Back to Journals » Infection and Drug Resistance » Volume 16
Potential Causes of Spread of Antimicrobial Resistance and Preventive Measures in One Health Perspective-A Review
Authors Endale H , Mathewos M , Abdeta D
Received 11 August 2023
Accepted for publication 24 October 2023
Published 8 December 2023 Volume 2023:16 Pages 7515—7545
DOI https://doi.org/10.2147/IDR.S428837
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Sandip Patil
Habtamu Endale,1 Mesfin Mathewos,2 Debela Abdeta3
1School of Veterinary Medicine, Wolaita Sodo University, Wolaita Sodo, Ethiopia; 2School of Veterinary Medicine, Wachemo University, Wachemo, Ethiopia; 3College of Veterinary Medicine and Agriculture, Addis Ababa University, Bishoftu, Ethiopia
Correspondence: Habtamu Endale, School of Veterinary Medicine, Wolaita Sodo University, P.O. Box 138, Wolaita Sodo, Ethiopia, Email [email protected]
Abstract: Antimicrobial resistance, referring to microorganisms’ capability to subsist and proliferate even when there are antimicrobials is a foremost threat to public health globally. The appearance of antimicrobial resistance can be ascribed to anthropological, animal, and environmental factors. Human-related causes include antimicrobial overuse and misuse in medicine, antibiotic-containing cosmetics and biocides utilization, and inadequate sanitation and hygiene in public settings. Prophylactic and therapeutic antimicrobial misuse and overuse, using antimicrobials as feed additives, microbes resistant to antibiotics and resistance genes in animal excreta, and antimicrobial residue found in animal-origin food and excreta are animals related contributive factors for the antibiotic resistance emergence and spread. Environmental factors including naturally existing resistance genes, improper disposal of unused antimicrobials, contamination from waste in public settings, animal farms, and pharmaceutical industries, and the use of agricultural and sanitation chemicals facilitatet its emergence and spread. Wildlife has a plausible role in the antimicrobial resistance spread. Adopting a one-health approach involving using antimicrobials properly in animals and humans, improving sanitation in public spaces and farms, and implementing coordinated governmental regulations is crucial for combating antimicrobial resistance. Collaborative and cooperative involvement of stakeholders in public, veterinary and ecological health sectors is foremost to circumvent the problem effectively.
Keywords: antimicrobial resistance, one health, antimicrobial resistance gene, environment, animal, wildlife
Introduction
Antimicrobial resistance (AMR), is the phenomenon by which microorganisms ability to resist the antibiotics’ effect and propagate in the presence of drugs that were effective against them, rendering treatments ineffective.1 The use of antibiotics, which were discovered over 70 years ago, has led to significant advancements in public and animal well-being and agriculture. However, throughout history, the emergence of resistant microbes has continuously challenged these medical innovations. This indicates that antimicrobial resistance is a natural, ancient, and widespread occurrence among bacteria in any biological system.2
Furthermore, it is worth noting that many antimicrobial substances share chemical similarities with naturally occurring compounds. Remarkably, genes responsible for AMR have been discovered from the permafrost, suggesting they existed long before humans developed the ability to produce antibacterial chemicals and use them on a large scale. Thus, the occurrence of AMR within bacterial populations can be considered a largely predictable phenomenon.3 Nevertheless, recent research focusing on modern commensal microbial genomes found in environment and humans has unveiled a significantly higher abundance of genes of antimicrobial-resistance than previously acknowledged. This finding suggests that various factors facilitate the rise and propagation of drug resistant bacteria.4
AMR was not created by humans, but rather promoted by applying evolutionary pressure on the ecosystem. The maintenance and distribution of resistance within bacterial populations depend on the organism’s lifestyle and the genomic root for resistance. AMR can be inherent, mutation-allied, or attained through horizontal gene handover between organisms.3 Bacteria can overcome the effect of one or more antimicrobial classes through either intrinsic, meaning it is naturally present in the bacteria or acquired (through transformation, transduction, or conjugation of mobile genetic elements like transposons and plasmids enabling numerous resistant genes incorporation into the genome or plasmid of receiver microbe) mechanisms.5 In the past few decades, pharmaceutical companies have decreased their interest in antibiotic research and development due to snags in clinical improvement and monitoring of scientific and financial subjects. Clinical evaluation of the effectiveness of new antimicrobials is problematic and expensive owing to the absence of rapid diagnostic tests, particularly when directing Gram-negative multidrug-resistant bacteria.6
Antimicrobial-resistant organisms are a major health crisis causing over 700,000 deaths annually across the globe, anticipated to increase to 10 million by 2050.7 AMR is a rising concern, especially in developing countries and is anticipated to upsurge in the coming years due to higher antimicrobial utilization in public and veterinary care. Antimicrobials are used for livestock farming at a significantly higher rate, 73–100% extra antimicrobials than human medicine, driven by a growing middle class’s demand for animal-based protein.8 According to a report by the antimicrobial resistance collaborators, E. coli was the leading cause of death associated with resistance to antimicrobial drugs in 2019 followed by Staphylococcus aureus, then by Klebsiella pneumoniae, Streptococcus pneumoniae, Acinetobacter baumannii, and Pseudomonas aeruginosa. These pathogens were responsible for approximately 929,000 deaths directly attributed to AMR globally and a total of 3.57 (2·62–4·78) million deaths owing to AMR with higher 27.3 (20.9–35.3) demises apiece 100,000 individuals in the Western part of Sub-Saharan Africa.8
A momentous increase in microbes resistant to one or more antibiotics is posing a great threat to the effectiveness of life-saving antibiotics worldwide. This necessitates the urgent need to identify the microbes that require more attention for drug development, given the severe consequences of multidrug resistance and the limited introduction of new drugs. WHO has prioritized certain pathogens based on their critical, high, and medium priorities. Critical-priority bacteria include Acinetobacter baumannii and Pseudomonas aeruginosa resistant against carbapenem, and Enterobacteriaceae resistant against carbapenem and third-generation cephalosporin.9 High-precedence bacteria include enterococcus faecium resistant against vancomycin, helicobacter pylori resistant against clarithromycin, and staphylococcus aureus resistant against methicillin, and shigella spp. resistant against fluoroquinolone, campylobacter spp. resistant against fluoroquinolone, Haemophilus influenzae resistant against ampicillin, and streptococcus pneumoniae resistant against penicillin (Figure 1).10
 |
Figure 1 Final ranking of antibiotic-resistant bacteria. Reprinted from Lancet Infect Dis. 18(3), Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 318–327, Copyright 2018, with permission from Elsevier.9 In the above figure, Mean (SD) pathogen weights derived by the software from the survey participants’ preferences and the segments represent the contribution of each criterion to each pathogen’s final weight. Abbreviations: PNS, penicillin non-susceptible; 3GCR, third-generation cephalosporin resistant; CR, carbapenem resistant; MR, methicillin resistant; VR, vancomycin resistant; ClaR, clarithromycin resistant; FQR, fluoroquinolone resistant; AmpR, ampicillin resistant. |
One health concept involves understanding the biological elements related to the evolution of AMR, including microorganisms, vectors, host organisms, environments, and cultural and socioeconomic factors that aid its spread. AMR is considered a significant one-health problem.11 Unification of the environment and human and animal territories play a role in the rise, progress, and blowout of AMR, posing a peril to human wellbeing. Resistance can spread across different microbiomes and alter bacterial population genetics, potentially altering the genetic makeup and population dynamics of microbiomes. Changes in habitats, such as contamination by antibiotics or antibiotic-resistant microbes can affect the assemblies of bacterial populations and hasten the propagation of AMR among the microbiomes.12
Genes responsible for AMR are carried by mobile genetic elements (MGEs) that can move among different species and clones of bacterial. These MGEs are present in various environments including human, animal, nourishment, mess, water and soil, forming the interconnected microbiome ecosystem. The proximity of densely populated areas with a high concentration of vertebrate animals increases the likelihood of frequent interactions and merging of microbiomes, leading to the spread of AMR.13,14 The decline in diversity and increase in interactions between animals and humans may lead to a repeated merging of similar types of microorganisms and the exchange of genes between microorganisms. This highlights the significance of One health in contending AMR, as AMR involves complex interactions among humans, animals and the milieu, providing various routes for the spread of drug residues and resistant bacteria.15
Statement of Problem
There are the latest research findings, insights, and recommendations regarding the distribution of antimicrobial resistance within humans, animals, the environment and wildlife and associated contributing factors and preventive measures. Therefore, there is a need for a comprehensive review that encompasses the contribution of humans, animals, the environment and wildlife for the propagation of antimicrobial resistance and preventive approaches in the One Health framework.
Objective of the Review
The current review aimed to overview an impression of the potential causes of the antimicrobial resistance spread and preventive measures from a One Health perspective.
Review Methodology
The manuscripts used for the current review were searched from reputable journals and Google Scholar by using keywords and phrases like antimicrobial resistance; antimicrobial resistance and one health; AMR and drug misuse and overuse in humans or animals; AMR and environment; antimicrobial resistance in the public settings and antimicrobial resistance in the wildlife; antimicrobial resistance and agrichemicals and preventive measure of antimicrobial resistance. Additionally, I have searched in Google Scholar by formulating sentences based on the specific ideas I was exploring. During the review, all possible efforts were made to use the most recent information.
Potential Causes of the Spread of Antimicrobial Resistance from One Health Perspective
AMR is influenced by factors like human practices (antimicrobial overuse and misuse, insufficient infection deterrence, and lack of awareness); animal-related practices (widespread antibiotic use in livestock and aquaculture, transmission through food chains and direct contact); environmental factors (release of antibiotics and resistant bacteria into water bodies, soil, and waste systems) and wildlife-related factors (transmission through contact with wildlife and habitat encroachment).16
Human-Related Causes of Antimicrobial Resistance
Inappropriate Antimicrobial Use in Human Medicine
Excessive and inappropriate use of antibiotics has played a significant role in the worldwide problem of antibiotic resistance. The available evidence indicates that our heavy reliance on antibiotics, along with the interconnected relationship between human health, animal farming, and animal health, has led to the emergence and dissemination of drug resistant bacteria and other organisms. This has resulted in a global epidemic where many commonly used antibiotics are becoming less effective in treating infections.17
The overuse of drugs, including self-medication, dispensing antibiotics without prescriptions, over-prescription, patients’ prospects and liability pressure, can cause the development of drug resistance. These practices promote the survival and proliferation of germs that are resistant to antimicrobials by destroying the susceptible ones, leading to the emergence of AMR. While antimicrobials can kill certain disease-causing germs, they also eliminate beneficial germs that protect our bodies from infections. As a result, the remaining antimicrobial-resistant microorganisms survive and multiply, carrying resistance traits in their DNA that can be transferred to other microorganisms. Unnecessary use of broad-spectrum antibiotics owing to the delayed or inaccurate identification of the etiology of the ailment further fuels the problem of AMR.18,19
There is an association between the use of certain antibiotics and resistance to antibiotics in which increased antibiotic use often increases antibiotic resistance, reducing antibiotic use will also reduce antibiotic resistance to some extent. Higher infection rates are seen in patients infected with bacteria resistant to bacteria from environments with high levels of antibiotics and multidrug-resistant (MDR) strains. The disease is treated more like an infection. Antibiotic use upsurges the menace of selection of resistant bacteria in the normal microbiota and raises the probability of the emergence and spread of resistant microbes when used longer.8
Although antibiotic resistance is seen as a result of antibiotic use, the interaction between these two factors is not a linear relationship. Both changes are involved in microbial antibiotics, and the development of drug resistance and the interaction between them is complex. From an epidemiological perspective, the occurrence, abundance and spread of AMR are influenced by many factors.20 According to21 the overprescribing of antibiotics is a significant issue, particularly in primary care settings mainly for infections caused by viruses. This in turn leads patients to re-attend medical facilities more frequently and use antibiotics excessively for self-limiting conditions, where they are not truly necessary. This refers to the act of using antibiotics inappropriately or too frequently, which can result in modifications/selection pressure within the bacteria which render the bacteria resistant to the antibiotics.
Microbes can mutate spontaneously or as a result of encounters with certain drugs, enabling them to amend or circumvent antimicrobial targets. Bacteria can undergo mutations that alter their cell walls or the enzymes responsible for breaking down antibiotics, for example; viruses can mutate to evade antiviral agents designed to target their replication cycle, while parasites can alter their metabolic pathways to avoid antiparasitic agents.22 Mutations or exchanges of genes horizontally upshot in a selection of variants in naturally sensitive bacterial populations with reduced antibiotic susceptibility. Additionally, mutations or genes responsible for resistance accrue in specific strains or clones of pathogenic bacteria and can subsist even when there are numerous drugs. In this process, the same antibiotic can select different microbial isolates resistant to more than one drug and different antimicrobials can also select microbes resistant to one drug (Figure 2).23
Another example shows that antibiotic overuse is a key contributive aspect to the development of antibiotic-resistant microbes like S. aureus resistant to methicillin and multidrug-resistant tuberculosis.24 Likewise, therapeutic and prophylactic excessive use of antiviral drugs such as oseltamivir (Tamiflu) for influenza facilitates the development of drug resistant influenza strains, including the H1N1pdm09 strain.25 Moreover, inducible genes, such as erythromycin resistance methylase (erm) genes, are present within mycobacteria and S. aureus which are synthesized only in the presence of certain types of antibiotics, resulting in the bacteria quickly developing resistance to those specific antibiotics.26
Inapt antimicrobial use in the community is identified as a foremost factor contributing to the development and propagation of MDR organisms that pose a solemn peril to public health.27 This occurs as the drug kills off the susceptible bacteria, leaving the resistant ones to grow unrestricted Antimicrobial misuse can happen in numerous customs, for example, taking antibiotics to relief viral infections like influenza, flu, or COVID-19.28 Besides, underutilization of antibiotics, ie, not appropriately prescribed or not taken as per the recommended dosage, can result in ineffective treatment outcomes and repeated infections. This, in turn, may necessitate the utilization of more antibiotics, which further fuel the rise and dissemination of AMR.29 As documented by,30 in the Southeast Asian region there is widespread usage of antibiotics without prescriptions from physicians (self-medication with antibiotics), increasing the likelihood of improper drug usage and subsequent appearance and distribution of drug-resistant microorganisms.
Another example demonstrates that S. aureus resistant to methicillin is also resistant to beta-lactams. However, S. aureus susceptible to methicillin remains vulnerable to cephalosporin like cefuroxime as well as beta-lactams containing beta-lactamase inhibitors like amoxicillin/clavulanate. It is imperative to consider that the use of these antimicrobials (cephalosporins, amoxicillin/clavulanate) possibly encourage resistance against methicillin via co-selection (Figure 3).17
Furthermore, the improper utilization of antibiotics, which includes the usage of broad-spectrum antibiotics that eliminate both harmful and beneficial bacteria within the body, can also actively contribute to the rise of AMR. Such antibiotic practices create an environment favorable growth of resistant bacteria by disrupting the normal bacterial flora. Consequently, the resistant bacteria are provided with an opportunity to multiply and spread.31 It is instigated that, the utilization of second-line antimicrobials such as macrolides and second-generation cephalosporins, in cases of acute self-limiting infections or infections that can be effectively treated by commonly used first-line antimicrobials like amoxicillin or penicillin alone, can generate selective pressure on microbes which inturn cause AMR.17
Under-prescription of antibiotics culminates in the administration and presence of subtherapeutic drug concentration in the body which in turn endorses the rise of AMR by favoring genetic modifications. Fluctuations in gene expression induced by antibiotics can result in increased virulence of bacteria and augmented mutagenesis and horizontal genetic transfer which further promote AMR and its spread among microbial populations.32 For instance, lower concentrations of drugs have been found to cause strain diversification in organisms like P. aeruginosa. Secondly, piperacillin and/or tazobactam at concentrations lower than MIC actuates wide genomic alteration in Bacteroides fragilis.33
The organisms created antimicrobial resistance in over said strategies spread to the environment and other non-human creatures through different implies. For instance, besides intestine organisms, which contain a wide cluster of resistance determinants, antimicrobials devoured by people are discharged into the environment in pee and fecal material contained in treated wastewater and sludge applied to the land.34 Past this, anthropogenic contamination presents antibiotic-resistant microbes (ARM) and antibiotic resistance genomes (ARGs) to the healthy setting, and resistance genomes that initially existed in natural microbes can be exchanged, by parallel genome exchange, to the microbes infecting humans.35 For instance, an investigation by Nepal et al34 demonstrated that the presence of resistance genes in crAssphage, a bacteriophage that’s uncommonly plenteous in and particular to human feces without clear signs of selection within the environment through the crAssphage arrangements is an indicator of human stool defilement.
