Back to Journals » Infection and Drug Resistance » Volume 17
Predicting Antibiotic Tolerance in hvKP and cKP Respiratory Infections Through Biofilm Formation Analysis and Its Resistance Implications
Authors Wen Z, Chen Y, Liu T, Han J, Jiang Y , Zhang K
Received 30 November 2023
Accepted for publication 26 March 2024
Published 17 April 2024 Volume 2024:17 Pages 1529—1537
DOI https://doi.org/10.2147/IDR.S449712
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Zhongwei Wen, Yiqiang Chen, Tangjuan Liu, Jiahui Han, Yuting Jiang, Ke Zhang
Department of Pulmonary and Critical Care Medicine, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, People’s Republic of China
Correspondence: Yiqiang Chen, The First Affiliated Hospital of Guangxi Medical University, Nanning, Guangxi Zhuang Autonomous Region, 530021, People’s Republic of China, Email [email protected]
Introduction: Respiratory infections are a major global health concern, with Klebsiella pneumoniae standing out due to its evolving antibiotic resistance. This study compares the resistance profiles of hypervirulent Klebsiella pneumoniae (hvKP) and classical Klebsiella pneumoniae (cKP), aiming to shed light on their clinical implications.
Methods: We analyzed 86 cases, comprising 42 hvKP and 44 cKP strains, using comprehensive antimicrobial susceptibility testing and clinical data evaluation to assess antibiotic tolerance and resistance mechanisms.
Results: Our findings reveal distinct resistance patterns between hvKP and cKP, highlighting the role of chromosomal mutations and plasmid-mediated gene transfer in conferring antibiotic resistance. Notably, hvKP strains exhibited unique resistance trends, including the production of extended-spectrum β-lactamases (ESBLs) and carbapenemases, differing from those of cKP.
Discussion: This research underscores the importance of continuous surveillance and the development of targeted therapies against antibiotic-resistant Klebsiella pneumoniae. It emphasizes the critical need for judicious antibiotic use and novel therapeutic approaches to combat respiratory infections caused by these increasingly resistant pathogens.
Keywords: Klebsiella pneumoniae, antibiotic resistance, hypervirulent Klebsiella pneumoniae, hvKP, classical Klebsiella pneumoniae, cKP, public health
Introduction
In the realm of infectious diseases, respiratory infections remain a formidable challenge, accounting for significant morbidity and mortality worldwide.1 Among the diverse array of pathogens implicated in respiratory tract infections, Klebsiella pneumoniae stands out due to its increasing prevalence and the remarkable ability to develop resistance against multiple antibiotic agents.2,3 K. pneumoniae can be classified into two major groups: hypervirulent K. pneumoniae (hvKP) and classical K. pneumoniae (cKP), each with distinct pathogenic profiles and clinical implications.4 Recent years have witnessed a surge in cases involving these two strains, underscoring the urgency to delve deeper into their resistance mechanisms and develop strategies to mitigate their impact on public health.5,6
hvKP, characterized by its hypermucoviscosity and the ability to cause invasive infections, has been associated with severe clinical manifestations such as liver abscesses, endophthalmitis, and meningitis.7 On the other hand, cKP is typically linked to hospital-acquired infections, exhibiting a predilection for individuals with compromised immune systems.8 Despite the differences in their virulence and clinical presentations, both hvKP and cKP pose significant challenges due to their capacity to acquire resistance to commonly used antibiotics.9
The emergence of antibiotic-resistant strains of K. pneumoniae has been documented globally, with alarming increases in the incidence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) isolates.10,11 This trend is particularly concerning given the limited therapeutic options available for treating infections caused by these resistant strains.10 In this context, understanding the antibiotic tolerance profiles of hvKP and cKP becomes paramount, as it holds the key to developing targeted interventions and optimizing antimicrobial therapy.
