Back to Journals » Cancer Management and Research » Volume 11
Polymorphisms in DNA repair pathway genes and ABCG2 gene in advanced colorectal cancer: correlation with tumor characteristics and clinical outcome in oxaliplatin-based chemotherapy
Authors Hu X, Qin W, Li S, He M , Wang Y, Guan S , Zhao H, Yao W, Wei M, Liu M, Wu H
Received 30 July 2018
Accepted for publication 9 November 2018
Published 27 December 2018 Volume 2019:11 Pages 285—297
DOI https://doi.org/10.2147/CMAR.S181922
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Xiaoyun Hu,1 Wenyan Qin,1 Shanqiong Li,1 Miao He,1 Yilin Wang,1 Shu Guan,2 Haishan Zhao,1 Weifan Yao,1 Minjie Wei,1 Mingyan Liu,1 Huizhe Wu1
1Department of Pharmacology, School of Pharmacy, Liaoning Key Laboratory of Molecular Targeted Anti-Tumor Drug Development and Evaluation, China Medical University, Shenyang 110122, China; 2Department of Breast Surgery, First Hospital of China Medical University, Shenyang 110001, China
Objective: Multiple factors are involved in oxaliplatin-resistant process in colorectal cancer (CRC) patients including decreased drug accumulation and enhanced capacity to repair and tolerate DNA damage. In the present study, we aimed to assess the impact of six single-nucleotide polymorphisms (SNPs) in DNA repair genes and ABCG2 gene on prognosis in advanced CRC patients treated with oxaliplatin-based chemotherapy.
Methods: In this study, 580 advanced CRC patients were recruited. Six SNPs of DNA repair genes (XPA rs10817938, XPA rs2808668, XPC rs2607775, and WRN rs1346044) and ABCG2 gene (rs2231142 and rs2622621) were genotyped by using the TaqMan assay.
Results: Regarding interaction with environmental factors, ABCG2 rs2231142 and the first-degree family history of cancer and XPC rs2607775 or ABCG2 rs2622621 and lymph node metastases status demonstrated significant interactions. Of these six SNPs, XPA rs10817938 CT/TT genotypes retained its significant association with longer overall survival (OS) (P=0.008) in CRC patients receiving oxaliplatin-based chemotherapy (n=580). Furthermore, a significantly better impact on the disease-free survival (DFS) (P=0.001) and OS (P<0.0001) was found in ABCG2 rs2231142CA/AA carriers. Furthermore, ABCG2 rs2622621 CG/GG genotype was verified to be an independent poor prognostic factor in DFS (P=0.010) and OS (P=0.030). In the stratification analysis, XPA rs10817938 CT/CC, rs2231142 CA/AA, and rs2622621 CC genotypes of ABCG2 were predictive of significantly better prognosis in the patients with tumor differentiation grade 3 (n=523), clinical stage IV (n=73), or lymph node-positive status (n=557). Additionally, multivariate logistic regression and multiple dimension reduction analysis consistently revealed that the combination of selected SNPs and five known risk factors showed a better prediction prognosis and represented the best model to predict CRC prognosis.
Conclusion: The current data indicated that the XPA gene and ABCG2 gene had significant interaction with environmental factors and prognosis, which could provide a comprehensive understanding of the implications of those SNPs in the prediction of prognosis in advanced CRC patients receiving oxaliplatin-based chemotherapy.
Keywords: DNA repair pathway, ABCG2, genetic variation, prognosis, oxaliplatin, colorectal cancer
Introduction
Globally, colorectal cancer (CRC) is the third leading cause of cancer-related death.1,2 Oxaliplatin-based chemotherapy acts as one of the most common therapeutic methods for CRC, which includes FOLFOX (oxaliplatin, 5-fluorouracil [5-FU], and folinic acid [FA]) and XELOX (oxaliplatin and capecitabine). Although it has become a standard regimen for CRC both as adjuvant therapy and as treatment for metastatic cancer, the response to oxaliplatin-based chemotherapy still varied wildly among individuals in clinical practice. The reported overall response rate was 40% in advanced CRC.3 Studies in human CRC cell lines confirmed that multiple factors are involved in the oxaliplatin resistance and clinical prognosis including decreased drug accumulation and enhanced capacity to repair and tolerate DNA damage.4,5 Therefore, identifying biomarkers associated with multidrug resistance and survival time has a long been a research goal in improving the clinical efficacy of oxaliplatin.