Inadequate Sanitation and Hygiene in the Public Settings
The destitute cleanliness and failure to take after disease avoidance and control measures have played a critical part in the spread and expansion of antibiotic-resistant microbe strains. Hospitals, in particular, have risen as noteworthy reservoirs of these resistant microbes. This will be ascribed to the recurrent and extensive use of drugs within hospitals and the nearness of patients, which creates more opportunities for the transmission of these microbes. Eventually, these variables contribute to the proceeded engendering and spread of microbial strains resistant to different antibiotics which inflict a serious risk to public wellbeing.36
Streptococcus-pyogenes resistant to sulfonamide was identified within public clinical settings in early the 19, S. pyogenes resistant to penicillin was noted in the 1940s and MDR microbes were reported in the 1950s.37 In certain occasions, the extent of patient-to-patient transmission is directly proportional to the extent of hospital environment contamination (Figure 4). Furthermore, inadequate sanitation of the health facility environment favours the persistence of these microbes in the environment (Figure 4).38 Each day, thousands of staff, visitors and patients from diverse parts reach hospitals, each harboring a set of the microbiome of diverse genotypes and phenotypes on their on/inside their bodies as well as clothing. Microbes can disseminate in case health settings do not practice satisfactory sanitary strategies and protocols to keep up setting hygiene and culminate in further development and spread of AMR.36
Moreover, the improperly released hospital water and effluents spread the antimicrobial-resistant organisms and antimicrobial-resistant genes to and encourage their event within the adjacent environment, and their procurement by humans and animals when uncovered in the environment (Figure 4).39 Hence, the presentation of diverse visitors and patients harboring diverse organisms with resistant genes besides insufficient sanitation and cleansing part in the spread of AMR from one place to another, humans and animals.40 In a study conducted in Vietnam, it was found that 83% of E. coli isolates from hospital wastewater showed resistance to at least one antibiotic, and 32% showed resistance to multiple drugs advocating that hospitals play a role in the spread of AMR. Another study in a Chinese teaching hospital investigated the transmission of MRSA in a pancreatic surgery ward and revealed the presence of MRSA (specifically the ST5-MRSA-II-T311 clone) in 20 strains isolated from both the environment and patients. This also instigates that the hospital environment and lack of proper hand hygiene could be risk factors for AMR spread.41
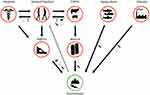 |
Figure 4 Schematic flow of antibiotic resistance from hotspots of evolution. Reprinted from Kraemer SA, Ramachandran A, Perron GG. Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms. 2019;7(6):180. Creative Commons.42 Notes: Antibiotic-resistant bacteria (ARBs) and antibiotic resistance genes (ARGs) from hotspots of evolution (hospital) transmit (red circles) to the environment (green circles) through sludge and different vectors (blue circles). |
Beyond this, after being discharged into the environment, resistant bacteria can reside on plants and be ingested by animals (Figure 4). Once inside the animals’ gut, they transfer the resistant gene, which then alters the structure of the microbiome community since gut contains different microorganisms.43 This renders the gastrointestinal tract an important source of ARGs that can be exchanged between strains of identical or dissimilar species through various mechanisms like phagocytic transduction, plasmid transfer, and natural transformation, which int urn shed in feces.44 According to45 some of the most common nosocomial infection-related pathogens with higher AMR, such as A. baumannii, S. aureus, P. aeruginosa and Enterobacteriaceae (E. Coli, K. pneumoniae), and are persevere in the hospital setting for an extended period.
It is shown that water bodies around the world massively reserve clinically significant antibiotic-resistant organisms and it is mainly attributed to defilement by anthropogenic discharges. Thus, the role of natural recreational waters in the acquisition and transmission of AMR is an area of increasing interest. It is demonstrated that bathing waters contain AMR organisms including Enterobacteriaceae producing New Delhi metallo-beta-lactamase (NDM) and E. coli resistant to cephalosporin.46 Due to the nature of swimming pools as public spaces for recreation, the likelihood of transmission of antimicrobial-resistant microorganisms through various contact points like the eyes, skin, nostrils, oral cavity, anus and vagina is high. A study conducted in central Ohio of the United States found that a significant proportion, 96%, of P. aeruginosa isolates obtained from swimming pools and hot tubs were resistant to multiple drugs.47 Another investigation from Zaria, Nigeria, reported recreational water to contain E. coli and K. pneumoniae strains harboring extended spectrum beta-lactamase genes (TEM, SHV and CTX-M).48
The other public settings with a high likelihood of spreading antimicrobial resistance include buses, railway stations, subway trains, and crowded university or school classrooms due to their high population density. This overcrowding hones the dissemination of different species of microbes with a great variety of general makeup and genetic exchange among them.40 Research conducted in Guangzhou, China depicted the presence of MRSA in railway and coach stations. Out of the samples collected, 39.21% tested positive for staphylococci, with 1.58% being identified as MRSA and 75.84% as multidrug-resistant staphylococci. These findings instigate that railways act as a hotspot for the distribution of MRSA between humans, the environment, and animals, and vice versa.49 Furthermore, an investigation in Chinese public buildings on airborne S. aureus, including MRSA, revealed varying levels of MRSA concentrations. The study reported mean total and respirable concentrations of airborne MRSA in different places, such as 24 and 17 CFU/m3 in university classrooms, 20 and 13 CFU/m3 in kindergarten, 3 and 16 CFU/m3 in the hotel, and 33 and 20 CFU/m3 in the movie theater. This study further supports the idea that public settings have a noteworthy contribution to the propagation of AMR.50
The aforementioned public settings comprise surfaces that are touched by many people carrying different resistant microbes to different antibiotics, which leads to the genetic allotment among microbes and becoming resistant to different drugs. Surfaces frequently touched by hands can act as a means of transmitting AMR from one individual to another if someone directly touches these surfaces or if bacteria on their skin is transferred to a surface and then touched by others.40 Furthermore, in crowded public places, the transmission of resistant bacteria can occur through different methods including the release of bacteria into the air from the skin, inhaling contaminated aerosols, or consuming tainted food.51
Using Antibiotic-Incorporated Cosmetics and Detergents, and Biocides
Everyday grooming and hygiene products like lotions, creams, shampoos, soaps, shower gels, toothpaste, and fragrances contain additives to preserve the product and have disinfection properties. However, these additives can enter ecosystems where they come into contact with microbial communities, potentially promoting the spread of resistance.52 Active substances found in many household detergents and cosmetics can contribute to antibiotic resistance. For example, triclocarban and triclosan, broad-spectrum antibacterial compounds found in a variety of cosmetic products, have been shown to contribute to AMR and promote the growth of resistant to both biocides and antibiotics. The low solubility of these compounds worsens contamination as they tend to build up and stay in surface waters and sediments for extended periods.53 Additionally, exposure to triclosan has led to the discovery of mutations in the enoyl-acyl carrier protein reductase gene in vulnerable bacteria, thereby to the presence of of genes responsible for AMR. It can facilitate the selection of cross-resistant and multidrug-resistant genes, thereby aiding in the spread of ARGs and portable genomic elements in bacterial communities.54
Biocides like chlorine, iodine, alcohols, etc., commonly found in disinfectants, are used to control infections in critical healthcare settings and are now being marketed for home use.55 Scientific studies have shown that the use and improper use of products containing biocides can potentially lead to antibiotic resistance in bacteria. Additionally, bacteria that are tolerant to biocides have been found to also possess resistance to antibiotics.56 This is due to that, biocides and antibiotics may show some resemblances in their mode of action and common mechanisms of microbial insusceptibility may work.57 Factors including the concentration of the biocide, contact time, pH, temperature, the presence of organic matter or other substances, and the characteristics of the microbes or entities being targeted can influence the effectiveness of biocides and aid in development resistance.58
For instance, an investigation on E. coli has shown that from 40 strains evolved in sub-inhibitory, constant concentrations of ten widespread biocides, 17 (43%) have low vulnerability to therapeutically pertinent antibiotics.59 High-touch and disinfected areas like office desks, ATM machines, public toilets, doorknobs and faucets as well as sinks are the risky areas where microbes develop biocide-induced resistance and transmit to humans.60
Animal-Related Causes of the Spread of Antimicrobial Resistance
Animals have an undeniable contribution to AMR spread as human antimicrobials are used in animals as a growth promotor and therapeutically, which has a significant effect on the development of AMR.61 Other variables that facilitate the dissemination of AMR in animals include globalization, worldwide travel and exchange, and contact with wildlife.62 Meanwhile, the misuse or overuse of antimicrobials in animal production has driven different ABR and ARGs, which can be exchanged among animals, the environment and people.63 These ARB and ARGs that emerged in farms can be transmitted to people through direct or backhanded contact with animals, manure, or products, and inhalation of bioaerosol that harbors them.39,64
Prophylactic and Therapeutic Antimicrobial Misuse in Animals and Antimicrobial Resistant Microbe and Resistance Gene in Animal-Derived Food
Antimicrobials prescribed for and crucial for human medicine are used in animals, including pets, aquaculture, apiculture, and farm animals, both prophylactically and therapeutically at higher rates.65 Antimicrobials misuse in animals and resultant AMR is a growing problem worldwide, affecting not only humans but also livestock along the entire food chain in almost every country.16 Genes responsible for antimicrobial resistance and drug resistant bacteria can spread from food-producing animals to people via close contact or consumption of animal products, especially if they are not handled or cooked properly. The United States centers for disease control and prevention estimates that approximately 1 in 5 cases of resistant infections are linked to pathogen found in animals and food of animal origin.66 Furthermore, the presence of drug resistant microbes and drug residue in animal waste is the main source of environmental AMR then humans through direct defilement and stimulation respectively.67 From the environment, animals get these resistant microorganisms through various routes and animals’ guts and tissue harbor a high population of ARB which is further disseminated to the environment as well as to humans and vice-versa.14 These pathogens access humans through handling and ingesting contaminated food items, or contact with animals, as well as through fishing in or drinking defiled water.62
Beyond this, extra-label use of antimicrobial ends in the higher or lower absorption, distribution, or excretion of the antimicrobial active principle. These are also related to the fact as the animal size increases, the ratio of body surface area to body weight decreases, resulting in slower physiological processes compared to smaller animals. These factors collectively influence the way drugs are absorbed, distributed, metabolized, and eliminated in different species.68 Administering antibiotics that are not efficient against the bacteria recorded on the label, or when the dose on the label is inadequate (due to AMR), upshots in unnecessary antibiotic exposure and fairing AMR.69 For instance, antibiotics including penicillins, lincosamides, quinolones, and combinations of sulfonamides with trimethoprim are used for most cases in animals without confirmatory diagnosis.70
It is crucial to note that consumers can potentially come into contact with or consume animal products carrying resistant bacteria, which adds to their exposure to such microorganisms.71 In a study on consumer exposure to antimicrobial-resistant bacteria in food at the Swiss retail level, it was found that out of 38,362 bacteria isolates and 122,438 samples of food tested, 22.9% of the bacterial isolates and 24.6% of the samples were identified as AMR positive. A significant portion of the samples (61.5%) and bacteria (70.2%) were discovered in meat products like cuts, ground pieces, or meatballs. These meat products also comprised the highest percentage (88.7%) of samples with AMR and the majority (60.1%) of AMR-positive bacteria detected. ARB includes enterococcus, campylobacter, salmonella, vibrio spp., listeria and Escherichia coli were isolated from animal products.65 Demonstrating that livestock products including meat, eggs and milk have been proven to be a key route of extraintestinal MDR pathogens, especially for E. coli and Salmonella, posing a potential health risk to consumers.72 For instance,73 reported that out of 152 Salmonella isolates from retail foods of animals, 92.8% of bacteria were MDR.
Furthermore,74 identified coagulase-negative staphylococcus (39.1%), followed by S. aureus (17.2%), S. hyicus (14.1%), S. intermedius (9.4%), and E. coli (9.4%) in cow, camels, and goats milk. E. coli isolates from both cows and goats were MDR against spectinomycin, vancomycin, doxycycline, ceftriaxone and penicillin. S. aureus isolates from all milk have MDR to penicillin G, spectinomycin, and clindamycin. In another investigation in Swiss, Enterobacteriaceae like Citrobacter, Cronobacter, Enterobacter, Klebsiella, and Shigella, E. coli and Salmonella were isolated from most types of milk, cheese, meat, plant and seafood products with a predominance of raw meat origin, particularly of beef, and poultry origin. These bacteria harbor blaACC, blaCMY, blaCTX, blaDHA, blaFOX, blaOXA, blaOXY, blaSHV, or blaTEM variants resistant to aminoglycosides, cephalosporins and penicillins.65
Presence of Antimicrobial Residue in Food of Animal Origin and Animals’ Excreta
Intensive production methods, which involve keeping large numbers of animals in small spaces under stressful conditions, have increased the risk of infectious diseases, necessitating antibiotics to combat them. Likewise, over time, antibiotics started being used extensively at lower doses for preventative and growth-promoting purposes in farm animals.75 Recently, antibiotic consumption in cattle and poultry has risen at an extraordinary rate globally, which is expected to rise further by 67% by the year 2030 in rapidly polluting and developing countries of the world.76
The use of antibiotics in animals can lead to the presence of antibiotic residues in food products like chicken, meat, milk, and eggs. These residues can still be present even after cooking the food, as the cooking heat may not eliminate all the antibiotic residues77 and pose a significant risk of public exposure when consuming animal originated food.78 Many veterinary drug dispensers lack sufficient knowledge about animal illnesses, drug usage, withdrawal periods, side effects, and proper methods of administration. This lack lack of adherence to withdrawal time for drugs used in animals, particularly concerning livestock products like milk and meat consumed by humans important cause for the presence of drug residue.79 These subinhibitory or sublethal concentrations of antibiotics present in food can enter the human body when consumed, either in raw or cooked form. Microbes within the human body are exposed to these concentrations, which can stimulate the development of various resistance mechanisms against the antibiotics present in the feed.35 In addition to this, sub-MIC of antimicrobials have been observed to potentially enhance the frequency of conjugation, which ultimately results in the transfer of genetic material through plasmids, facilitating the spread of antibiotic resistance by mechanism.80
For instance, an investigation by81 sub-MIC concentration different drugs on the handover of ARGs carried in the plasmid to Escherichia coli from K. pneumoniae. The study utilized K. pneumoniae strain SW1780 with plasmid carried MDR gene PSW1780-KPC as the donor strain, and Escherichia coli strain J53 as the recipient. They treated K. pneumoniae SW1780 with subinhibitory concentrations of ciprofloxacin, amikacin, meropenem and cefotaxime. The study demonstrated a substantial increase in the resistance gene handover frequencies upon treatment of K. pneumoniae SW1780 with these subinhibitory concentrations of the above antibiotics.