A plethora of studies have shed light on the resistance mechanisms employed by K. pneumoniae, revealing a complex interplay of chromosomal mutations and plasmid-mediated gene transfer.12 In particular, the production of extended-spectrum β-lactamases (ESBLs) and carbapenemases has been identified as a major driver of antibiotic resistance in these strains.13 These enzymes confer resistance to a wide range of β-lactam antibiotics, including penicillins, cephalosporins, and carbapenems, severely limiting the treatment options and contributing to the high mortality rates associated with these infections.14,15
In this study, we endeavor to unravel the intricate landscape of antibiotic tolerance in hvKP and cKP respiratory infections, with a particular focus on their resistance profiles and the underlying molecular mechanisms. By leveraging clinical data and conducting comprehensive antimicrobial susceptibility testing, we aim to provide a nuanced understanding of the resistance patterns exhibited by these strains and draw implications for clinical practice. Our findings underscore the need for continuous surveillance, judicious use of antibiotics, and the development of novel therapeutic agents to curb the spread of antibiotic-resistant K. pneumoniae and improve patient outcomes.
Methods
Clinical Sample Collection and Classification
The study involved collecting 86 strains of Klebsiella pneumoniae (KP) isolated from inpatients with respiratory tract infections at the First Affiliated Hospital of Guangxi Medical University between January 2019 and January 2021. All strains underwent identification and drug sensitivity testing using the VITEK-2 Compact system. The strains were initially classified into two groups based on the positive string test: hypervirulent Klebsiella pneumoniae (hvKP) as the observation group and classic Klebsiella pneumoniae (cKP) as the control group. Clinical data of patients infected with hvKP were analyzed to identify the risk factors and clinical characteristics associated with hvKP infections.
Identification of Klebsiella Pneumoniae Strains
In this study, the clinical isolates were subjected to meticulous microbiological procedures to ensure precise identification of Klebsiella pneumoniae strains. Following overnight incubation at 37°C, single colonies were isolated and identified using an automated rapid microbial mass spectrometry system. This state-of-The-art technology guarantees a high level of accuracy, essential for the reliability of subsequent analyses.16
Preservation and Revival of Bacterial Strains
To maintain the viability and integrity of the Klebsiella pneumoniae strains, a systematic preservation protocol was employed. Single colonies were cultured on MH agar/broth plates and in MH broth, followed by storage in a −80°C ultra-low temperature freezer with a specific preservation fluid. The revival process was meticulously carried out, involving streaking the bacteria onto blood agar plates and incubating them under optimal conditions, ensuring the strains remained true to their original form for the experimental analyses.
Antimicrobial Susceptibility Testing
The VITEK-2 Compact system from bioMérieux, France, was utilized for the antimicrobial susceptibility testing, encompassing a comprehensive range of antibiotics. The interpretation of results was strictly based on the 2019 Clinical and Laboratory Standards Institute (CLSI) guidelines, ensuring the data’s relevance and accuracy.17 This thorough approach is crucial for understanding the resistance patterns of the Klebsiella pneumoniae strains and guiding effective clinical treatment.
Mucoviscosity String Test and Clinical Data Collection
The mucoviscosity string test was conducted to differentiate hvKP from cKP strains, following established protocols and criteria.18 Simultaneously, a comprehensive collection of clinical data from patients was undertaken, ensuring the inclusion of a wide range of relevant parameters. This data collection process was exhaustive, with stringent inclusion and exclusion criteria, ensuring the study’s validity and reliability.
Crystal Violet Assay for the Evaluation of KP Biofilm Formation Ability
In-Vitro Biofilm Model Establishment: Klebsiella pneumoniae (KP) strains were revived from −80°C deep freeze storage and streaked onto MH agar plates using a sterile inoculating loop. After overnight incubation at 37°C, a single colony was transferred into 5mL of MH broth and incubated at 37°C with shaking (220 rpm) for 18 hours to form a bacterial suspension. The concentration of the bacterial suspension was adjusted to OD600=0.01 using MH broth. 200μL of the adjusted suspension was added to each well of a 96-well plate, with three replicates per strain. Sterile MH broth served as the negative control. The 96-well plate was then incubated at 37°C for 24 hours.