The nucleotide excision repair (NER) system is one of the major DNA repair systems to remove a wide range of helix distorting lesions of those formed by various environmental mutagens and certain chemotherapeutic agents from DNA.6–11 XPA, XPC, and DNA unwinding-related proteins such as XPB and XPD belong to the protein members of the NER signaling pathway.12,13 To date, hundreds of genetic variations of these genes are founded (https://genome-cancer.ucsc.edu/), which may alter protein expression and the DNA repair capacity and thus play critical roles in carcinogenesis or response to chemotherapeutic agents.14–17 Previous studies provided evidence about the association of NER single nucleotide polymorphisms (SNPs) with cancer risk and mainly focused on SNPs such as XPA A23G, XPC Ala499Val, XPC Lys939Gln, and XPD Lys751Gln.18–23 However, little information is available about the impact of NER SNPs XPA rs10817938, XPA rs2808668, and XPC rs2607775 on clinical outcomes, especially in CRC. Furthermore, WRN protein plays a crucial role in DNA replication and the maintenance of genome stability.24 Inactivating mutations in WRN gene such as Cys1367Arg (rs1346044) was associated with accelerating aging, led to Werner syndrome, and increased susceptibility to multiple cancers.25–28 Therefore, it is speculated that above functional genetic polymorphisms may correlate with clinical outcomes in advanced CRCs patients with oxaliplatin-based chemotherapy.
Beside the function of DNA repair systems, ABC family of drug efflux transporters plays a vital role in pumping out of cancer cells more than 80% of currently used chemotherapeutic drugs.29 Specifically, the ABCG2 subfamily (BCRP) has been shown to be involved in the multidrug resistance associated to anticancer agents such as adriamycin, mitoxantrone, and platinum drugs.30,31 Immunohistochemistry staining showed that BCRP expression was significantly associated with shortened survival; meanwhile, blood BCRP concentrations were significantly higher in chemoresistance advanced non-small cell lung cancer treated with platinum-based chemotherapy.32,33 Our previous study found a remarkably better response to anthracycline-based chemotherapy among patients who carrying ABCG2 rs2231142 AA genotype.34 The SNP rs2231142 located in the ATP-binding region between Walker A and B motifs and is charged amino acid substitution (Gln141Lys, Q141K). The expression of ABCG2 protein is significantly lower in human placenta with rs2231142 A allele.35 Another study indicated that the ABCG2 rs2231142 polymorphism is associated with reducing the plasma membrane BCRP expression and the transport function in the model cells.36 These data suggested that the ABCG2 SNPs may alter the expression and transporter activity of ABCG2 protein, which is useful for predicting the individualized response variability to anticancer chemotherapy.
Therefore, we expanded our previous studies by analyzing additional potentially functional six SNPs in DNA repair genes and multiple dimension reduction (MDR)-associated genes (XPA rs10817938, XPA rs2808668, XPC rs2607775, WRN rs1346044, ABCG2 rs2231142, and ABCG2 rs2622621) for their correlation with clinical outcomes in CRC patients treated with oxaliplatin-based chemotherapy. The data could provide evidence that these genetic variations may influence clinical outcomes in advanced CRC patients receiving oxalipatin-based chemotherapy, as well as screening the most significantly functional SNP for predicting CRC prognosis.
Methods
Patients and clinical information
This hospital-based retrospective study was performed at China Medical University (Shenyang, China). This study acquired the approval of the Medical Ethics Committee of China Medical University. A total of 580 histopathologically confirmed advanced CRC patients were consecutively recruited from 2009 to 2015 at the First Hospital and the Shengjing Hospital of China Medical University. All of the patients received the oxaliplatin-based chemotherapy including FOLFOX4 regimen (n=464) and XELOX regimen (n=116). The inclusion criteria for CRC patients were as follows: 1) availability of complete clinical data and follow-up status; 2) patients with clinical stages III and IV; and 3) patients who underwent oxaliplatin-based chemotherapy. The exclusion criteria were as follows: 1) incomplete clinical data; 2) blood samples for genotyping were not available; 3) patients who underwent only radiation therapy; 4) other types of cancer and cancers with unknown primary sites; 5) did not receive the FOLFOX6 and XELOX regimens. Clinicopathological data were collected including age at diagnosis, gender, first-degree family history of cancer, smoking status, prime cancer (rectal cancer or colon cancer), tumor size, tumor differentiation, pathological grade, and lymph node metastases. Moreover, the preoperative serum levels of tumor markers (B-mg, AFP, CEA, CA12-5, CA15-3, and CA19-9) were acquired from the interviewer-administered health risk questionnaires and medical records. Nonsmokers were defined as individuals who had <100 cigarettes in lifetime. BMI was calculated from self-reported height and weight. The patients were administered FOLFOX and XELOX regimens for at least 2–3 cycles and were followed up clinically for each month until an occurrence of recurrence and death. The last follow-up date was May 2018. With a median follow-up of 52 months, 291 disease-free survival (DFS) events and 275 OS events were observed in these advanced CRC patients. All enrolled patients signed the written informed consent form according to the relevant regulations. After interview, 5 mL of blood samples were collected for further SNPs’ genotyping.