Antibiotic residues are significantly shed in to and disposed of with farm waste, which poses selection pressure on the environmental microbes. Several antimicrobials administered to the animals are poorly absorbed from the gut of the animals, and as much as 90% of the main antimicrobial active compound is excreted in urine and up to 75% in feces,82 quantities varying among animal species. Animal waste containing such residue is applied to the land and, in turn, contaminates air, water, and soils near both storage and field application sites. There AMR emerged, as microbes were found everywhere and exposed to sub-MIC.83
For instance, tetracyclines, are among the most frequently used antibiotics in poultry farming owing to their broad spectrum action, for prophylactic, therapeutic, or growth promotion roles.84 However, due to poor absorption, active ingredients of tetracyclines can accumulate in muscle, feces and urine. In an investigation by Mahmoud et al85 in Egypt in broiler farms, high concentrations of chlortetracycline 6.05μg/g and 5.9μg/g and oxytetracycline 2.47μg/g and 1.33μg/g was recovered from chicken litter and droppings, respectively. Slightly lower amounts of tetracycline and doxycycline were found: 1.9 μg/g in litter and 0.46 μg/g in droppings for tetracycline, and 0.87 μg/g and 0.02 μg/g for doxycycline.
Antimicrobial Resistant Microbe and Resistance Gene in Animals’ Excreta
Irrational use (like under prescription, lower frequency, inappropriate route) of antimicrobials in animals results in the development of AMR in organisms inhabiting animals’ gut and on their body surface.86 These organisms advance and spread to the people and environment through direct contact and fecal defilement. Gut antibiotic-resistant microbes are excreted in defecation and can spread within the environment and conceivably enter other animals and farmworkers.71 Antibiotics not only affect pathogenic bacteria but also affect the resident microbiota making animal gut rich with ARM and ARGs associated with microbial pathogenicity and diversity.87 This defilement permits the simple selection and exchange of genes related to antimicrobial resistance inside the microbial community of that specific environment. As a result, these ARGs can be transmitted to people and other situations through the utilization of a sullied diet and introduction to squander materials individually.63 For instance, 210 fecal samples were collected from farm animals and the farmer and analyzed the spread of MCR-associated with plasmids among E. coli by multiplex PCR, 18,4 and 1 colistin-resistant E. coli isolates carrying mcr-1 (including 13) with 33 virulence factors were isolated from calves, pigs and farmer respectively and. This instigates that, mcr-1 gene was transferred from the calf to the farmer, thus farm is a reservoir of ARB with zoonotic potential.88
It is demonstrated that farm animals are a reservoir of ARB (such as E. coli, K. pneumonia, Salmonella, S. aureus, etc.) and ARGs that reaches peoples via close contact with farm animals.64 Farm bioaerosols generated during farming practices have also been regarded as a reservoir of ARB and ARGs can be inhaled into the respiratory tract of animals and humans.89 Another author,90 demonstrated that animal waste and slaughterhouses are a reservoir of MDR bacteria, indicating an occupational risk to workers. In addition to this, ARB and ARGs from animal bioaerosols could easily be transmitted from animal farms to the environmental dust through the natural wind.91 AMR persists in animal wastes since animal waste contains resistant pathogens and stable antimicrobial agents.92 Geographic clustering of large-scale livestock farms is highly associated with pollution of soils and irrigation water transferring to and the presence of antimicrobial-resistant bacteria in the food crops, fruits, and vegetables and subsequent transmission to humans upon consumption.93 Fertilization of soil by raw animal squander (fertilizer) or discharge of farmhouse squander into streams are chief route of spread of ARGs and AMR organisms in new fruit and vegetables and aquatic food (Figure 5).62 People acquire these AMR organisms and ARGs as they consume plant-derived foods, especially those eaten raw and aquatic food (Figure 5).94
 |
Figure 5 Spread of ARB and ARGs from animal faeces to the environment and human health. Reprinted from Iwu CD, Korsten L, Okoh AI. The incidence of antibiotic resistance within and beyond the agricultural ecosystem: a concern for public health. MicrobiologyOpen. 2020;9(9):e1035. © 2020 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd. Creative Commons.32 |
Use of Antimicrobials as a Feed Additive in Livestock
Antibiotics are extensively used in animal farming as feed additives to enhance the utilization of nutrients, promote growth and improve overall animal productivity at concentrations lower than the therapeutic dose.95 These antibiotics inflict selection pressure triggering the synthesis of resistant genes in the bacteria and creating a suitable environment for the emergence of AMR since sub-therapeutic antibiotic doses are applied to the feed.61 Furthermore, as mentioned earlier in this document, residual antimicrobial substances can accumulate in animal tissues such as beef, pork, and poultry, as well as in products like eggs and milk which reach microbes in humans.77 In certain instances, contact with sub-MIC of antibiotics can lead to the production of reactive oxygen species. This can potentially contribute to an escalation in mutation rates and the emergence of multidrug-resistant mutants. Additionally, antibiotics in animal feedstuff can hasten the handover of genes through bacteriophages, thereby promoting the dissemination of ARGs. It is worth mentioning that sub-MIC of antibiotics also foster exchange of gene horizontally, a chief means responsible for the exchange of ARGs.32
Environment-Related Cause of the Spread of Antimicrobial Resistance
Environment as a Source of Naturally Existing ARG and AMRM for Human and Animal Pathogens
With respect to the take-up of novel resistance components, water, soil and other environments with profoundly variable biological specialties give an unmatched genetic pool with differing qualities that incredibly surpasses that of the human and domestic animal microbiota.96 Indeed, the most striking feature of the environmental microbiome is, that it is incredibly diverse, offering a wide array of genes that pathogens may acquire to counteract antibiotic effects.42 Antibiotic resistance genes are thought to originate from environmental bacteria that produce and release antibacterial substances to impact other microbes they compete with for resources.97 The recent discovery of antibiotic resistance genes in ancient permafrost sediments suggests that AMR existed naturally even earlier than the use of modern antibiotics. This natural resistance may have contributed appearance and dissemination of AMR in disease-causing microbes showing that milieu is the main source of drug resistant microorganisms.98
Most ARGs did not solely originate in clinical or agricultural sceneries, instead, they evolved in the natural environment where naturally occurring antibiotics derived from soil bacteria and fungi existed. These antibiotics, like penicillin, streptomycin, tetracycline, and chloramphenicol, allowed bacteria to compete for limited resources.40 Since antibiotic-producing microorganisms are comparable to other microorganisms they compete with, there’s a chance they can be hurt by the harmful metabolic compounds created by others. To safeguard themselves, they have evolved defense mechanisms.99 ARGs likely came from antibiotic-secreting microbes and entered into other bacteria through horizontal gene transfer. AMR can also arise from mutations in target sites or pre-existing genes that encode enzymes or are transported via efflux pumps, eventually becoming resistance mechanisms.40
Improper Disposal of Unused and Expired Antimicrobials
Indecorous disposal and/or handling of unused drugs is one of the pathways through which antibiotics can disseminate in the water and other parts of the environment where microorganisms can encounter through various ways such as the release of sewage from urban, industry, animal husbandry, plant production and household.100 The rise in antibiotic use is repeatedly associated with the free availability, accessibility, and incongruous use of antibiotics and the reason for developing antibiotic resistance.101
Many products that are given to patients or clients are not fully used, resulting in large amounts of unused or expired medicines. This has become a global issue that has caught the attention of policymakers, healthcare professionals, pharmaceutical companies, and the general community.102 Improper disposal of pharmaceuticals is a significant issue because users often lack knowledge about proper disposal methods, leading them to flush or throw away their unused and expired medication. This in turn ends up in landfills, water supplies and drains that lead to contamination of the environment.103 This subinhibitory or sublethal concentration in the expired medicines and the containers or in unexpired ones but deteriorated by environmental factors triggers environmental microbes to develop all possible resistance machinery against the antibiotics.104
Environmental Contamination by Wastes from Public Settings, Animal Farms and Pharmaceutical Industries
Wastewater contains diverse contaminants including pharmaceuticals, personal care products, disinfectants, and pesticides, collected from residences, industries, and medical facilities by urban wastewater systems (Figure 6).105 These antibiotics and related chemical constituents in the wastewater pose a foremost health risk owing to their ability to persuade AMR since their microorganism comes in contact with those substances.106 Different studies showed that wastewaters are hotspot reservoirs for both antimicrobials and clinically relevant antimicrobial-resistant bacteria and resistance genes demonstrating that wastewater is a significant vehicle for the AMR spread.107 As mentioned earlier in this manuscript, these AMR microbes and ARGs spread to the environment, animals and humans through different direct or indirect contact and food chains (Figure 6).62,94
 |
Figure 6 Transmission pathways of AMRM and ARGs from wastewater hotspots to the other environment, animals and humans. Reprinted from J Environ Chem Eng; 8(1), Gwenzi W, Musiyiwa K, Mangori L. Sources, behaviour and health risks of antimicrobial resistance genes in wastewaters: a hotspot reservoir. 102220, Copyright 2020, with permission from Elsevier.110 |
Long-term use of manure and waste as fertilizer can lead to plants absorbing antibiotics from treated soil, contaminated water, or raw manure. This can expose humans to lower concentrations of antibiotics throughout the food chain.108 In vegetables like forage, corn, wheat, and peanuts, there is a possibility of them absorbing and storing antibiotics when they are fertilized with animal manure instigating that antibiotics present in the manure can be taken up by these crops and retained within their tissues (Figure 7). These low concentration antibiotics can increase development and persistence of ARGs in human and animal pathogens.109
Furthermore, AMR microorganisms and ARGs in the wastewater and raw manure can also be transferred to plants. These microorganisms inhabit the intercellular spaces or cells of various tissues and organs of healthy plants at a certain stage, or all stages, and are called plant endophytes. These AMR microbes and ARGs are obtained by humans and animals in the food chain (Figure 7).111 Another author112 it was found that the endophytic bacteria present in celery, Chinese cabbage, and cucumber plants, which were cultivated using chicken manure, exhibited a notable level of antibiotic resistance. Among the antibiotics tested, the bacteria showed the highest rate of resistance against cefalexin.
 |
Figure 7 Transmission loop of AMRM and ARG among agronomic production, plants, animals, and human systems. Reprinted from Yan Z, Xiong C, Liu H, et al. Sustainable agricultural practices contribute significantly to one health. J Sustain Agric Environ. 2022;1(3):165–176. © 2022 The Authors. Journal of Sustainable Agriculture and Environment published by Global Initiative of Crop Microbiome and Sustainable Agriculture and John Wiley & Sons Australia, Ltd. Creative Commons.111 AMR microorganism and ARG in wastewater, raw manure, or contaminated water adds to the leaf and flowers (the nectar and pollens), fruits, and seeds phyllosphere microbiomes and reach humans via raw foods. |
The burial service industry, involving thanatopraxy care, burial and incineration may be flourishing worldwide commerce and closely connected to well-being care including in the arrangement, capacity, transport and consequent burial in cemeteries.113 In societies around the world, the predominance of infectious and incessant maladies caused by hurtful microorganisms incredibly contributes to human sickness and mortality. To combat these infections, different therapeutic drugs such as antimicrobial, antiretrovirals, and antifungals are utilized based on the particular pathogens included.114
In another way, as someone passes away, the process of thanatopraxy care including the use of embalming fluids containing various synthetic substances like personal care products, pharmaceuticals, antimicrobials, disinfectants, colognes and moisturizers provoke and/or disseminate AMR.113 Wastewaters from funeral parlors and leachates from cemeteries/gravesites may also serve as a hotspot reservoir of ARGs. In addition is shown that cemeteries contribute to groundwater contamination by non-steroidal anti-inflammatory drugs/analgesics (salicylic acid, nimesulide) and psychiatric drugs (carbamazepine, fluoxetine).115
Human cadavers can potentially contain traces of pharmaceuticals, particularly antibiotics administered during treatment for various infections, contributing to the development of AMR. Consequently, solid waste materials like used bandages, wipes, and gloves, as well as wastewater and airborne particles from facilities involved in thanatopraxy, may carry a diverse range of pharmaceuticals, AMR bacteria, and ARGs (Figure 8).116 Moreover, the interment and possible decay of human bodies in cemeteries result in the discharge of leachates containing medications, resistant microbes, and ARGs to the surroundings (Figure 8).117
 |
Figure 8 General routes of transmission of AMR microbes and ARG in the funeral industry. Reprinted from Sci Total Environ; 749, Gwenzi W. The ‘thanato-resistome’-The funeral industry as a potential reservoir of antibiotic resistance: early insights and perspectives. 141120, Copyright 2020, with permission from Elsevier.116 |
Use of Agricultural Chemicals
Agrichemicals part within the spread of AMR could be a multifaceted issue that has earned noteworthy consideration in later times. The utilization of substances like pesticides, fertilizers, rodenticides, weed executioners, and insecticides in agriculture has been found to have both coordinated and backhanded impacts on the improvement of AMR. Pesticides, particularly utilized to combat pests that harm crops straightforwardly contribute to the determination of antibiotic-resistant microbes by adjusting the microbial environments shown in soil and water.118
Microorganisms that can degrade pesticides acquire and utilize various mechanisms including membrane transport systems, efflux pumps, enzymes, and a genetic composition comprising catabolic genes encoded in both plasmids and chromosomes for the degradation process.119 Genes carrying these specific traits can be transferred to microbial organisms, culminating in the development of AMR with clinical implications. Additionally, in induced mutations, the altered active site of the enzyme affected by the mutation can result in the degradation of both pesticides and antibiotics, creating cross-resistance between these two substances.120 For instance121 reported that upon continued exposure to sub-lethal levels of herbicides including Kamba, Roundup, and 2.4-D, wild-type bacterial species E. coli and S. typhimurium, become resistant to various drugs such as ampicillin, tetracycline, ciprofloxacin, kanamycin, and chloramphenicol. This highlights the potential link between herbicide exposure and the development of antibiotic resistance in bacteria.
Toxic metals found in metal-contaminated soils preferentially support the growth of metal-resistant bacteria, though, there is an increasing concern about co-selecting antibiotic-resistant traits, leading to the proliferation of antibiotic resistance in the environment. Additionally, low levels of biocides trigger mutations responsible for AMR akin to antibiotics.122 The co-existence of ARGs on the extrachromosomal genome improves their resilience and transmission throughout pathogens or microbial communities. One such example is the co-resistance to both benzalkonium chloride (a biocide) and antibiotics in the bacterium S. aureus. This co-resistance leads to a significantly advanced tolerance against antibiotics, making the strain eight times more resistant compared to the wild-type strain sensitive to these agents.123 In another study,124 suggested certain insecticide (Monocrotophos) degrading bacteria isolates belonging to the Bacillus species such as B. cereus, B. firmus, and B. thuringiensis showed resistance against various antibiotics including chloramphenicol, ampicillin, cefotaxime, streptomycin, and tetracycline.