Crystal Violet Staining for Biofilm Detection
After incubation, the supernatant in the 96-well plate was discarded, and the wells were washed three times with sterile distilled water to remove planktonic bacteria. Following drying at room temperature, each well was stained with 0.1% crystal violet for 10 minutes. Excess stain was removed by washing gently with sterile distilled water. After air drying, 200μL of 95% ethanol was added to each well to dissolve the stain, and the plate was left for 10 minutes. The optical density (OD) at 550nm was measured three times using a microplate reader. According to the standards in the referenced literature, a well was considered positive for biofilm formation if the OD value was greater than or equal to the sum of the negative control’s OD value plus three times the standard deviation of the negative control. All experiments were repeated independently three times.
Scanning Electron Microscopy (SEM) Observation of KP Biofilm Formation
The preparation of biofilm samples and SEM observation followed the protocols outlined in the referenced literature,19 with slight modifications.
2.4.1 Biofilm Preparation: KP strains were revived and cultured as described in section 2.3.1. After adjusting the bacterial suspension to OD600=0.1 using MH broth, 2mL of the suspension was added to each well of a 24-well plate, along with a piece of polyvinyl chloride (PVC) carrier. Three replicates were prepared for each strain. The 24-well plate was incubated at 37°C for 24 and 72 hours. The PVC carriers with biofilm were washed three times with sterilized PBS (PH=7.4) to remove planktonic bacteria. The biofilm was then fixed with 2.5% glutaraldehyde for 2 hours at 4°C. After washing three times with sterilized PBS (PH=7.4), the samples were dehydrated in a series of ethanol solutions of increasing concentration (50%, 70%, 80%, 90%, and 100%, with three changes at 100% and one change at each of the other concentrations) for 10 minutes each. The samples were then coated with gold in a vacuum and observed under a scanning electron microscope.
Statistical Analysis
The statistical analyses in this study were conducted using the SPSS 22.0 software package. Quantitative data conforming to a normal distribution were expressed as mean ± standard deviation 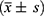 and comparisons between two groups were performed using the t-test. Non-normally distributed data were represented by median and interquartile range [M (P25-P75)], and the Mann–Whitney U-test was applied for intergroup analysis. Categorical data were described using frequencies and percentages, and comparisons were made using the chi-square test or Fisher’s exact test, as appropriate. All tests were two-tailed, and a P-value of less than 0.05 was considered statistically significant.
and comparisons between two groups were performed using the t-test. Non-normally distributed data were represented by median and interquartile range [M (P25-P75)], and the Mann–Whitney U-test was applied for intergroup analysis. Categorical data were described using frequencies and percentages, and comparisons were made using the chi-square test or Fisher’s exact test, as appropriate. All tests were two-tailed, and a P-value of less than 0.05 was considered statistically significant.
Results
Demographic and Clinical Characteristics
The study analyzed 86 cases of Klebsiella pneumoniae (KP) infections, comprising 42 cases of hypervirulent Klebsiella pneumoniae (hvKP) and 44 cases of classic Klebsiella pneumoniae (cKP). The gender distribution across the groups showed no significant difference, with 68 males and 18 females in the total KP group, 35 males and 7 females in the hvKP group, and 33 males and 11 females in the cKP group (P=0.342). The age distribution was also similar across the groups, with a median age of 54.65 years in the KP group, 54.38 years in the hvKP group, and 54.91 years in the cKP group (P=0.707). In terms of clinical presentation, a significant difference was observed in the incidence of community-acquired pneumonia, with 35 cases in the KP group, of which 12 were hvKP and 23 were cKP (P=0.025). However, hospital-acquired pneumonia was more prevalent in the hvKP group, with 30 cases compared to 21 in the cKP group, indicating a higher hospital-associated infection rate in patients with hvKP (Table 1).
