 as a therapeutic target and a prognostic marker in breast cancer. Additionally, the PPI network may offer potential associations between proteins related to TRIM58 and chemosensitivity, which requires further validation. For example, UBE2H, which was shown to have close interaction with TRIM58 in PPI network analysis, was reported to be related with drug resistance and poor prognosis in cancer.44,45 And OR2W3, another TRIM58 interacted protein, was reported to be potentially correlated to breast cancer progression.46 TRIM58 is likely to affect tumor sensitivity to treatment through the above interacting proteins; or TRIM58, as the downstream effector molecule of the above gene, could realize the regulation of therapeutic response. Future in-depth study into the interaction between TRIM58 and the above genes is likely to find the underlying mechanism that TRIM58 indicates poor chemotherapy response.
Although our neoadjuvant trials have an insufficient follow-up period to perform survival analysis, the results from the Kaplan–Meier Plotter database analyses indicated a significantly worse OS with increased TRIM58 expression, which may be clinically significant. These findings are supported by those of Han and Xu et al, where TRIM58 expression was associated with prognosis of lung cancer patients.47,48 Although TRIM58 may indicate a better prognosis in other tumors than in breast cancer, it still indicates that TRIM58 affects the biological behavior of cancer cells in various tumors by different mechanisms. TRIM58 remained significant in predicting OS in the subgroup analysis, namely HorR+ subgroup, HER2+ subgroup and HER2- subgroup, except for the HorR− subgroup. Even so, HorR- subgroup still showed a trend of higher expression of TRIM58 indicating worse OS, which is very likely to be consistent with the expected results if the sample size is sufficient. In the HER2+ subgroup, the HR of different TRIM58 expression groups was as high as 2.06, much higher than the other subgroups, suggesting that TRIM58 is a stronger prognostic indicator in HER2-positive breast cancer. Therefore, TRIM58 serves as a prognostic marker and a promising target molecule in breast cancer, especially in HER2+ breast cancer.
There are several limitations of our study. First, owing to the short starting time, the sample size was small, resulting in the failure to perform subgroup analyses. Moreover, due to the short follow-up time, we were not able to conduct long-term survival analysis in our study cohort, which is pending future follow-up and analyses. As a supplement, we analyzed the online Kaplan‒Meier Plotter database with a larger sample size and a longer follow-up time.
Conclusion
In conclusion, our study revealed that TRIM58 is not only a novel biomarker for predicting pCR in patients receiving NAT, but it is also a prognostic indicator for breast cancer, especially in HER2+ breast cancer. Increased TRIM58 expression is associated with smaller tumor size. Potential pathways of chemoresistance shared between TRIM58 and cytochrome P450, UBE2H and OR2W3 were identified. These results may help further identify candidate responders and guide treatment strategies. The mechanisms of TRIM58 in chemoresistance need to be elucidated in future studies.
Ethics Approval and Informed Consent
The research protocol was approved by the Ethics Committee of Shenzhen Second People’s Hospital Ethics Committee of Shenzhen Second People’s Hospital, Shenzhen University (approval ID 20220810003). Ethics Committee of Shenzhen Second People’s Hospital exempted this study from informed consent requirements. Informed consent is exempted from this study for the following reasons: the study used patient data with identifiable information obtained from previous clinical treatment; the subject can no longer be located; the research project does not involve personal privacy and commercial interests; this study is of medical scientific value; and without exemption of informed consent, this study cannot be carried out. To protect patient data confidentiality, medical record information is anonymized and only access to researchers. The methods were performed in accordance with the principles stated in the Declaration of Helsinki.
Acknowledgments
The authors appreciate Director Xia Liu for technical support in pathological film reading and Zhen-han Deng for advice on study design.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Number: 81902682, 82273397, 82172356, 81972003); Guangdong Basic and Applied Basic Research Foundation (Grant Number: 2021A1515011122, 2021A1515012144, 2020A1515111165); Shenzhen Science and Technology Project (Grant Number: JCYJ20210324103003010); Science, Technology and Innovation Commission of Shenzhen Municipality (Grant Number: JCYJ20180507182025817); Clinical Research Project of Shenzhen Second People’s Hospital (Grant Number: 20223357015).