Role of Wild Life and Insects in the Spread of Antimicrobial Resistance
As human populations expand and natural habitats shrink, wildlife increasingly comes into contact with humans and their livestock. ARGs and AMRB found in wildlife indicate the presence of effluence caused by human activities rather than natural selection. Nevertheless, once antibiotic resistance emerges in wildlife, it plays a role in spreading it across various ecosystems at two levels.34 Firstly, in non-migrating organisms, the transfer of antibiotic resistance mainly occurs within local environments. Non-migrating organisms such as cockroaches, fleas, or rats, which frequently live alongside humans, can serve as reservoirs and carriers of antibiotic resistance, contributing to its spread.14 Secondly, migratory faunae, like fish, gulls, or turtles may take part in the distribution of AMR across various geographic areas, even between different continents.125
Although wildlife may not receive antibiotic treatments, various ecosystems (human and domestic animals) and microbiomes including the human and animal gut, can still serve as potential sources of antibiotic-resistant genes (ARGS) since ARGS evolved earlier in the clinical application.126 The existence of antimicrobial-resistant bacteria in wildlife can be attributed to two main factors. One is the acquisition of resistance genes through pollution caused by human activities, which results in the contamination of their natural ecosystems. The other factor is the emergence of resistance due to natural selection pressure, where bacteria develop resistance in response to environmental challenges.127 With increasing human populations, wild animals are compelled to scavenge contaminated resources, leading to exposure to human pathogen pollution. Both resistant bacteria and antimicrobials from humans and domesticated animals are released into the environment. This pollution, including fecal matter from fields, can flow into estuaries, coastal waters, and beaches, posing a critical point of exposure for marine animals, waders, and seabirds to ARB and ARGs. As the wildlife feeds dead animals or their discarded remains, they obtain ARB and ARGs found in that animal tissue (figure 9).128
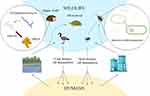 |
Figure 9 Contribution of wildlife to the spread of antibiotic resistance. Reprinted from Laborda P, Sanz-García F, Ochoa-Sánchez LE, et al. Wildlife and antibiotic resistance. Front Cell Infect Microbiol. 2022;12:568. Creative Commons.196 |
Wild fowls such as open-bill storks and brown-headed gulls can carry and spread ARB and ARGs in their fecal droppings which poses a hazard of spreading ARB and ARGs to household and rummaging winged creatures, as well as cultivate situations (Figure 9).129 For instance,130 reported ESBL-producing E. coli in Swedish wild gulls shared similar ESBL-containing plasmids with isolates from humans, livestock, and surface water. This indicates that the existence of ESBL-producing E. coli in gulls is probably due to anthropogenic pollution. In addition131 documented that wildlife harbor high levels of ARGs and antibiotic-resistant microbes compared to domestic animals. Suggesting that wildlife in contact with domestic animals or living in anthropized environments act as potential reservoirs and spreaders of antibiotic-resistant microorganisms and genes in rural or urban areas.
Various types of insects commonly found in households, such as cockroaches, houseflies, ants, and mosquitoes, as well as household rodents like mice, rats, and raccoons, have been identified as carriers and spreaders of AMR.132 According to133 insects circulate AMR from antimicrobial sinks like concentrated animal feeding operations and healthcare settings to other surroundings that include healthy human communities (Figure 9). The same authors reported that house flies carry multidrug-resistant pathogenic E. coli and it was highest in flies sampled from dairy farms (70.67%) followed by from food centers (65.33%), dustbins (64%) and the area close to the hospital (44%). Another investigation by134 demonstrates that household cockroaches may carry drug-resistant isolates, including MDR blaCTX-M-15-, extensively drug-resistant (XDR) blaOXA-48- and XDR blaNDM-1-producing enterobacteria. They convey this bacterium to humans as they move about freely but are usually found breeding in bathrooms, toilets and cupboards for storing food.
Preventive Measures of Spread of Antimicrobial Resistance from One Health Approach
Preventive Measures for Human-Related Causes of the Spread of AMR
Antimicrobial Stewardship and Community Awareness Campaign
Antimicrobial stewardship (AMS) is a strategy implemented in healthcare organizations to ensure the responsible and effective use of antimicrobial drugs. It aims to preserve the effectiveness of these drugs in the future while promoting and protecting public health. AMS has been highly successful in promoting the appropriate use of antimicrobials through evidence-based interventions.135 Initially, it was created to organize interventions aimed at enhancing the use of antimicrobial agents by selecting the right ones, and determining the correct dosage, method of administration, and duration of therapy while prioritizing favorable patient outcomes. The “one health” approach, vaccination guidelines, healthcare professionals’ training, and public awareness about antimicrobial resistance should be driven by evidence-based policies.136
Interventions for improving appropriate antibiotic use and management of infectious diseases include guidelines publication, educational sessions for prescribing antibiotics, educational sessions on infectious disease diagnosis and management, review of prescribing data, local interviews conducted by pharmacists, and distribution of messages through TV, radio, and other mass media platforms.137 Antimicrobial Stewardship involves reducing antimicrobial usage to minimize resistance and costs, while also increasing awareness among the public and educating healthcare professionals on the responsible use of antimicrobials as part of an Antimicrobial Stewardship Program (ASP). These strategies can be implemented worldwide to help combat AMR and save currently available antimicrobials effective for the future as few new drugs are available.136 While the antimicrobial stewardship campaign had initially reduced antibiotic use, it was found that without ongoing educational efforts, the usage reverted to previous levels, especially in developing countries. Therefore, it is crucial to expand these interventions on a national scale to ensure sustained impact and behavioral changes in antibiotic prescribing practices.17
Environmental Sanity and Hygiene in Public Settings and Human Residential Area
To decrease the emergence of new resistant strains, we need to enhance hygiene practices, prevent infection spread, and upgrade healthcare systems while improving living conditions. These measures can reduce the need for antimicrobials, thereby mitigating the risk of antimicrobial resistance.16 Scientific evidence indicates that maintaining clean hospital environments, along with promoting hand hygiene, leads to decreased surface contamination and subsequently reduces the transmission of microbes to patients. Thus, the lessening of surface contamination seems a very significant point toward the deterrence of MDR bacteria spread and the management of nosocomial infections which hasten the spread of AMR.138 Existing evidence shows that conventional chemical sanitation can contribute to the selection of drug resistant strains of microbes.139
Hence, automatic cleaning systems, such as “no-touch” technologies and the use of ultraviolet (UV) light or hydrogen peroxide, have been developed to enhance cleaning practices in hospitals. For instance, portable automated units emitting UV light have been proven effective in decontaminating hospital rooms by eliminating Clostridium difficile spores, MRSA, and vancomycin-resistant Enterococcus spp.140
To ensure ecological and human health, a combination of preventive and control methods is necessary, which can involve both soft and hard engineering solutions. Soft engineering measures may include implementing occupational health and safety procedures, raising public and worker awareness through training campaigns, and educational initiatives to address potential health risks. Additionally, formulating a national strategy, establishing policy and regulatory frameworks, developing monitoring and control systems, conducting training workshops at national and regional levels, and mobilizing resources such as technical experts, funding, and laboratory equipment are also vital in this process.141 Hard engineering approaches for the prevention of the spread of AMR, may involve using antimicrobial coatings or materials that hinder bacterial growth in healthcare facilities, installing advanced filtration systems to remove antibiotic residues from healthcare facility wastewater, or designing specialized ventilation systems to minimize the transmission of resistant pathogens. These approaches aim to decline the dissemination of resistant bacteria and prevent the development of new strains that are resistant to different drugs.142 To avert the emergence and dissemination of AMR, hospitals and medical centers wastewaters should be treated by advanced methods like hybrid and multistage treatments, including supercritical water oxidation. Mainly on-site treatment, reduce ARGs and active antibiotic reaching the surrounding wastewater.143
Ensuring sanity and hygiene in human residential areas is essential for preventing the spread of infectious diseases and reducing the need for antimicrobial treatment. Proper waste management, clean water supply and sanitation, promoting good personal hygiene practices, disease surveillance and control, and disease-carrying vectors, were some of the measures involved in public hygiene and sanity. By adopting and promoting these practices, we can contribute to global efforts in combating antimicrobial resistance and safeguarding public health.144 The main environmental health practices that help prevent AMR include obtaining hygienic water, sanitation and hygiene (WASH); proper rid of, treatment and discarding of human excreta; practicing good personal hygiene like washing hands with soap at crucial times; correct disposal of solid waste, including unused and expired antimicrobials and implementing proper food hygiene measures.144
Improving Community Health and Using Alternatives to Antimicrobials
Promoting the development and utilization of vaccines and alternative treatments against antibiotic-resistant bacteria is crucial in reducing the number of patients infected with serious antibiotic-resistant infections. By investing in these advancements, such as phage therapy, probiotics, antibodies, and lysins, we can decrease the reliance on traditional antimicrobial treatments. This approach will not only help combat antibiotic resistance but also limit the number of patients requiring antimicrobial therapies.16
Vaccines play a pivotal role in reducing AMR, although they are overlooked in this context. However, their positive impact in combating AMR has been widely proven. Many vaccines not only provide direct protection to the vaccinated individuals but also contribute to herd immunity, which safeguards unvaccinated individuals and those who cannot be vaccinated within a population. This approach significantly minimizes disease transmission in the overall population. By preventing diseases through vaccination, the reliance on antibiotics decreases, leading to a reduction in AMR.145 Contrary to expectations, viral vaccines help decrease antibacterial resistance. For instance, influenza vaccines not only prevent the flu but also reduce the risk of secondary bacterial infections such as pneumonia or ear infections. This demonstrates the significance of recognizing the positive effects of vaccines in combating worldwide antimicrobial resistance (AMR). Encouraging scientists to create vaccines that may not yield financial profits but are valuable in addressing AMR is essential for public welfare and effectively combating the issue.146
One approach is the use of probiotics and prebiotics, which can promote the growth of normal microflora in the gut. This can be addressed by using probiotics, microorganisms found in certain foods and supplements that offer health benefits when consumed in adequate amounts. They help prevent antimicrobial resistance by maintaining a healthy balance of gut microflora and impeding the growth of harmful bacteria that may develop antibiotic resistance. Probiotics also boost the immune response, reducing the risk of infections and potentially decreasing the reliance on antibiotics.147 Prebiotics, on the other hand, are plant fibers that nourish beneficial bacteria already present in the gut and contribute to healthy gut microbiota. Prebiotics indirectly aid in preventing AMR by increasing the abundance of beneficial bacteria. Combining the use of probiotics and prebiotics, known as synbiotics, can have a synergistic effect in preventing antimicrobial resistance.148
Antimicrobial peptides (AMPs), naturally synthesized by organisms, including microbes, to defend against pathogens, have demonstrated their effectiveness in combating microbial pathogens. These peptides possess the ability to eradicate a broad spectrum of bacteria and offer a potential alternative to traditional antibiotics.149 Small molecules, non-peptide organic molecules that are synthetic or obtained from natural product extracts also have drug-like properties that can interact with biological molecules, including protein and nucleic acids, and can alter their normal functions. By doing so, they treat microbial infections as antimicrobials.150 They have a wide array of activity, minimal toxicity, and the ability to disrupt bacterial membranes in unique ways. This makes them promising candidates to combat AMR and encourages their widespread application.149
Other alternatives include phage therapy, immunotherapy, use of plant extracts like essential oils. Phage therapy is a type of treatment that utilizes bacteriophages, which are viruses that specifically target and eliminate bacteria. This alternative approach has been utilized in certain countries for many years and it holds promising potential as a substitute for antibiotics. Another potential alternative to antibiotics is immunotherapy, where the body’s immune system is harnessed to combat infections. By enhancing the body’s natural defense mechanisms, immunotherapy can potentially offer an alternative solution to antibiotic treatment. Additionally, essential oils have been found to possess antimicrobial properties. This discovery suggests that they could also be explored as an alternative to traditional antibiotics in the future.151
Improving Food Safety
The transmission of antibiotic-resistant bacteria from food-producing animals to humans through various means such as direct contact, handling, or consuming their products is a significant threat to public health. To combat AMR, it is crucial to improve food safety measures. Since food safety is closely linked to overall health, by enhancing the safety of our food, we can effectively diminish AMR spread and encourage responsible drug use.152 Addressing AMR requires a comprehensive approach involving multiple stakeholders such as governments, healthcare providers, veterinarians, farmers, and the general public. The monitoring of antimicrobial residues in animal food at various levels, including farms, slaughterhouses, and the food industry. This sets maximum residue limits for medicines used in food animals, ensuring the safety of the final products consumed by humans.153
Growing vegetables on unpolluted soil is a proactive approach to prevent antimicrobial resistance (AMR). By cultivating crops in soil that is free from pollutants and harmful substances, we can curtail the risk of contamination and the proliferation of resistant bacteria in the edible tissues of crops.144 Avoiding using untreated animal manure as fertilizer for crops also aids in the prevention of AMR. This is because plants can absorb and store antimicrobial-resistant microbes and genes in their tissues, potentially transmitting them to animals through the food chain. By taking this precautionary step, we can minimize the risk of exposing both humans and animals to antimicrobial resistance microbes and genes.154
Governmental Policies on the Dispensing and Usage of Antimicrobials
The absence of proper regulations regarding the sale of antimicrobials contributes to the problem of accessibility and misuse. Many developing countries allow over-The-counter sales of these drugs without requiring prescriptions, facilitating the emergence of antimicrobial-resistant bacteria. To address this issue, governments should establish regulations for the responsible use of drugs for human and animal healthcare. It is crucial for them to monitor prescription practices, sales, and distribution of antibiotics to ensure their responsible usage.155 To address the problem of over-The-counter antibiotic dispensing and unnecessary prescriptions, governments need to enforce policies and regulations. Alongside this, emphasis should be placed on expanding the workforce of professionals specialized in infectious diseases, including communicable disease experts, pharmacists, microbiologists, epidemiologists, nurses, veterinarians and infection control experts. The government needs to allocate resources for training these individuals to effectively combat AMR.16
Limiting over-The-counter sales of antibiotics and making them available only with a prescription can significantly decrease the misuse of drugs and the development of AMR. To achieve the best results, it is important to involve all stakeholders and consistently reinforce the message through comprehensive interventions. Evaluating the policies during implementation is crucial to measure their effectiveness, identify areas of improvement, and enhance the intervention strategies. Studying the impact of policies can be done through rigorous methods like interrupted time series analyses and mystery client surveys to track their effectiveness.156 Recent predictions indicate that antimicrobial resistance may impede the attainment of sustainable development goals due to its detrimental effects on human health, social well-being, and economic progress. To combat this issue, a collective and coordinated response involving multiple sectors is imperative. Governments have implemented various policies to combat the excessive and inappropriate use of antimicrobials, to reduce AMR.157
International cooperation, that is Governments collaborate with other countries, organizations, and stakeholders working together to tackle antimicrobial resistance (AMR) globally. This collaboration includes sharing information, exchanging successful approaches, and coordinating efforts to prevent the global transmission of drug-resistant infections. Health system strengthening involves governments’ efforts to develop robust healthcare systems that prioritize infection prevention and control by promoting good hygiene practices and ensuring proper sanitation standards.158
Preventive Measures for Animal-Related Causes of the Spread of AMR
Antimicrobial Stewardship and Community Awareness Campaign
Antimicrobial stewardship programs in animal husbandry effectively reduce AMR while maintaining productivity. It is crucial to monitor AMR in livestock and study the AMR patterns of specific bacterial strains to develop policies promoting the responsible use of antimicrobials in animals.159 Standardizing the testing methods for AMR and establishing consistent interpretation criteria in veterinary medicine is crucial. This will support the use of antimicrobials in animals by following scientific indications and promoting responsible antimicrobial use (AMU).160