 |
Table 1 Baseline Characteristics of 86 KP-Infected Patients with Respiratory System Infection |
Underlying Diseases and Risk Factors in Klebsiella Pneumoniae Infections
In a comparative analysis of 86 patients with Klebsiella pneumoniae infections, encompassing 42 cases of hypervirulent Klebsiella pneumoniae (hvKP) and 44 cases of classic Klebsiella pneumoniae (cKP), the study revealed distinct differences in the prevalence of certain comorbidities (Table 2). Notably, diabetes mellitus, malignant tumors, post-surgical complications within one week, and hypoproteinemia were more prevalent in the hvKP group. Specifically, diabetes mellitus was found in 18 cases of hvKP compared to 9 in cKP, malignant tumors in 12 hvKP patients versus 5 in cKP, post-surgery complications within one week in 17 hvKP cases against 9 in cKP, and hypoproteinemia in 11 hvKP patients compared to 4 in cKP, all showing statistically significant differences (P<0.05). These findings highlight the increased association of certain underlying diseases and postoperative complications with hypervirulent Klebsiella pneumoniae infections, suggesting a trend towards more severe health outcomes in the hvKP patient group.
 |
Table 2 Analysis of Underlying Diseases and Risk Factors in Patients with hvKP and cKP in Respiratory |
Inflammatory Indices in Respiratory System Infection Patients with hvKP and cKP
In the analysis of 86 patients clinically diagnosed with Klebsiella pneumoniae (KP) infections, comprising 42 hvKP and 44 cKP cases, the study evaluated various inflammatory indices. The assessment revealed that there were no significant statistical differences in body temperature, respiration rate, heart rate, white blood cell count (WBC), neutrophil percentage (N%), and procalcitonin levels between patients with hvKP and cKP respiratory system infections. The average body temperature was similar between the hvKP (37.37±0.97°C) and cKP (37.20±0.86°C) groups, as were the respiration rates (21.55±5.70 for hvKP vs 20.68±2.12 for cKP), heart rates (91.76±20.13 for hvKP vs 90.18±17.70 for cKP), WBC counts (10.58±6.19 for hvKP vs 10.67±6.96 for cKP), neutrophil percentages (74.60±14.11% for hvKP vs 75.54±13.42% for cKP), and procalcitonin levels (3.26±6.14 for hvKP vs 2.98±6.14 for cKP). These findings suggest that the inflammatory response in hvKP and cKP infections presents similarly in respiratory system infections (Table 3).
 |
Table 3 The Levels of Inflammatory Indexes in Patients with hvKP and cKP in Respiratory System Infection |
Prognosis Analysis in Respiratory System Infections
In the study of 86 patients with Klebsiella pneumoniae (KP) infections, it was observed that patients with hypervirulent Klebsiella pneumoniae (hvKP) experienced significantly longer hospital stays, averaging 30.80±21.89 days, compared to classic Klebsiella pneumoniae (cKP) patients, who averaged 19.60±13.03 days (P<0.05). The overall mortality rate among these patients was 5.81% (5/86), with a slightly higher rate in the hvKP group (7.14%, 3/42) than in the cKP group (4.55%, 2/44). Furthermore, the incidence of septic shock, severe pneumonia, ICU admission, mechanical ventilation, tracheal intubation or tracheostomy, and fungal coinfections was significantly higher in the hvKP group compared to the cKP group, indicating a more severe clinical course in hvKP infections (Table 4).
 |
Table 4 Analysis of the Prognosis of Patients with Respiratory System Infection hvKP and cKP |
Fungal and Other Bacterial Infections
Analysis of fungal infections revealed that 21 out of 42 hvKP patients and 12 out of 44 cKP patients had fungal coinfections, with a significantly higher prevalence in the hvKP group. Additionally, among patients with hvKP, 21 had solo KP infections, while 13 had only fungal coinfections. This contrasts with the cKP group, where 32 had solo KP infections and 5 had only fungal coinfections. These findings suggest a higher complexity in the infection patterns in the hvKP group compared to the cKP group (Table 5).