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Siegel R, Miller K, Fuchs H, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33. doi:10.3322/caac.21654
2. Perou C, Sørlie T, Eisen M, et al. Molecular portraits of human breast tumours. Nature. 2000;406(6797):747–752. doi:10.1038/35021093
3. Leon-Ferre R, Hieken T, Boughey J. The landmark series: neoadjuvant chemotherapy for triple-negative and HER2-positive breast cancer. Ann Surg Oncol. 2021;28(4):2111–2119. doi:10.1245/s10434-020-09480-9
4. Yee D, DeMichele A, Yau C, et al. Association of event-free and distant recurrence-free survival with individual-level pathologic complete response in neoadjuvant treatment of stages 2 and 3 breast cancer: three-year follow-up analysis for the I-SPY2 adaptively randomized clinical trial. JAMA Oncology. 2020;6(9):1355–1362. doi:10.1001/jamaoncol.2020.2535
5. Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–172. doi:10.1016/s0140-6736(13)62422-8
6. Gianni L, Pienkowski T, Im Y, et al. 5-year analysis of neoadjuvant pertuzumab and trastuzumab in patients with locally advanced, inflammatory, or early-stage HER2-positive breast cancer (NeoSphere): a multicentre, open-label, Phase 2 randomised trial. Lancet Oncol. 2016;17(6):791–800. doi:10.1016/s1470-2045(16)00163-7
7. Caudle A, Gonzalez-Angulo A, Hunt K, et al. Impact of progression during neoadjuvant chemotherapy on surgical management of breast cancer. Ann Surg Oncol. 2011;18(4):932–938. doi:10.1245/s10434-010-1390-8
8. Koklesova L, Liskova A, Samec M, et al. Genoprotective activities of plant natural substances in cancer and chemopreventive strategies in the context of 3P medicine. EPMA J. 2020;11(2):261–287. doi:10.1007/s13167-020-00210-5
9. Crigna A, Samec M, Koklesova L, et al. Cell-free nucleic acid patterns in disease prediction and monitoring-hype or hope? EPMA J. 2020;11(4):603–627. doi:10.1007/s13167-020-00226-x
10. Mazurakova A, Koklesova L, Samec M, et al. Anti-breast cancer effects of phytochemicals: primary, secondary, and tertiary care. EPMA J. 2022;13(2):315–334. doi:10.1007/s13167-022-00277-2
11. Hatakeyama S. TRIM proteins and cancer. Nat Rev Cancer. 2011;11(11):792–804. doi:10.1038/nrc3139
12. Jamal A, Swarnalatha M, Sultana S, Joshi P, Panda S, Kumar V. The G1 phase E3 ubiquitin ligase TRUSS that gets deregulated in human cancers is a novel substrate of the S-phase E3 ubiquitin ligase Skp2. Cell Cycle. 2015;14(16):2688–2700. doi:10.1080/15384101.2015.1056946
13. Kim H, Frederick D, Levesque M, et al. Downregulation of the ubiquitin ligase RNF125 underlies resistance of melanoma cells to BRAF inhibitors via JAK1 deregulation. Cell Rep. 2015;11(9):1458–1473. doi:10.1016/j.celrep.2015.04.049
14. Koepp D, Schaefer L, Ye X, et al. Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science. 2001;294(5540):173–177. doi:10.1126/science.1065203
15. Ozato K, Shin D, Chang T, Morse H. TRIM family proteins and their emerging roles in innate immunity. Nat Rev Immunol. 2008;8(11):849–860. doi:10.1038/nri2413
16. Napolitano L, Meroni G. TRIM family: pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life. 2012;64(1):64–71. doi:10.1002/iub.580
17. Zhang W, Cui Q, Qu W, Ding X, Jiang D, Liu H. TRIM58/cg26157385 methylation is associated with eight prognostic genes in lung squamous cell carcinoma. Oncol Rep. 2018;40(1):206–216. doi:10.3892/or.2018.6426
18. Liu M, Zhang X, Cai J, et al. Downregulation of TRIM58 expression is associated with a poor patient outcome and enhances colorectal cancer cell invasion. Oncol Rep. 2018;40(3):1251–1260. doi:10.3892/or.2018.6525
19. Liu X, Long Z, Cai H, Yu S, Wu J. TRIM58 suppresses the tumor growth in gastric cancer by inactivation of β-catenin signaling via ubiquitination. Cancer Biol Ther. 2020;21(3):203–212. doi:10.1080/15384047.2019.1679554
20. Du X, Wang J, Zhang D, et al. viaAUF1 Promotes Proliferation and Invasion of Thyroid Cancer Downregulation of ZBTB2 and Subsequent TRIM58. Front Oncol. 2021;11:681736. doi:10.3389/fonc.2021.681736
21. Gan Y, Cao C, Li A, et al. TRIM58Silencing of the gene by aberrant promoter methylation is associated with a poor patient outcome and promotes cell proliferation and migration in clear cell renal cell carcinoma. Front Mol Biosci. 2021;8:655126. doi:10.3389/fmolb.2021.655126
22. Wang J, Yang F, Zhuang J, Huo Q, Li J, Xie N. TRIM58 inactivates p53/p21 to promote chemoresistance via ubiquitination of DDX3 in breast cancer. Int J Biochem Cell Biol. 2022;143:106140. doi:10.1016/j.biocel.2021.106140