Recent studies have emphasized the importance of including susceptibility testing for bacteria, which are typically not assessed for AMR profiling, in AMR surveillance systems. This should be done while taking into account the epidemiological and microbiological data relevant to particular countries or animal production sectors like fish farming, for a comprehensive understanding of AMR patterns.161 To prevent the irresponsible utilization and spread of AMR in agriculture, particularly in livestock and aquaculture, it is crucial to reduce their usage. This includes banning the practice of antimicrobials as growth enhancers for farm animals in countries where such regulations are not already in place. By doing so, we can safeguard the environment and address the global concern of antimicrobial resistance.16
To effectively tackle the global challenge of antibiotic resistance (AMR) and ensure food safety, it is crucial to create awareness at a national level. Involving the entire community, including stakeholders like veterinarians, livestock producers, and consumers, is crucial. By actively engaging these stakeholders, the campaign can effectively promote preventive measures and address the spread of AMR from production to consumption.162 Promoting awareness about the importance of reducing antibiotic usage and avoiding food contamination at every stage, from production to consumption, is crucial in reducing the spread of AMR. This can be attained through implementing strong regulations, offering incentives for compliance, and nurturing a supportive social structure. These can viably bring almost a discernible change in behavior regarding antimicrobial use.163 Regulating the use of antibiotics in legislation and optimizing their usage is crucial to controlling the emergence and spread of AMR. The World Organisation for Animal Health (OIE) suggests strengthening veterinary legislation and enforcement policies to ensure adherence to regulations encouraging judicious and responsible utilization of critically important, highly important, and important antimicrobial agents in veterinary medicine.164
Cohesive Surveillance Systems and Risk Identification in Hotspot Areas
To effectively tackle the public health hazard of AMR, assessing the extent of resistance in various stages of the food chain, like farms, abattoirs, markets, and suppliers is paramount. This is particularly important in low-income countries with limited information about AMR, antibiotic usage, and foodborne diseases. Surveillance data can provide valuable insights to decision-makers, enabling the creation of evidence-based policies and proper resource allocation. This is particularly relevant in preventing and containing AMR throughout the food chain.129
Beyond this, Next-generation sequencing (NGS) technologies are increasingly being utilized in the field of antimicrobial resistance (AMR) due to their ability to monitor resistant microorganisms, AMR factors, and mobile genetic elements. NGS can also provide insights into the mechanisms behind AMR and its spread through the food chain.165 NGS technology has greatly improved the ability to track bacterial clones, including those that are resistant to multiple drugs. It also helps in identifying new antibiotic-resistant genes and the genetic elements that carry them, such as plasmids. NGS plays a crucial role in assessing risks and monitoring AMR, while also enhancing our understanding of how AMR spreads between pathogens and commensal bacteria from different sources like the environment, animals, and humans.166
Improving Animal Health and Using Alternatives to Antimicrobials
The current unparalleled increase in global food demands enthused livestock producers to highly depend on the usage of antibiotics to produce large quantities of animal protein at low cost. Improving animal welfare and health has the potential to diminish this over-dependence on antibiotics without affecting productivity and costs.167 To prevent infections in animals, it is recommended to use vaccines and practice good biosecurity. This can help decrease the need for antibiotics and increase productivity. By immunizing animals with vaccines on time and effectively, the occurrence of diseases is reduced, consequently reducing the reliance on antimicrobials for prophylaxis or treatment.168
Implementing biosecurity measures in animal production campaigns effectively reduces the occurrence of diseases among farm animals, leading to a decreased need for antimicrobial usage. Consequently, this helps reduce the presence of antimicrobial-resistant bacteria on farms and minimizes their potential spread to the environment and humans.169 An integrated approach is used to address this issue, which includes practices like good agricultural, hygiene, and veterinary practices, as well as hazard analysis and critical control points (HACCP) procedures. By following these measures and assessing and managing microbiological risks, the need for antibiotics to treat healthy animals can be minimized. In case antimicrobial resistance occurs, effective barriers such as good agricultural and hygiene practices and HACCP-based measures help prevent its transmission, ensuring food safety, security, and public health.170
Development and use of effective and commercially available alternatives such as herbal plant extracts, probiotics, enzymes, antimicrobial peptides, phages, etc., to antibiotics have been applied by animal farmers to antibiotics to reduce antibiotic use in food animals in livestock production campaigns.171 Among them, some herbal plant extracts (such as tannins, saponins, essential oils, etc.) and probiotics are regarded as effective natural alternatives to antibiotics due to their various functions including antimicrobial activity, antioxidant activity, anti-inflammatory, immunomodulation, etc.,172 For instance, it was suggested that garlic oils are highly effective in treating bacterial infections. The study demonstrated that garlic extracts have strong antibacterial properties against various bacteria including enterococcus faecalis, S. aureus, P. aeruginosa, K. pneumonia and E. coli. Additionally, garlic extracts showed positive interactions with antibiotics such as gentamicin and ciprofloxacin. These bioactive compounds found in garlic can be used in animals to enhance gut health and digestive performance, thereby serving as an alternative treatment to improve animal health.173
Studies have shown that the moringa oleifera plant has various beneficial effects such as anti-coccidial and antibacterial activities, enhanced feed intake and efficiency, improved immune system response, and increased carcass production. The powdered leaves of this plant have particularly strong anti-coccidial properties and could potentially be a useful preventive measure against avian coccidiosis in chickens.174 Integrating moringa oleifera plant in poultry production can significantly decrease disease occurrence, boost growth, and provide antioxidants, thus reducing the need for antimicrobials. This instigates, that utilizing medicinal plants in poultry farming can result in healthier animals, increased production, and decreased reliance on antibiotics.175
Another alternative to minimize the use of antimicrobial drugs in animal-origin food and combat antimicrobial resistance is the utilization of antimicrobial peptides (AMPs). These small molecules, which are part of a diverse and plentiful group of biomolecules, exhibit significant biological activity against bacteria, viruses, and fungi, offering promising effects in reducing the presence of harmful microorganisms and drug residues in food.176 AMPs such as cecropins, bacteriocins, and defensins are essential components of the innate immune system found in animals, and play a crucial role in defense mechanisms, resembling the innate immunity seen in animals. These peptides offer potential therapeutic applications as they possess the ability to effectively combat various microorganisms, including drug-resistant fungi, viruses, and bacteria.177
Farm Animal Manure and Farm Wastewater Management
Anaerobic digestion (AD) is a cost-effective and efficient method of treating manure that effectively reduces pathogens. It has the added benefit of degrading antibiotics present in manure, thereby reducing their concentration and potentially decreasing the proliferation of antibiotic-resistant bacteria.178 In addition to this, it significantly diminish the presence of antimicrobial-resistance gene in manure, by up to 90%. The higher temperatures involved in anaerobic digestion can further enhance its effectiveness in reducing antibiotic-resistant bacteria, making thermophilic anaerobic digestion more efficient in this regard compared to mesophilic anaerobic digestion.179
Composting is a manure treatment method that reduces active drugs in dung through temperature-based non-biological methods like adsorption and degradation, rather than biotic methods. It can effectively decrease populations of antibiotic-resistant bacteria and ARG compared to other methods like anaerobic digestion or stockpiling, but this effect depends on the type of manure and the duration of composting. Throughout composting the maximum half-lives of major antibiotics are up to 16 days, but this can sometimes take weeks depending on the specific antibiotics and their interactions with one another.180 For instance, an investigation by181 found that composting had a significant impact on reducing the presence of enteric bacteria in litter from broiler chickens. This reduction included bacteria carrying antibiotic resistance. The study also revealed that most of the targeted genetic markers decreased after composting, except for sul1, inl1, incw, and erm(f), which remained stable.
Preventive Measures of Environmental and Agricultural Causes of the Spread of AMR
Proper waste management in households, public areas, and livestock farms is crucial in preventing AMR spread by reducing the release of unused antimicrobials and residue and resistant microbes to the environment. It is equally important to ensure the proper treatment and disposal of waste from pharmaceutical industries, leftover and expired antimicrobials, as well as disinfection and sanitation chemicals. Failing to do so can create subinhibitory concentrations and selection pressure, promoting the development of AMR in microbes.81 Properly treating wastewater is crucial to prevent the environmental dissemination of AMR. Wastewater serves as a significant source and breeding ground for both antimicrobials and clinically significant antimicrobial-resistant bacteria.107
Studies have shown that reintroducing indigenous microbes to agricultural soils can help decrease and eliminate antimicrobial-resistant microbes and ARGS in the agricultural environment. This approach has gained attention as a potential solution for combating AMR and ARGS in farming. A recent study by182 revealed that reintroducing native microorganisms into agricultural soils can help increase soil microbial biodiversity and potentially address the issue of ARGS. The authors suggested that the presence of various indigenous microorganisms in the soil was associated with a decrease in the proliferation and selection of ARGs. The basic mechanisms include interfering with the available resources, antibiotic degradation, producing natural antimicrobials (can inhibit the growth of bacteria resistant to the antimicrobial) and limitation of genetic exchange (reducing encountering probability with genetically compatible bacteria).183
Preventive Measures for Wildlife and Insect-Related Causes of the Spread of AMR
The key strategy to prevent the spread of AMR in wildlife is minimizing the antimicrobial resistant microbes and ARGs transmission from animals and humans to an environment accessible by wildlife. These sources are the primary contributors to AMR in wildlife.110 In addition to this, the spread of AMR or ARGs between humans, animals, and wildlife, can be avoided by focusing on two key aspects. Firstly, ensuring the proper and safe disposal of dead animals is crucial. Secondly, efforts should be made to expand the separation between wildlife and livestock, as well as humans, to minimize interactions that could lead to the transmission of AMR or ARGs.127
Challenges and Strategies for Addressing AMR in Remote Areas and Low and Middle-Income Countries
Challenges for Addressing AMR in Remote Areas and Low and Middle-Income Countries
AMR is a worldwide problem that affects developed as well as developing countries. However, remote regions and low as well as middle-income countries face specific challenges in addressing AMR due to limited resources, poor infrastructure, and inadequate surveillance systems.184 The fundamental sources of AMR in these regions are complicated and associated with the practices of medical staff, patients’ attitudes towards antibiotic use, and the distribution of antibiotics to the general public. These causes could include ineffective prescribing practices, poor patient education, a lack of adequate diagnostic resources, the dispensing of drugs without a prescription, lack of functional drug regulatory systems, and the usage of antibiotics for purposes other than treating humans, such as animal husbandry.185
Limited access to healthcare services: Remote areas and developing countries often lack adequate healthcare infrastructure and resources, including diagnostic facilities and skilled healthcare professionals. This hampers the timely diagnosis and appropriate management of infections, leading to the overuse and misuse of antibiotics.186 Addressing this challenge requires improving access to healthcare services, including strengthening healthcare systems, training healthcare workers, and establishing diagnostic capabilities in remote areas through telemedicine or mobile health initiatives.187
Inadequate awareness and education: Lack of awareness about AMR and appropriate antibiotic use is prevalent in many remote areas and developing countries. Public education campaigns are crucial to promote understanding and behavior change.188 These campaigns can focus on raising understanding about the shortcomings of AMR, promoting responsible antibiotic use, and emphasizing the importance of hygiene practices. Utilizing community health workers and local leaders can be effective in disseminating information and ensuring its understanding and acceptance within these communities.189
Limited surveillance systems: Surveillance systems for monitoring AMR and antibiotic consumption are often underdeveloped or nonexistent in remote areas and developing countries. Establishing robust surveillance systems is foremost for understanding the local epidemiology of AMR, identifying emerging resistance patterns, and guiding appropriate antibiotic prescribing practices. Strengthening laboratory capacities, implementing antimicrobial stewardship programs, and encouraging data sharing and collaboration at national and international levels are key strategies in this regard.184,190
Socio-economic factors: Socio-economic factors, such as poverty, inadequate sanitation, and overcrowding are involved in the spread of AMR in remote areas and developing countries. These factors can lead to increased transmission of infections and limited access to clean water and sanitation facilities, which are essential for infection prevention and control.191 Addressing these challenges requires multi-sectoral collaborations involving healthcare, sanitation, agriculture, and education sectors to improve living conditions, promote hygiene practices, and ensure access to clean water and sanitation facilities.192
Weak regulatory frameworks: Developing countries face challenges in implementing and enforcing regulations on antibiotic use, quality, and availability due to limited resources, weak governance, corruption, informal healthcare providers, nonprescription sales, and difficulties in ensuring the quality of drugs.155 Inadequate staffing, training, and infrastructure hinder effective regulation and monitoring, while weak governance and corruption undermine implementation. Informal providers may not follow guidelines, leading to inappropriate antibiotic use, and unregulated over-The-counter sales contribute to misuse. Weak regulatory systems struggle to control substandard or counterfeit antibiotics, exacerbating the problem of antimicrobial resistance.193
Strategies for Addressing AMR in Remote Areas and Low and Middle-Income Countries
Addressing AMR in remote regions or developing countries requires a multi-disciplinary approach involving a wide range of partners, including governments, regulatory agencies, healthcare workers, researchers, and the general public. The One Health approach is significant in this respect because it brings together numerous segments and stakeholders to cooperate within the plan and execution of programs, approaches, enactment, and inquiries to achieve improved public health results.155
The conceivable methodologies helping to address AMR in further and creating country incorporates, conducting a worldwide public awareness campaign to teach the open approximately the harm caused by AMR and energize best hones among the common populace, healthcare specialists, and policymakers to check the advancement and spread of drug-resistant contaminations.16 The other one is establishing guidelines for AMR surveillance to ensure all countries implement integrated surveillance, which will cover the use and consumption of antimicrobials in human and animal populations. These guidelines will provide a clear understanding of how AMR spreads in different settings and specific areas.16
Building a solid research facility foundation, which helps create up-to-date data on the AMR design inside populaces and empowers doctors to form informed decisions on the resistance or vulnerability pattern of the pathogen isolated from a patient is among the key instruments to address AMR. Creating an integrated approach including a wide run of accomplices to constrain the hazard of AMR and minimize its impact on human and animal wellbeing, particularly in developing countries. The involvement of government regulatory organizations is vital in this respect.184
Encouraging commitments that focus on at least one of the five commitment areas: tracking and data, infection prevention and control, antimicrobial stewardship, research and development, and improving international collaboration and capacities. The One Health approach integrates various and decisive sectors and stakeholders involved in public, terrestrial and aquatic animal and plant and environmental health as well as food and feed production to collaborate in designing and implementing the programs, strategies, legislation, and research to come up with better public health.194 Collaborative work with worldwide organizations, such as the World Health Organization (WHO), the Food and Agriculture Organization (FAO), and the World Organization for Animal Health (OIE), to create and actualize programs, arrangements, and enactment to address AMR.195
Conclusion and Recommendations
The factors within the one health framework, including the public, animals and the milieu, take part in hastening the spread of antimicrobial resistance. To efficiently confront this issue, one health approach is essential. It requires a comprehensive strategy involving governments, healthcare professionals, veterinarians, farmers, environmentalists, and the general population. By preventing and controlling resistance in hotspot areas, both regionally and globally, the threat of antimicrobial resistance can be reduced. Integrated involvement of different stakeholders, including human health professionals, veterinarians, environmentalists, those in the food chain, and the whole public is necessary. Additionally, creating awareness among the general population, particularly those in remote areas of developing countries, about the causes and risks of antimicrobial resistance and how to combat it, is vital in preventing its spread.
Abbreviations
AMR-antimicrobial resistance, AMRM-antimicrobial resistant microbes, ARGs-antimicrobial resistance genes, MGEs-mobile genetic elements, MDR-multidrug resistance, MRSA-methicillin resistant staphylococcus aureus, CFU -colony-forming unit, MGEs-mobile genetic elements, spp-species.