 |
Table 5 Fungal Infections of hvKP and cKP |
Antibiotic Resistance Analysis in hvKP and cKP
We conducted a comprehensive analysis to compare the drug resistance profiles between hvKP and cKP strains isolated from respiratory system infections. The analysis, presented in Table 6, highlights significant differences in resistance rates to various antibiotics. Notably, hvKP strains showed lower resistance percentages compared to cKP across most antibiotics tested. For instance, resistance to Ceftriaxone was observed in 14.29% of hvKP isolates compared to 38.64% in cKP, indicating a statistically significant difference (P=0.011). Similarly, notable differences were found in resistance to Cefuroxime, Ceftazidime, and Gentamicin, with P-values of 0.003, <0.001, and 0.005, respectively.
 |
Table 6 Analysis of the Difference of Drug Resistance Between hvKP and cKP in Respiratory System Infection |
Biofilm Formation and Virulence Genes Distribution in hvKP and cKP
Our comprehensive analysis highlighted the distinct biofilm formation capabilities and virulence gene distribution between hvKP and cKP strains. Notably, biofilm formation was observed in 69.77% of the 86 clinically isolated KP strains, with hvKP demonstrating significantly higher biofilm formation rates (85.71%) compared to cKP (54.54%). SEM results underscored that hvKP strains not only formed denser and more cohesive biofilms but also exhibited a more complex extracellular matrix, suggesting enhanced survival and antibiotic resistance mechanisms. These observations were corroborated by the dominance of capsule serotype K2 and the prevalence of virulence genes such as rmpA, iutA, and PEG-344 in hvKP strains, pointing to a robust virulence factor expression in these strains. Specifically, the rmpA gene was notably more expressed in hvKP strains under biofilm-forming conditions, highlighting a potential link between biofilm formation capabilities, virulence gene expression, and the observed increase in antibiotic tolerance (Figure 1).
Discussion
This study presents a nuanced exploration of the clinical and microbiological disparities between hvKP and cKP strains in respiratory infections, offering valuable insights into their pathogenesis, resistance patterns, and implications for patient outcomes. Our findings underscore the heightened virulence of hvKP, as evidenced by longer hospitalization durations, increased severity of infections, and higher mortality rates compared to cKP. This aligns with emerging literature, highlighting hvKP’s role in exacerbating respiratory infections, a trend that is increasingly reported in clinical settings globally.20
A pivotal aspect of our study was the examination of biofilm formation capabilities of these strains. Biofilms are known to play a critical role in the persistence and resistance of bacterial infections.21 Our observations using Scanning Electron Microscopy (SEM) revealed distinct biofilm architectures between hvKP and cKP strains at different growth phases. The denser and more cohesive biofilm structure of hvKP, especially noted in the 72-hour cultures, can be correlated with the increased antibiotic tolerance and virulence seen in these strains. These findings contribute to the growing body of evidence suggesting that biofilm formation is a key virulence factor in hvKP strains, a feature that complicates treatment strategies and underscores the need for targeted therapeutic approaches.22 In the realm of antibiotic resistance, our study adds to the intricate narrative of the evolving resistance patterns in K. pneumoniae. While hvKP and cKP showed similar resistance patterns to a range of antibiotics, such as Amikacin and Ciprofloxacin, significant differences were noted in their susceptibility to other antibiotics including Amoxicillin/Clavulanic Acid, Ceftazidime, and Piperacillin. Interestingly, hvKP strains showed lower resistance rates to these antibiotics compared to cKP strains. This observation suggests that the mechanisms underlying antibiotic resistance may differ between hvKP and cKP. Rather than indicating a straightforward measure of resistance, the lower resistance rates in hvKP strains could reflect unique genetic or phenotypic traits that influence how these strains acquire or express resistance to antibiotics. This distinction underlines the importance of designing antibiotic therapies that are specifically tailored to the strain of K. pneumoniae involved in the infection, acknowledging the complex landscape of antibiotic resistance that varies not only across bacterial species but also within strains of the same species. The clinical implications of our findings are significant. The increased virulence and complex resistance patterns of hvKP necessitate a more aggressive and tailored approach to treatment.6 Early identification and differentiation of hvKP from cKP in clinical specimens should be prioritized to facilitate appropriate therapeutic interventions. Moreover, our results highlight the urgent need for novel therapeutic agents and strategies, particularly those targeting biofilm formation and antibiotic resistance mechanisms in hvKP.