23. Hammond ME, Hayes DF, Dowsett M, et al. American Society of Clinical Oncology/College Of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010;28(16):2784–2795. doi:10.1200/JCO.2009.25.6529
24. Wolff A, Hammond M, Hicks D, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. Arch Pathol Lab Med. 2014;138(2):241–256. doi:10.5858/arpa.2013-0953-SA
25. Glazyrin A, Shen X, Blanc V, Eliason JF. Direct detection of herceptin/trastuzumab binding on breast tissue sections. J Histochem Cytochem. 2007;55(1):25–33. doi:10.1369/jhc.6A7017.2006
26. Lánczky A, Győrffy B. Web-based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Internet Res. 2021;23(7):e27633. doi:10.2196/27633
27. Huang DW, Sherman B, Lempicki R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
28. Huang DW, Sherman B, Lempicki R. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi:10.1093/nar/gkn923
29. Szklarczyk D, Gable A, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi:10.1093/nar/gky1131
30. Weigelt B, Peterse JL, Van ‘t Veer LJ. Breast cancer metastasis: markers and models. Nat Rev Cancer. 2005;5(8):591–602. doi:10.1038/nrc1670
31. Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat Rev Cancer. 2003;3(6):453–458. doi:10.1038/nrc1098
32. Foulkes WD, Reis-Filho JS, Narod SA. Tumor size and survival in breast cancer--a reappraisal. Nat Rev Clin Oncol. 2010;7(6):348–353. doi:10.1038/nrclinonc.2010.39
33. Wo JY, Chen K, Neville BA, Lin NU, Punglia RS. Effect of very small tumor size on cancer-specific mortality in node-positive breast cancer. J Clin Oncol. 2011;29(19):2619–2627. doi:10.1200/JCO.2010.29.5907
34. Zheng Y, Wang X, Fan L, Shao Z. Breast cancer-specific mortality in small-sized tumor with stage IV Breast cancer: a population-based study. Oncologist. 2021;26(2):e241–e250. doi:10.1002/onco.13567
35. Yu KD, Jiang YZ, Chen S, et al. Effect of large tumor size on cancer-specific mortality in node-negative breast cancer. Mayo Clinic Proceedings. 2012;87(12):1171–1180. doi:10.1016/j.mayocp.2012.07.023
36. Hatakeyama S. TRIM family proteins: roles in autophagy, immunity, and carcinogenesis. Trends Biochem Sci. 2017;42(4):297–311. doi:10.1016/j.tibs.2017.01.002
37. Azuma K, Ikeda K, Suzuki T, Aogi K, Horie-Inoue K, Inoue S. TRIM47 activates NF-κB signaling via PKC-ε/PKD3 stabilization and contributes to endocrine therapy resistance in breast cancer. Proc Natl Acad Sci U S A. 2021;118(35). doi:10.1073/pnas.2100784118
38. Xing L, Tang X, Wu K, Huang X, Yi Y, Huan J. TRIM27 functions as a novel oncogene in non-triple-negative breast cancer by blocking cellular senescence through p21 ubiquitination. Mol Ther Nucleic Acids. 2020;22:910–923. doi:10.1016/j.omtn.2020.10.012
39. Ye R, AiErken N, Kuang X, et al. Tripartite motif-containing 3 (TRIM3) enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogenesis. 2021;10(9):60. doi:10.1038/s41389-021-00350-x
40. McFadyen M, McLeod H, Jackson F, Melvin W, Doehmer J, Murray G. Cytochrome P450 CYP1B1 protein expression: a novel mechanism of anticancer drug resistance. Biochem Pharmacol. 2001;62(2):207–212. doi:10.1016/s0006-2952(01
41. Marsh S, Somlo G, Li X, et al. Pharmacogenetic analysis of paclitaxel transport and metabolism genes in breast cancer. Pharmacogenomics J. 2007;7(5):362–365. doi:10.1038/sj.tpj.6500434
42. Bray J, Sludden J, Griffin M, et al. Influence of pharmacogenetics on response and toxicity in breast cancer patients treated with doxorubicin and cyclophosphamide. Br J Cancer. 2010;102(6):1003–1009. doi:10.1038/sj.bjc.6605587
43. Luo B, Yan D, Yan H, Yuan J. Cytochrome P450: implications for human breast cancer. Oncol Lett. 2021;22(1):548. doi:10.3892/ol.2021.12809
44. Zhu Y, Wang W, Song Z, et al. MET-UBE2H fusion as a novel mechanism of acquired EGFR resistance in lung adenocarcinoma. J Thorac Oncol. 2018;13(10):e202–e204. doi:10.1016/j.jtho.2018.05.009
45. Yen M, Wu K, Liu Y, et al. Ubiquitin conjugating enzyme E2 H (UBE2H) is linked to poor outcomes and metastasis in lung adenocarcinoma. Biology. 2021;10(5):378. doi:10.3390/biology10050378
46. Masjedi S, Zwiebel L, Giorgio T. Olfactory receptor gene abundance in invasive breast carcinoma. Sci Rep. 2019;9(1):13736. doi:10.1038/s41598-019-50085-4
47. Xu M, Gong J. Prognostic signature, immune features, and therapeutic responses of a novel ubiquitination-related gene signature in lung adenocarcinoma. J Oncol. 2022;2022:2524649. doi:10.1155/2022/2524649
48. Han Y, Li Y. Comprehensive exploration of M2 macrophages and its related genes for predicting clinical outcomes and drug sensitivity in lung squamous cell carcinoma. J Oncol. 2022;2022:1163924. doi:10.1155/2022/1163924
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.