Acknowledgment
The authors acknowledge Wolaita Sodo University.
Author Contributions
All authors have made a significant contribution to this manuscript, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This work was not supported by any funding source or institution.
Disclosure
All authors declared that there is no conflict of interest upon publication of this manuscript.
References
1. WHO. Antimicrobial Resistance; 2021.
2. D’Costa VM, King CE, Kalan L, et al. Antibiotic resistance is ancient. Nature. 2011;477(7365):457–461. doi:10.1038/nature10388
3. Baker S, Thomson N, Weill F-X, et al. Genomic insights into the emergence and spread of antimicrobial-resistant bacterial pathogens. Science. 2018;360(6390):733–738. doi:10.1126/science.aar3777
4. Andersson DI, Balaban NQ, Baquero F, et al. Antibiotic resistance: turning evolutionary principles into clinical reality. FEMS Microbiol Rev. 2020;44(2):171–188. doi:10.1093/femsre/fuaa001
5. Luepke KH, Suda KJ, Boucher H, et al. Past, present, and future of antibacterial economics: increasing bacterial resistance, limited antibiotic pipeline, and societal implications. Pharmacotherapy. 2017;37(1):71–84. doi:10.1002/phar.1868
6. Jensen US, Muller A, Brandt CT, et al. Effect of generics on price and consumption of ciprofloxacin in primary healthcare: the relationship to increasing resistance. J Antimicrob Chemother. 2010;65(6):1286–1291. doi:10.1093/jac/dkq093
7. WHO, Interagency Coordination Group on Antimicrobial Resistance. No time to wait: securing the future from drug-resistant infections, Report to the secretary-general of the united nations; 2019. 1: p. 28.
8. Murray CJ, Ikuta KS, Sharara F, et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399(10325):629–655. doi:10.1016/S0140-6736(21)02724-0
9. Tacconelli E, Carrara E, Savoldi A, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect Dis. 2018;18(3):318–327. doi:10.1016/S1473-3099(17)30753-3
10. Khan ZA, Siddiqui MF, Park S. Current and emerging methods of antibiotic susceptibility testing. Diagnostics. 2019;9(2):49. doi:10.3390/diagnostics9020049
11. Wernli D, Jørgensen PS, Morel CM, et al. Mapping global policy discourse on antimicrobial resistance. BMJ Glob Health. 2017;2(2):e000378. doi:10.1136/bmjgh-2017-000378
12. Hwengwere K, Paramel Nair H, Hughes KA, et al. Antimicrobial resistance in Antarctica: is it still a pristine environment? Microbiome. 2022;10(1):1–13. doi:10.1186/s40168-022-01250-x
13. Peixoto RS, Harkins DM, Nelson KE. Advances in microbiome research for animal health. Ann Rev Animal Biosci. 2021;9(1):289–311. doi:10.1146/annurev-animal-091020-075907
14. Baquero F, Coque TM, Martínez J-L, et al. Gene transmission in the one health microbiosphere and the channels of antimicrobial resistance. Front Microbiol. 2019;10:2892. doi:10.3389/fmicb.2019.02892
15. Rillig MC, Mansour I. Microbial ecology: community coalescence stirs things up. Curr Biol. 2017;27(23):R1280–R1282. doi:10.1016/j.cub.2017.10.027
16. Velazquez ME, Galarde-López M, Carrillo-Quiróz B, et al. Antimicrobial resistance: one Health approach. Vet World. 2022;15(3):743. doi:10.14202/vetworld.2022.743-749
17. Karakonstantis S, Kalemaki D. Antimicrobial overuse and misuse in the community in Greece and link to antimicrobial resistance using methicillin-resistant S. aureus as an example. J Infect Public Health. 2019;12(4):460–464. doi:10.1016/j.jiph.2019.03.017
18. Mancuso G, Midiri A, Gerace E, et al. Bacterial antibiotic resistance: the most critical pathogens. Pathogens. 2021;10(10):1310. doi:10.3390/pathogens10101310
19. Manohar P, Loh B, Leptihn S. Will the overuse of antibiotics during the Coronavirus pandemic accelerate antimicrobial resistance of bacteria? Infect Microbes Dis. 2020;2(3):87. doi:10.1097/IM9.0000000000000034
20. Cantón R, Horcajada JP, Oliver A, et al. Inappropriate use of antibiotics in hospitals: the complex relationship between antibiotic use and antimicrobial resistance. Enferm Infecc Microbiol Clin. 2013;31:3–11. doi:10.1016/S0213-005X(13)70126-5
21. Sulis G, Daniels B, Kwan A, et al. Antibiotic overuse in the primary health care setting: a secondary data analysis of standardised patient studies from India, China and Kenya. BMJ Glob Health. 2020;5(9):e003393. doi:10.1136/bmjgh-2020-003393
22. Naveed M. Antibiotics resistance mechanism. In: Antibiotics and Antimicrobial Resistance Genes in the Environment. Elsevier; 2020:292–312.
23. Tao S. The spread of antibiotic resistance genes in vivo model. Can J Infect Dis Med Microbiol. 2022;2022. doi:10.1155/2022/3348695
24. Laxminarayan R, Matsoso P, Pant S, et al. Access to effective antimicrobials: a worldwide challenge. Lancet. 2016;387(10014):168–175. doi:10.1016/S0140-6736(15)00474-2
25. Smyk JM, Szydłowska N, Szulc W, et al. Evolution of Influenza Viruses—Drug Resistance, Treatment Options, and Prospects. Int J Mol Sci. 2022;23(20):12244. doi:10.3390/ijms232012244
26. Van Duin D, Paterson DL. Multidrug-resistant bacteria in the community: trends and lessons learned. Infect Dis Clin. 2016;30(2):377–390. doi:10.1016/j.idc.2016.02.004
27. Subramaniam G, Girish M. Antibiotic resistance—A cause for reemergence of infections. Indian J Pediatr. 2020;87(11):937–944. doi:10.1007/s12098-019-03180-3
28. Vinoth R, Kumar RS, Venkateswaramurthy N. Misuse of Antibiotic during COVID 19 Outbreaks. J Drug Deliv Ther. 2021;11(6–S):181–187. doi:10.22270/jddt.v11i6-S.5102
29. WHO. Antimicrobial Resistance: Global Report on Surveillance. Geneva: World Health Organization; 2015.
30. Nepal G, Bhatta S. Self-medication with antibiotics in WHO Southeast Asian Region: a systematic review. Cureus. 2018;10(4). doi:10.7759/cureus.2428
31. Rappuoli R, Young P, Ron E, et al. Save the microbes to save the planet. A call to action of the International Union of the Microbiological Societies (IUMS). One Health Outlook. 2023;5(1):1–5. doi:10.1186/s42522-023-00077-2
32. Iwu CD, Korsten L, Okoh AI. The incidence of antibiotic resistance within and beyond the agricultural ecosystem: a concern for public health. Microbiologyopen. 2020;9(9):e1035. doi:10.1002/mbo3.1035
33. Ventola CL. The antibiotic resistance crisis: part 1: causes and threats. Pharm Ther. 2015;40(4):277.
34. Karkman A, Pärnänen K, Larsson DJ. Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments. Nat Commun. 2019;10(1):80. doi:10.1038/s41467-018-07992-3
35. Stanton IC, Bethel A, Leonard AFC, et al. Existing evidence on antibiotic resistance exposure and transmission to humans from the environment: a systematic map. Environ Evid. 2022;11(1):1–24. doi:10.1186/s13750-022-00262-2
36. Almagor J, Temkin E, Benenson I, et al. The impact of antibiotic use on transmission of resistant bacteria in hospitals: insights from an agent-based model. PLoS One. 2018;13(5):e0197111. doi:10.1371/journal.pone.0197111
37. Kumar M, Sarma DK, Shubham S, et al. Futuristic non-antibiotic therapies to combat antibiotic resistance: a review. Front Microbiol. 2021;12:609459. doi:10.3389/fmicb.2021.609459
38. Chia PY, Sengupta S, Kukreja A, et al. The role of hospital environment in transmissions of multidrug-resistant gram-negative organisms. Antimicrob Resist Infect Control. 2020;9(1):1–11. doi:10.1186/s13756-020-0685-1
39. Smith RP, May HE, AbuOun M, et al. A longitudinal study reveals persistence of antimicrobial resistance on livestock farms is not due to antimicrobial usage alone. Front Microbiol. 2023;14:700. doi:10.3389/fmicb.2023.1070340
40. Cave R, Cole J, Mkrtchyan HV. Surveillance and prevalence of antimicrobial resistant bacteria from public settings within urban built environments: challenges and opportunities for hygiene and infection control. Environ Int. 2021;157:106836. doi:10.1016/j.envint.2021.106836
41. Huang L, Liu C, Li Z, et al. Characteristics of Virulent ST5-SCC mec II Methicillin-Resistant Staphylococcus aureus Prevalent in a Surgery Ward. Infect Drug Resist. 2023;Volume 16:3487–3495. doi:10.2147/IDR.S410330
42. Kraemer SA, Ramachandran A, Perron GG. Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms. 2019;7(6):180. doi:10.3390/microorganisms7060180
43. He Y, Yuan Q, Mathieu J, et al. Antibiotic resistance genes from livestock waste: occurrence, dissemination, and treatment. NPJ Clean Water. 2020;3(1):4. doi:10.1038/s41545-020-0051-0
44. Dafale NA, Srivastava S, Purohit HJ. Zoonosis: an emerging link to antibiotic resistance under “one health approach”. Indian J Microbiol. 2020;60:139–152. doi:10.1007/s12088-020-00860-z
45. D’Accolti M, Soffritti I, Mazzacane S, et al. Fighting AMR in the healthcare environment: microbiome-based sanitation approaches and monitoring tools. Int J Mol Sci. 2019;20(7):1535. doi:10.3390/ijms20071535
46. Hooban B, Fitzhenry K, Cahill N, et al. A point prevalence survey of antibiotic resistance in the Irish environment, 2018–2019. Environ Int. 2021;152:106466. doi:10.1016/j.envint.2021.106466
47. Ma L, Yang H, Guan L, et al. Risks of antibiotic resistance genes and antimicrobial resistance under chlorination disinfection with public health concerns. Environ Int. 2022;158:106978. doi:10.1016/j.envint.2021.106978
48. Atta H, Idris SM, Gulumbe BH, et al. Detection of extended spectrum beta-lactamase genes in strains of Escherichia coli and Klebsiella pneumoniae isolated from recreational water and tertiary hospital waste water in Zaria, Nigeria. Int J Environ Health Res. 2022;32(9):2074–2082. doi:10.1080/09603123.2021.1940884
49. Lin J, Peng Y, Ou QT, et al. A molecular epidemiological study of methicillin-resistant Staphylococci environmental contamination in railway stations and coach stations in Guangzhou of China. Lett Appl Microbiol. 2017;64(2):131–137. doi:10.1111/lam.12700
50. Li X, Qiu Y, Yu A, et al. Characteristics of airborne Staphylococcus aureus (including MRSA) in Chinese public buildings. Aerobiologia. 2015;31(1):11–19. doi:10.1007/s10453-014-9342-6
51. Sivaraman G, Muneeb KH, Sudha S, et al. Fish-borne methicillin resistant Staphylococcus haemolyticus carrying atypical staphylococcal cassette chromosome mec (SCCmec) elements. Gene Rep. 2021;22:100982. doi:10.1016/j.genrep.2020.100982
52. Wang Y, Li G, Zhu Q, et al. Occurrence of parabens, triclosan and triclocarban in paired human urine and indoor dust from two typical cities in China and its implications for human exposure. Sci Tot Environ. 2021;786:147485. doi:10.1016/j.scitotenv.2021.147485
53. Liao C, Shi J, Wang X, et al. Occurrence and distribution of parabens and bisphenols in sediment from northern Chinese coastal areas. Environ Pollut. 2019;253:759–767. doi:10.1016/j.envpol.2019.07.076
54. Wang Z, Gao J, Wang S, et al. Triclocarban shifted the microbial communities and promoted the spread of antibiotic resistance genes in nitrifying granular sludge system. Bioresour Technol. 2022;347:126429. doi:10.1016/j.biortech.2021.126429
55. Exner M, Bhattacharya S, Gebel J, et al. Chemical disinfection in healthcare settings: critical aspects for the development of global strategies. GMS Hyg Infect Control. 2020;15. doi:10.3205/dgkh000371
56. Chawla J, Shrivastav A. Antimicrobial Tolerance in Biofilms, in Application of Biofilms in Applied Microbiology. Elsevier; 2022:235–255.
57. Elekhnawy E, Sonbol F, Abdelaziz A, et al. Potential impact of biocide adaptation on selection of antibiotic resistance in bacterial isolates. Future J Pharm Sci. 2020;6(1):1–10. doi:10.1186/s43094-020-00119-w
58. Iniguez MM, Avila-Novoa MG, Gutierrez-Lomeli M. Resistance of pathogenic and spoilage microorganisms to disinfectants in the presence of organic matter and their residual effect on stainless steel and polypropylene. J Glob Antimicrob Resist. 2018;14:197–201. doi:10.1016/j.jgar.2018.04.010
59. Merchel B, Wang X, Tagkopoulos I. Biocide-induced emergence of antibiotic resistance in Escherichia coli. Front Microbiol. 2021;12:640923. doi:10.3389/fmicb.2021.640923
60. Mahnert A, Moissl-Eichinger C, Zojer M, et al. Man-made microbial resistances in built environments. Nat Commun. 2019;10(1):968. doi:10.1038/s41467-019-08864-0
61. Pokharel S, Shrestha P, Adhikari B. Antimicrobial use in food animals and human health: time to implement ‘One Health’approach. Antimicrob Resist Infect Control. 2020;9:1–5. doi:10.1186/s13756-020-00847-x
62. Koch N, Islam NF, Sonowal S, et al. Environmental antibiotics and resistance genes as emerging contaminants: methods of detection and bioremediation. Curr Res Microbial Sci. 2021;2:100027. doi:10.1016/j.crmicr.2021.100027
63. Liu Z, Wang K, Zhang Y, et al. High prevalence and diversity characteristics of blaNDM, mcr, and blaESBLs harboring multidrug-resistant Escherichia coli from chicken, pig, and cattle in China. Front Cell Infect Microbiol. 2022;11:1364.
64. Founou LL, Founou RC, Essack SY. Antimicrobial resistance in the farm-to-plate continuum: more than a food safety issue. Future Sci OA. 2021;7(5):FSO692. doi:10.2144/fsoa-2020-0189
65. Jans C, Sarno E, Collineau L, et al. Consumer exposure to antimicrobial resistant bacteria from food at Swiss retail level. Front Microbiol. 2018;9:362. doi:10.3389/fmicb.2018.00362
66. Berman TS, Barnett-Itzhaki Z, Berman T, et al. Antimicrobial resistance in food-producing animals: towards implementing a one health based national action plan in Israel. Isr J Health Policy Res. 2023;12(1):18. doi:10.1186/s13584-023-00562-z
67. Rhouma M, Archambault M, Butaye P. Antimicrobial Use and Resistance in Animals from a One Health Perspective. Vet Sci. 2023;10:319.