In conclusion, our study elucidates distinct characteristics of hvKP and cKP strains in respiratory infections, emphasizing the critical role of biofilm formation in hvKP’s heightened virulence and antibiotic tolerance. We have uncovered significant clinical and microbiological disparities, marking hvKP’s association with more severe infections and outcomes. The differential antibiotic resistance patterns observed necessitate a nuanced approach to treatment, highlighting the importance of early strain identification for effective therapy. Our findings advocate for intensified research into targeted treatments, especially against biofilm-associated virulence and resistance mechanisms. Ultimately, addressing these challenges is vital for improving patient outcomes and combating the public health threat posed by these adaptable pathogens.
Data Sharing Statement
The original data supporting the conclusions of this article will be made available by the Dr.Zhongwei Wen, without undue reservation.
Ethics Statement
This research was conducted in full accordance with ethical principles, including the World Medical Association Declaration of Helsinki (as revised in 2013) concerning human rights. Ethics Committee Approval: The study was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (Approval Number: 2022-E404-05). Informed Consent: Informed consent was obtained from all individual participants involved in the study. Participants were informed about the purpose of the research, the procedures to be undertaken, potential risks and benefits, and the confidentiality of their data. They were also informed of their right to withdraw from the study at any time without penalty. Data Handling and Confidentiality: All data collected during this study are stored securely and are accessible only to the research team. Personal identifiers have been removed to ensure the confidentiality and privacy of participants.
Disclosure
The authors declare no conflict of interest.
References
1. Wahab S, Ahmad I, Irfan S, Siddiqua A, Usmani S, Ahmad MP. Pharmacological efficacy and safety of Glycyrrhiza glabra in the treatment of respiratory tract infections. Mini Reviews in Med Chem. 2022;22(11):1476–1494. doi:10.2174/1389557521666210927153001
2. Hobson CA, Pierrat G, Tenaillon O, et al. Klebsiella pneumoniae carbapenemase variants resistant to ceftazidime-avibactam: an evolutionary overview. Antimicrob Agents Chemother. 2022;66(9):e0044722. doi:10.1128/aac.00447-22
3. David S, Reuter S, Harris SR, et al. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Rev Microbiol. 2019;4(11):1919–1929. doi:10.1038/s41564-019-0492-8
4. Russo TA, MacDonald U. The galleria mellonella infection model does not accurately differentiate between hypervirulent and classical Klebsiella pneumoniae. mSphere. 2020;5(1). doi:10.1128/mSphere.00850-19
5. Liu C, Du P, Xiao N, Ji F, Russo TA, Guo J. Hypervirulent Klebsiella pneumoniae is emerging as an increasingly prevalent K. pneumoniae pathotype responsible for nosocomial and healthcare-associated infections in Beijing, China. Virulence. 2020;11(1):1215–1224. doi:10.1080/21505594.2020.1809322
6. Wang Y, Hua M, Wang J, et al. Clonal dissemination of multidrug-resistant and hypervirulent Klebsiella pneumoniae clonal complex in a Chinese hospital. Pathogens. 2022;11(10):1202. doi:10.3390/pathogens11101202