68. Nair AB, Jacob S. A simple practice guide for dose conversion between animals and human. J Basic Clin Pharm. 2016;7(2):27. doi:10.4103/0976-0105.177703
69. Papich MG. Antimicrobial agent use in small animals what are the prescribing practices, use of PK‐PD principles, and extralabel use in the United States? J Vet Pharmacol Ther. 2021;44(2):238–249. doi:10.1111/jvp.12921
70. Nhung NT, Cuong N, Thwaites G, et al. Antimicrobial usage and antimicrobial resistance in animal production in Southeast Asia: a review. Antibiotics. 2016;5(4):37. doi:10.3390/antibiotics5040037
71. Kim J, Hwang BK, Choi H, et al. Characterization of mcr-1-harboring plasmids from pan drug-resistant Escherichia coli strains isolated from retail raw chicken in South Korea. Microorganisms. 2019;7(9):344. doi:10.3390/microorganisms7090344
72. Díaz-Jiménez D, García-Meniño I, Fernández J, et al. Chicken and Turkey meat: consumer exposure to multidrug-resistant Enterobacteriaceae including mcr-carriers, uropathogenic E. coli and high-risk lineages such as ST131. Int J Food Microbiol. 2020;331:108750. doi:10.1016/j.ijfoodmicro.2020.108750
73. Deng W, Quan Y, Yang S, et al. Antibiotic Resistance in Salmonella from Retail Foods of Animal Origin and Its Association with Disinfectant and Heavy Metal Resistance. Microb Drug Resist. 2018;24(6):782–791. doi:10.1089/mdr.2017.0127
74. Balemi A, Gumi B, Amenu K, et al. Prevalence of mastitis and antibiotic resistance of bacterial isolates from CMT positive milk samples obtained from dairy cows, camels, and goats in two pastoral districts in Southern Ethiopia. Animals. 2021;11(6):1530. doi:10.3390/ani11061530
75. Prevention, E.C.f.D, et al. ECDC/EFSA/EMA second joint report on the integrated analysis of the consumption of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from humans and food‐producing animals: joint Interagency Antimicrobial Consumption and Resistance Analysis (JIACRA) Report. EFSA J. 2017;15(7):e04872. doi:10.2903/j.efsa.2017.4872
76. Ahmad I, Malak HA, Abulreesh HH. Environmental antimicrobial resistance and its drivers: a potential threat to public health. J Glob Antimicrob Resist. 2021;27:101–111. doi:10.1016/j.jgar.2021.08.001
77. Bacanlı M, Başaran N. Importance of antibiotic residues in animal food. Food Chem Toxicol. 2019;125:462–466. doi:10.1016/j.fct.2019.01.033
78. Canton L, Lanusse C, Moreno L. Rational pharmacotherapy in infectious diseases: issues related to drug residues in edible animal tissues. Animals. 2021;11(10):2878. doi:10.3390/ani11102878
79. Tufa TB, Gurmu F, Beyi AF, et al. Veterinary medicinal product usage among food animal producers and its health implications in Central Ethiopia. BMC Vet Res. 2018;14(1):1–7. doi:10.1186/s12917-018-1737-0
80. Zhang Y, Gu AZ, Cen T, et al. Sub-inhibitory concentrations of heavy metals facilitate the horizontal transfer of plasmid-mediated antibiotic resistance genes in water environment. Environ Pollut. 2018;237:74–82. doi:10.1016/j.envpol.2018.01.032
81. Ding M, Ye Z, Liu L, et al. Subinhibitory antibiotic concentrations promote the horizontal transfer of plasmid-borne resistance genes from Klebsiellae pneumoniae to Escherichia coli. Front Microbiol. 2022;13:1017092. doi:10.3389/fmicb.2022.1017092
82. Shao S, Hu Y, Cheng J, et al. Research progress on distribution, migration, transformation of antibiotics and antibiotic resistance genes (ARGs) in aquatic environment. Crit Rev Biotechnol. 2018;38(8):1195–1208. doi:10.1080/07388551.2018.1471038
83. Checcucci A, Trevisi P, Luise D, et al. Exploring the animal waste resistome: the spread of antimicrobial resistance genes through the use of livestock manure. Front Microbiol. 2020;11:1416. doi:10.3389/fmicb.2020.01416
84. Ljubojević D, Pelić M, Puvača N, et al. Resistance to tetracycline in Escherichia coli isolates from poultry meat: epidemiology, policy and perspective. Worlds Poult Sci J. 2017;73(2):409–417. doi:10.1017/S0043933917000216
85. Mahmoud MA, Abdel-Mohsein HS. Hysterical tetracycline in intensive poultry farms accountable for substantial gene resistance, health and ecological risk in Egypt-manure and fish. Environ Pollut. 2019;255:113039. doi:10.1016/j.envpol.2019.113039
86. Tóth AG, Tóth I, Rózsa B, et al. Canine saliva as a possible source of antimicrobial resistance genes. Antibiotics. 2022;11(11):1490. doi:10.3390/antibiotics11111490
87. Rothrock J, James M, Castleberry L, et al. Antibiotic resistance, antimicrobial residues, and bacterial community diversity in pasture-raised poultry, swine, and beef cattle manures. J Anim Sci. 2021;99(8):kab144. doi:10.1093/jas/skab144
88. Vines J, Cuscó A, Napp S, et al. Transmission of similar mcr-1 carrying plasmids among different Escherichia coli lineages isolated from livestock and the farmer. Antibiotics. 2021;10(3):313. doi:10.3390/antibiotics10030313
89. Aworh MK, Abiodun-Adewusi O, Mba N, et al. Prevalence and risk factors for faecal carriage of multidrug resistant Escherichia coli among slaughterhouse workers. Sci Rep. 2021;11(1):13362. doi:10.1038/s41598-021-92819-3
90. Bai H, He L-Y, Wu D-L, et al. Spread of airborne antibiotic resistance from animal farms to the environment: dispersal pattern and exposure risk. Environ Int. 2022;158:106927. doi:10.1016/j.envint.2021.106927
91. Gwenzi W, Shamsizadeh Z, Gholipour S, et al. The air-borne antibiotic resistome: occurrence, health risks, and future directions. Sci Total Environ. 2022;804:150154. doi:10.1016/j.scitotenv.2021.150154
92. Zhou G, Qiu X, Wu X, et al. Horizontal gene transfer is a key determinant of antibiotic resistance genes profiles during chicken manure composting with the addition of biochar and zeolite. J Hazard Mater. 2021;408:124883. doi:10.1016/j.jhazmat.2020.124883
93. Hernando AS, Coque TM, Baquero F, et al. Defining and combating antibiotic resistance from One Health and Global Health perspectives. Nat Microbiol. 2019;4(9):1432–1442. doi:10.1038/s41564-019-0503-9
94. Freitag C, Michael GB, Li J, et al. Occurrence and characterisation of ESBL-encoding plasmids among Escherichia coli isolates from fresh vegetables. Vet Microbiol. 2018;219:63–69. doi:10.1016/j.vetmic.2018.03.028
95. Tian M, He X, Feng Y, et al. Pollution by antibiotics and antimicrobial resistance in livestock and poultry manure in China, and countermeasures. Antibiotics. 2021;10(5):539. doi:10.3390/antibiotics10050539
96. Larsson DJ, Flach C-F. Antibiotic resistance in the environment. Nat Rev Microbiol. 2022;20(5):257–269. doi:10.1038/s41579-021-00649-x
97. Smith WP, Wucher BR, Nadell CD, Foster KR. Bacterial defences: mechanisms, evolution and antimicrobial resistance. Nat Rev Microbiol. 2023;21:519–534.
98. Mulder M Antimicrobial Use and Antimicrobial Resistance in Community-Acquired Urinary Tract Infections; 2022.
99. Dame ZT, Rahman M, Islam T. Bacilli as sources of agrobiotechnology: recent advances and future directions. Green Chem Lett Rev. 2021;14(2):246–271. doi:10.1080/17518253.2021.1905080
100. Anwar M, Iqbal Q, Saleem F. Improper disposal of unused antibiotics: an often overlooked driver of antimicrobial resistance. Expert Rev Anti Infect Ther. 2020;18(8):697–699. doi:10.1080/14787210.2020.1754797
101. Wall B, Mateus AL, Marshall L, et al., Drivers, dynamics and epidemiology of antimicrobial resistance in animal production. Food and Agriculture Organization of the United Nations, 2016.
102. Ayele Y, Mamu M. Assessment of knowledge, attitude and practice towards disposal of unused and expired pharmaceuticals among community in Harar city, Eastern Ethiopia. J Pharm Policy Pract. 2018;11:1–7. doi:10.1186/s40545-018-0155-9
103. Bashaar M, et al. Disposal practices of unused and expired pharmaceuticals among general public in Kabul. BMC Public Health. 2017;17:1–8. doi:10.1186/s12889-016-3954-4
104. Stanton IC, Murray AK, Zhang L, et al. Evolution of antibiotic resistance at low antibiotic concentrations including selection below the minimal selective concentration. Commun Biol. 2020;3(1):467. doi:10.1038/s42003-020-01176-w
105. Wang K, Zhuang T, Su Z, et al. Antibiotic residues in wastewaters from sewage treatment plants and pharmaceutical industries: occurrence, removal and environmental impacts. Sci Tot Environ. 2021;788:147811. doi:10.1016/j.scitotenv.2021.147811
106. Madikizela LM, Nuapia YB, Chimuka L, et al. Target and Suspect Screening of Pharmaceuticals and their Transformation Products in the Klip River, South Africa, using Ultra‐High–Performance Liquid Chromatography–Mass Spectrometry. Environ Toxicol Chem. 2022;41(2):437–447. doi:10.1002/etc.5265
107. Ulvi A, Aydın S, Aydın ME. Fate of selected pharmaceuticals in hospital and municipal wastewater effluent: occurrence, removal, and environmental risk assessment. Environ Sci Pollut Res. 2022;29(50):75609–75625. doi:10.1007/s11356-022-21131-y
108. Zalewska M, Błażejewska A, Czapko A, et al. Antibiotics and antibiotic resistance genes in animal manure–consequences of its application in agriculture. Front Microbiol. 2021;12:640. doi:10.3389/fmicb.2021.610656
109. Zhao F, Yang L, Chen L, et al. Bioaccumulation of antibiotics in crops under long-term manure application: occurrence, biomass response and human exposure. Chemosphere. 2019;219:882–895. doi:10.1016/j.chemosphere.2018.12.076
110. Gwenzi W, Musiyiwa K, Mangori L. Sources, behaviour and health risks of antimicrobial resistance genes in wastewaters: a hotspot reservoir. J Environ Chem Eng. 2020;8(1):102220. doi:10.1016/j.jece.2018.02.028
111. Yan Z, Xiong C, Liu H, et al. Sustainable agricultural practices contribute significantly to one health. J Sustain Agric Environ. 2022;1(3):165–176. doi:10.1002/sae2.12019
112. Yang Q, Ren S, Niu T, et al. Distribution of antibiotic-resistant bacteria in chicken manure and manure-fertilized vegetables. Environ Sci Pollut Res. 2014;21(2):1231–1241. doi:10.1007/s11356-013-1994-1
113. Varlet V, Bouvet A, Cadas H, et al. Toward safer thanatopraxy cares: formaldehyde‐releasers use. J Anat. 2019;235(5):863–872. doi:10.1111/joa.13047
114. Paiga P, Delerue C. Determination of pharmaceuticals in groundwater collected in five cemeteries’ areas (Portugal). Sci Tot Environ. 2016;569:16–22. doi:10.1016/j.scitotenv.2016.06.090
115. Silva RBPD, Campos MC, Silva LS, et al. Concentration of heavy metals in soils under cemetery occupation in Amazonas, Brazil. Soil Sediment Contam. 2020;29(2):192–208.
116. Gwenzi W. The ‘thanato-resistome’-The funeral industry as a potential reservoir of antibiotic resistance: early insights and perspectives. Sci Total Environ. 2020;749:141120. doi:10.1016/j.scitotenv.2020.141120
117. Abia ALK, Ubomba-Jaswa E, Schmidt C, et al. Where did they come from—multi-drug resistant pathogenic Escherichia coli in a cemetery environment? Antibiotics. 2018;7(3):73. doi:10.3390/antibiotics7030073
118. Miller SA, Ferreira JP, LeJeune JT. Antimicrobial use and resistance in plant agriculture: a one health perspective. Agriculture. 2022;12(2):289. doi:10.3390/agriculture12020289
119. Ramakrishnan B, Venkateswarlu K, Sethunathan N, et al. Local applications but global implications: can pesticides drive microorganisms to develop antimicrobial resistance? Sci Tot Environ. 2019;654:177–189. doi:10.1016/j.scitotenv.2018.11.041
120. Rangasamy K, Athiappan M, Devarajan N, et al. Pesticide degrading natural multidrug resistance bacterial flora. Microb Pathog. 2018;114:304–310. doi:10.1016/j.micpath.2017.12.013
121. Kurenbach B, Marjoshi D, Amábile-Cuevas CF, et al. Sublethal exposure to commercial formulations of the herbicides dicamba, 2, 4-dichlorophenoxyacetic acid, and glyphosate cause changes in antibiotic susceptibility in Escherichia coli and Salmonella enterica serovar Typhimurium. MBio. 2015;6(2):e00009–15. doi:10.1128/mBio.00009-15
122. Lu J, Jin M, Nguyen SH, et al. Non-antibiotic antimicrobial triclosan induces multiple antibiotic resistance through genetic mutation. Environ Int. 2018;118:257–265. doi:10.1016/j.envint.2018.06.004
123. Zhao Y, Cocerva T, Cox S, et al. Evidence for co-selection of antibiotic resistance genes and mobile genetic elements in metal polluted urban soils. Sci Tot Environ. 2019;656:512–520. doi:10.1016/j.scitotenv.2018.11.372
124. Rangasamy K, Athiappan M, Devarajan N, et al. Emergence of multi drug resistance among soil bacteria exposing to insecticides. Microb Pathog. 2017;105:153–165. doi:10.1016/j.micpath.2017.02.011
125. Zeballos-Gross D, Rojas-Sereno Z, Salgado-Caxito M, et al. The role of gulls as reservoirs of antibiotic resistance in aquatic environments: a scoping review. Front Microbiol. 2021;12:703886. doi:10.3389/fmicb.2021.703886
126. Ruppe E, Ghozlane A, Tap J, et al. Prediction of the intestinal resistome by a three-dimensional structure-based method. Nat Microbiol. 2019;4(1):112–123. doi:10.1038/s41564-018-0292-6
127. Di Lallo G, D’Andrea MM, Sennati S, et al. Evidence of another anthropic impact on Iguana delicatissima from the Lesser Antilles: the presence of antibiotic resistant enterobacteria. Antibiotics. 2021;10(8):885. doi:10.3390/antibiotics10080885
128. Vittecoq M, Godreuil S, Prugnolle F, et al. Antimicrobial resistance in wildlife. J Appl Ecol. 2016;53(2):519–529. doi:10.1111/1365-2664.12596
129. Khan SA, Imtiaz MA, Sayeed MA, et al. Antimicrobial resistance pattern in domestic animal-wildlife-environmental niche via the food chain to humans with a Bangladesh perspective; a systematic review. BMC Vet Res. 2020;16:1–13. doi:10.1186/s12917-020-02519-9
130. Atterby C, Börjesson S, Ny S, et al. ESBL-producing Escherichia coli in Swedish gulls—a case of environmental pollution from humans? PLoS One. 2017;12(12):e0190380. doi:10.1371/journal.pone.0190380
131. Lee S, Fan P, Liu T, et al. Transmission of antibiotic resistance at the wildlife-livestock interface. Commun Biol. 2022;5(1):585. doi:10.1038/s42003-022-03520-8
132. Desvars-Larrive A, Ruppitsch W, Lepuschitz S, et al. Urban brown rats (Rattus norvegicus) as possible source of multidrug-resistant Enterobacteriaceae and meticillin-resistant Staphylococcus spp., Vienna, Austria, 2016 and 2017. Eurosurveillance. 2019;24(32):1900149. doi:10.2807/1560-7917.ES.2019.24.32.1900149
133. Sobur A, Haque ZF, Sabuj AA, et al. Molecular detection of multidrug and colistin-resistant Escherichia coli isolated from house flies in various environmental settings. Future Microbiol. 2019;14(10):847–858. doi:10.2217/fmb-2019-0053
134. Obeng-Nkrumah N, et al. Household cockroaches carry CTX-M-15-, OXA-48-and NDM-1-producing enterobacteria, and share beta-lactam resistance determinants with humans. BMC Microbiol. 2019;19:1–11. doi:10.1186/s12866-018-1372-8
135. Seifert JD. Antimicrobial Stewardship and its Effects on Resistant Organisms. 2022.
136. Majumder MAA, Rahman S, Cohall D, et al. Antimicrobial stewardship: fighting antimicrobial resistance and protecting global public health. Infect Drug Resist. 2020;Volume 13:4713–4738. doi:10.2147/IDR.S290835
137. Donà D, Barbieri E, Daverio M, et al. Implementation and impact of pediatric antimicrobial stewardship programs: a systematic scoping review. Antimicrob Resist Infect Control. 2020;9:1–12.