7. Walker KA, Miller VL. The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Curr Opin Microbiol. 2020;54:95–102. doi:10.1016/j.mib.2020.01.006
8. Nobrega DB, Calarga AP, Nascimento LC, et al. Molecular characterization of antimicrobial resistance in Klebsiella pneumoniae isolated from Brazilian dairy herds. J Dairy Sci. 2021;104(6):7210–7224. doi:10.3168/jds.2020-19569
9. Tang M, Kong X, Hao J, Liu J. Epidemiological characteristics and formation mechanisms of multidrug-resistant hypervirulent Klebsiella pneumoniae. Front Microbiol. 2020;11:581543. doi:10.3389/fmicb.2020.581543
10. Bassetti M, Righi E, Carnelutti A, Graziano E, Russo A. Multidrug-resistant Klebsiella pneumoniae: challenges for treatment, prevention and infection control. Exp Rev Anti-Infective Ther. 2018;16(10):749–761. doi:10.1080/14787210.2018.1522249
11. Li L, Yu T, Ma Y, et al. The genetic structures of an Extensively Drug Resistant (XDR) Klebsiella pneumoniae and its plasmids. Front Cell Infect Microbiol. 2018;8:446. doi:10.3389/fcimb.2018.00446
12. Kidd TJ, Mills G, Sá-Pessoa J, et al. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med. 2017;9(4):430–447. doi:10.15252/emmm.201607336
13. Sawa T, Kooguchi K, Moriyama K. Molecular diversity of extended-spectrum β-lactamases and carbapenemases, and antimicrobial resistance. J Intensive Care. 2020;8(1):13. doi:10.1186/s40560-020-0429-6
14. Bonomo RA. β-lactamases: a focus on current challenges. Cold Spring Harb Perspect Med. 2017;7(1):a025239. doi:10.1101/cshperspect.a025239
15. Frère JM, Sauvage E, Kerff F. From ”an enzyme able to destroy penicillin” to carbapenemases: 70 years of beta-lactamase misbehaviour. Curr Drug Targets. 2016;17(9):974–982. doi:10.2174/1389450116666151001112859
16. Kil K-S, Darouiche RO, Hull RA, Mansouri MD, Musher DM. Identification of a Klebsiella pneumoniae strain associated with nosocomial urinary tract infection. J Clin Microbiol. 1997;35(9):2370–2374. doi:10.1128/jcm.35.9.2370-2374.1997
17. Humphries R, Bobenchik AM, Hindler JA, Schuetz AN. Overview of changes to the clinical and laboratory standards institute performance standards for antimicrobial susceptibility testing, M100. J Clin Microbiol. 2021;59(12). doi:10.1128/JCM.00213-21
18. Russo TA, MacDonald U, Hassan S, et al. An assessment of siderophore production, mucoviscosity, and mouse infection models for defining the virulence spectrum of hypervirulent Klebsiella pneumoniae. Msphere. 2021;6(2). doi:10.1128/mSphere.00045-21.
19. Relucenti M, Familiari G, Donfrancesco O, et al. Microscopy methods for biofilm imaging: focus on SEM and VP-SEM pros and cons. Biology. 2021;10(1):51. doi:10.3390/biology10010051
20. Wei T, Zou C, Qin J, et al. Emergence of hypervirulent ST11-K64 Klebsiella pneumoniae poses a serious clinical threat in older patients. Front Public Health. 2022;10:765624. doi:10.3389/fpubh.2022.765624
21. Kaushik V, Tiwari M, Tiwari V. Interaction of RecA mediated SOS response with bacterial persistence, biofilm formation, and host response. Int J Biol Macromol. 2022;217:931–943. doi:10.1016/j.ijbiomac.2022.07.176
22. Shui J, Luo L, Xiang YG, Shi GM, Wu JL, Pan JH. 高毒力肺炎克雷伯菌生物被膜形成能力及耐药性分析 [Analysis of biofilm-forming ability and drug resistance for Hypervirulent Klebsiella pneumoniae]. Zhonghua yu fang yi xue za zhi. 2023;57(9):1452–1457. Chinese. doi:10.3760/cma.j.cn112150-20220929-00938
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