138. Kiersnowska Z, Lemiech-Mirowska E, Michałkiewicz M, et al. Hand hygiene as the basic method of reducing Clostridium difficile infections (CDI) in a hospital environment. Ann Agri Environ Med. 2021;28(4):535–540. doi:10.26444/aaem/131121
139. Bock L, Wand M, Sutton J. Varying activity of chlorhexidine-based disinfectants against Klebsiella pneumoniae clinical isolates and adapted strains. J Hosp Infect. 2016;93(1):42–48. doi:10.1016/j.jhin.2015.12.019
140. Weber DJ, Kanamori H, Rutala WA. ‘No touch’technologies for environmental decontamination: focus on ultraviolet devices and hydrogen peroxide systems. Curr Opin Infect Dis. 2016;29(4):424–431. doi:10.1097/QCO.0000000000000284
141. Sanganyado E, Gwenzi W. Antibiotic resistance in drinking water systems: occurrence, removal, and human health risks. Sci Tot Environ. 2019;669:785–797. doi:10.1016/j.scitotenv.2019.03.162
142. Bassegoda A, Ivanova K, Ramon E, et al. Strategies to prevent the occurrence of resistance against antibiotics by using advanced materials. Appl Microbiol Biotechnol. 2018;102(5):2075–2089. doi:10.1007/s00253-018-8776-0
143. Paulus GK, Hornstra LM, Alygizakis N, et al. The impact of on-site hospital wastewater treatment on the downstream communal wastewater system in terms of antibiotics and antibiotic resistance genes. Int J Hyg Environ Health. 2019;222(4):635–644. doi:10.1016/j.ijheh.2019.01.004
144. Musoke D, Namata C, Lubega GB, et al. The role of Environmental Health in preventing antimicrobial resistance in low-and middle-income countries. Environ Health Prev Med. 2021;26(1):1–6. doi:10.1186/s12199-021-01023-2
145. Jansen KU, Knirsch C, Anderson AS. The role of vaccines in preventing bacterial antimicrobial resistance. Nat Med. 2018;24(1):10–19. doi:10.1038/nm.4465
146. Jansen KU, Anderson AS. The role of vaccines in fighting antimicrobial resistance (AMR). Hum Vaccin Immunother. 2018;14(9):2142–2149. doi:10.1080/21645515.2018.1476814
147. Palai S, Derecho CM, Kesh SS, et al. Prebiotics, probiotics, synbiotics and its importance in the management of diseases. Func Foods Nutraceut. 2020;173–196.
148. Anadon A, Ares I, Martínez-Larrañaga MR, et al. Prebiotics and probiotics in feed and animal health. Nutraceut Vet Med. 2019;261–285.
149. Yan Y, Li Y, Zhang Z, et al. Advances of peptides for antibacterial applications. Biointerfaces. 2021;202:111682. doi:10.1016/j.colsurfb.2021.111682
150. Helmy YA, Taha-Abdelaziz K, Hawwas HA, et al. Antimicrobial Resistance and Recent Alternatives to Antibiotics for the Control of Bacterial Pathogens with an Emphasis on Foodborne Pathogens. Antibiotics. 2023;12(2):274.
151. Uchil RR, Kohli GS, KateKhaye VM, Swami OC. Strategies to combat antimicrobial resistance. J Clin Diagn Res. 2014;8(7):ME01.
152. Serwecinska L. Antimicrobials and antibiotic-resistant bacteria: a risk to the environment and to public health. Water. 2020;12(12):3313. doi:10.3390/w12123313
153. Florez-Cuadrado D, et al. Antimicrobial resistance in the food chain in the European Union. Adv Food Nutr Res. 2018;86:115–136.
154. Christou A, Agüera A, Bayona JM, et al. The potential implications of reclaimed wastewater reuse for irrigation on the agricultural environment: the knowns and unknowns of the fate of antibiotics and antibiotic resistant bacteria and resistance genes–a review. Water Res. 2017;123:448–467. doi:10.1016/j.watres.2017.07.004
155. Ayukekbong JA, Ntemgwa M, Atabe AN. The threat of antimicrobial resistance in developing countries: causes and control strategies. Antimicrob Resist Infect Control. 2017;6(1):1–8. doi:10.1186/s13756-017-0208-x
156. Jacobs TG, Robertson J, van den Ham HA, et al. Assessing the impact of law enforcement to reduce over-The-counter (OTC) sales of antibiotics in low-and middle-income countries; a systematic literature review. BMC Health Serv Res. 2019;19(1):1–15. doi:10.1186/s12913-019-4359-8
157. Bank W. Drug-Resistant Infections: A Threat to Our Economic Future. World Bank; 2017.
158. WHO. Antimicrobial Resistance Multi-Partner Trust Fund annual report 2021. 2022: Food & Agriculture Org.
159. More SJ. European perspectives on efforts to reduce antimicrobial usage in food animal production. Ir Vet J. 2020;73(1):2. doi:10.1186/s13620-019-0154-4
160. Fernandes V, Cunha E, Nunes T, et al. Antimicrobial Resistance of Clinical and Commensal Escherichia coli Canine Isolates: profile Characterization and Comparison of Antimicrobial Susceptibility Results According to Different Guidelines. Vet Sci. 2022;9(6):284. doi:10.3390/vetsci9060284
161. Mahmoud SF, Fayez M, Swelum AA, et al. Genetic Diversity, Biofilm Formation, and Antibiotic Resistance of Pseudomonas aeruginosa Isolated from Cow, Camel, and Mare with Clinical Endometritis. Vet Sci. 2022;9(5):239. doi:10.3390/vetsci9050239
162. Mazinska B, Struzycka I, Hryniewicz W, Gupta V. Surveys of public knowledge and attitudes with regard to antibiotics in Poland: did the European Antibiotic Awareness Day campaigns change attitudes? PLoS One. 2017;12(2):e0172146. doi:10.1371/journal.pone.0172146
163. Mathew P, Sivaraman S, Chandy S. Communication strategies for improving public awareness on appropriate antibiotic use: bridging a vital gap for action on antibiotic resistance. J Family Med Prim Care. 2019;8(6):1867. doi:10.4103/jfmpc.jfmpc_263_19
164. OIE, W.O.f.A.H. Terrestrial Animal Health Code. Paris: World Organization for Animal Health; 2016.
165. Hygiene DOF, Bergšpica I, Kaprou G, et al. Identification of risk factors and hotspots of antibiotic resistance along the food chain using next‐generation sequencing. EFSA J. 2020;18(Suppl 1):e181107. doi:10.2903/j.efsa.2020.e181107
166. Ruppe E, Bengtsson-Palme J, Charretier Y, Schrenzel J. How next-generation sequencing can address the antimicrobial resistance challenge. AMR Control. 2019;20:60–66.
167. Kasimanickam V, Kasimanickam M, Kasimanickam R. Antibiotics use in food animal production: escalation of antimicrobial resistance: where are we now in combating AMR? Med Sci. 2021;9(1):14. doi:10.3390/medsci9010014
168. Rahman MM, Alam Tumpa MA, Zehravi M, et al. An overview of antimicrobial stewardship optimization: the use of antibiotics in humans and animals to prevent resistance. Antibiotics. 2022;11(5):667. doi:10.3390/antibiotics11050667
169. Huber N, Andraud M, Sassu EL, et al. What is a biosecurity measure? A definition proposal for animal production and linked processing operations. One Health. 2022;15:100433. doi:10.1016/j.onehlt.2022.100433
170. Postma M, Backhans A, Collineau L, et al. Evaluation of the relationship between the biosecurity status, production parameters, herd characteristics and antimicrobial usage in farrow-to-finish pig production in four EU countries. Porcine Health Manag. 2016;2(1):1–11. doi:10.1186/s40813-016-0028-z
171. Callaway TR, Lillehoj H, Chuanchuen R, et al. Alternatives to antibiotics: a symposium on the challenges and solutions for animal health and production. Antibiotics. 2021;10(8):1024. doi:10.3390/antibiotics10050471
172. Farha A, Yang QQ, Kim G, et al. Tannis as an alternative to antibiotics. Food Biosci. 2020;38:100751.
173. Magrys A, Olender A, Tchorzewska D. Antibacterial properties of Allium sativum L. against the most emerging multidrug-resistant bacteria and its synergy with antibiotics. Arch Microbiol. 2021;203(5):2257–2268. doi:10.1007/s00203-021-02248-z
174. Khan RU, Khan A, Naz S, et al. Potential applications of Moringa oleifera in poultry health and production as alternative to antibiotics: a review. Antibiotics. 2021;10(12):1540. doi:10.3390/antibiotics10121540
175. Abdel-Tawab H, Abdel-Haleem HM, Abdel-Baki A-AS, et al. Anticoccidial and antioxidant activities of Moringa oleifera leaf extract on murine intestinal eimeriosis. Acta Parasitol. 2020;65(4):823–830. doi:10.2478/s11686-020-00219-w
176. Magana M, Pushpanathan M, Santos AL, et al. The value of antimicrobial peptides in the age of resistance. Lancet Infect Dis. 2020;20(9):e216–e230. doi:10.1016/S1473-3099(20)30327-3
177. Dicks LM, Dreyer L, Smith C, et al. A review: the fate of bacteriocins in the human gastro-intestinal tract: do they cross the gut–blood barrier? Front Microbiol. 2018;9:2297. doi:10.3389/fmicb.2018.02297
178. Marutescu LG, Jaga M, Postolache C, et al. Insights into the impact of manure on the environmental antibiotic residues and resistance pool. Front Microbiol. 2022;3489.
179. Gurmessa B, Pedretti EF, Cocco S, et al. Manure anaerobic digestion effects and the role of pre-and post-treatments on veterinary antibiotics and antibiotic resistance genes removal efficiency. Sci Tot Environ. 2020;721:137532. doi:10.1016/j.scitotenv.2020.137532
180. Oliver JP, Gooch CA, Lansing S, et al. Invited review: fate of antibiotic residues, antibiotic-resistant bacteria, and antibiotic resistance genes in US dairy manure management systems. J Dairy Sci. 2020;103(2):1051–1071. doi:10.3168/jds.2019-16778
181. Subirats J, Murray R, Scott A, et al. Composting of chicken litter from commercial broiler farms reduces the abundance of viable enteric bacteria, Firmicutes, and selected antibiotic resistance genes. Sci Total Environ. 2020;746:141113. doi:10.1016/j.scitotenv.2020.141113
182. Klumper U, Recker M, Zhang L, et al. Selection for antimicrobial resistance is reduced when embedded in a natural microbial community. ISME J. 2019;13(12):2927–2937. doi:10.1038/s41396-019-0483-z
183. Chen Q-L, An X-L, Zheng B-X, et al. Loss of soil microbial diversity exacerbates spread of antibiotic resistance. Soil Ecol Lett. 2019;1(1–2):3–13. doi:10.1007/s42832-019-0011-0
184. Iskandar K, Molinier L, Hallit S, et al. Surveillance of antimicrobial resistance in low-and middle-income countries: a scattered picture. Antimicrob Resist Infect Control. 2021;10(1):1–19. doi:10.1186/s13756-021-00931-w
185. Sharma A, Singh A, Dar MA, et al. Menace of antimicrobial resistance in LMICs: current surveillance practices and control measures to tackle hostility. J Infect Public Health. 2022;15(2):172–181. doi:10.1016/j.jiph.2021.12.008
186. Jaeger FN, Bechir M, Harouna M, et al. Challenges and opportunities for healthcare workers in a rural district of Chad. BMC Health Serv Res. 2018;18(1):1–11. doi:10.1186/s12913-017-2799-6
187. Loosli K, Davis A, Muwonge A, et al. Addressing antimicrobial resistance by improving access and quality of care—a review of the literature from East Africa. PLoS Negl Trop Dis. 2021;15(7):e0009529. doi:10.1371/journal.pntd.0009529
188. Simegn W, Moges G, Aslam MS. Awareness and knowledge of antimicrobial resistance and factors associated with knowledge among adults in Dessie City, Northeast Ethiopia: community-based cross-sectional study. PLoS One. 2022;17(12):e0279342. doi:10.1371/journal.pone.0279342
189. Chukwu EE, Oladele DA, Enwuru CA, et al. Antimicrobial resistance awareness and antibiotic prescribing behavior among healthcare workers in Nigeria: a national survey. BMC Infect Dis. 2021;21(1):1–12. doi:10.1186/s12879-020-05689-x
190. Gulumbe BH, Haruna UA, Almazan J, et al. Combating the menace of antimicrobial resistance in Africa: a review on stewardship, surveillance and diagnostic strategies. Biol Proced Online. 2022;24(1):1–13. doi:10.1186/s12575-022-00182-y
191. Wozniak TM, Cuningham W, Ledingham K, et al. Contribution of socio-economic factors in the spread of antimicrobial resistant infections in Australian primary healthcare clinics. J Glob Antimicrob Resist. 2022;30:294–301. doi:10.1016/j.jgar.2022.06.005
192. Allel K, García P, Labarca J, et al. Socioeconomic factors associated with antimicrobial resistance of Pseudomonas aeruginosa, Staphylococcus aureus, and Escherichia coli in Chilean hospitals (2008–2017). Rev Panam Salud Publica. 2020;44. doi:10.26633/RPSP.2020.30
193. Mambula G, Nanjebe D, Munene A, et al. Practices and challenges related to antibiotic use in paediatric treatment in hospitals and health centres in Niger and Uganda: a mixed methods study. Antimicrob Resist Infect Control. 2023;12(1):67. doi:10.1186/s13756-023-01271-7
194. Chua AQ, Verma M, Hsu LY, et al. An analysis of national action plans on antimicrobial resistance in Southeast Asia using a governance framework approach. Lancet Reg Health–Western Pac. 2021;7:100084. doi:10.1016/j.lanwpc.2020.100084
195. Gongal G, Ofrin RH, de Balogh K. Operationalization of One Health and tripartite collaboration in the Asia-Pacific region. WHO South-East Asia J Public Health. 2020;9(1):21–25.
196. Laborda P, Sanz-García F, Ochoa-Sánchez LE, et al. Wildlife and antibiotic resistance. Front Cell Infect Microbiol. 2022;12:568.
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