SNP selection and genotyping
Due to the huge number of SNPs in the human genome, we searched the National Center for Biotechnology Information dbSNP database (http://www.ncbi.nlm.nih.gov/projects/SNP) and HapMap data (http://www.broadinstitute.org/mpg/haploview), in order to minimize the number of potentially functional SNPs. The tag SNPs were selected separately using the following criteria: 1) the minor allele frequency >5% in Chinese Han population; 2) potentially functional SNPs as predicted by SNPinfo software (http://snpinfo.niehs.nih.gov/snpinfo/snpfunc.htm); and 3) not investigated the clinical outcomes in the published genome-wide association studies of CRC. Ultimately, a total of six SNPs in DNA repair pathway and MDR-associated genes (XPA rs10817938, XPA rs2808668, XPC rs2607775, WRN rs1346044, ABCG2 rs2231142, and ABCG2 rs2622621) were selected for the present study. Location and characterization of the selected SNPs are shown in Table S1.
Genomic DNA was isolated from a leukocyte cell pellet of each blood sample using the Tiangen DNA Blood Mini Kit (Tiangen Biotech Co., Ltd, Beijing, China) and performed the TaqMan assay for SNPs’ genotyping. The information about assay conditions, probes, and primers was available upon request. We labeled SNP allele-specific probes with the fluorescent dyes VIC and FAM by using the TaqMan SNP Genotyping Assays on the ABI 7500 Fast Real-Time PCR platform (Thermo Fisher Scientific, Waltham, MA, USA). The genotyping rates of these SNPs were all above 90%. For quality control, at least 10% of the samples were randomly selected for repeated genotyping for confirmation. Some samples were further confirmed by the DNA sequencing analysis. The concordance rate of these repeated samples reached 100%, indicating that the genotyping method and results were reliable.
Statistical analysis
All data analysis was carried out using the SPSS 16.0 software package (SPSS Inc., Chicago, IL, USA). All statistical tests were two sided, and P-value <0.05 was regarded as statistically significant. Associations between genetic polymorphisms and CRC and clinical variables were analyzed by OR and 95% CI using unconditional logistic regression models. The OR and 95% CI were assessed for per-allele, dominant, and recessive models with adjustment for age, gender, body weight, and smoking status. DFS was measured from surgery until an occurrence of recurrence, death, or last known follow-up. Overall survival (OS) was defined as the time between the surgery and death or last known follow-up. The survival curves were generated by using the Kaplan–Meier method, and log-rank test was used to estimate the associations of the DFS or OS with SNPs. Multivariate Cox proportional hazards regression models were applied to obtain the adjusted HR and 95% CI for evaluating the independent prognostic value of each genotypes and clinical variables. The high-order interactions assessed between the SNPs and clinicopathological parameters’ interaction by using the MDR analysis.
Results
Characteristics of CRC patients
Overall, the demographics, risk exposure, and serological biomarkers of the advanced CRC patients are summarized in Table S2. Briefly, the mean age at diagnosis was 59 years (range, 21–82 years; mean) for CRC patients. Most of the cases were in grade 3 (poor, n=523) according to tumor differentiation classification, in clinical stages III (n=507) and IV (n=73) and had lymph node metastases (n=557). All the patients underwent oxaliplatin-based chemotherapy followed by FOLFOX (80.0%) and XELOX (20.0%) treatments. To minimize possible residual confounding effects, the variables of age, gender, body weight, first-degree family history of cancer, and smoking status were then adjusted in the subsequent multivariate logistic regression and multivariate Cox regression analyses.
Interaction between environmental factors, clinicopathological parameters, and genetic factors
To assess the clinical utility of the genotypes, the interactive effects of DNA repair pathway and MDR-associated gene SNPs and the environmental factors, clinicopathological parameters, and serological biomarkers were explored using the Chi-squared test and unconditional logistic regression adjusted by age, gender, body weight, and smoking status. Because body weight, smoking, and first-degree family history of cancer were the environmental higher risk factors for CRC, we also stratified our analyses by these factors of CRC patients. There was no evidence of a statically significant increase in the environmental factors of body weight, smoking patients with all six SNPs even in the adjusted logistic regression model (data shown as in Table S3). However, there was a significantly increased frequency (58.7%) in the ABCG2 rs2231142 CC genotype in the CRC patients who had the first-degree family history of cancer compared with the CA/AA genotypes (45.1%) (adjusted OR =1.82; 95% CI =1.05–3.14, P=0.033) (Figure 1A and B and Table S3). We further observed a significant difference between lymph node metastases status and the XPC rs2607775 and ABCG2 rs2231142 and rs2622621. The frequency (74.7%) of XPC rs2607775 CG/GG genotypes in patients with lymph node metastases was significantly higher than that in patients without lymph node metastases (64.5%) (adjusted OR =0.61; 95% CI =0.41–0.92, P=0.018) (Figure 1A and B and Table S3). Moreover, in the patients with lymph node metastases who had higher frequency of ABCG2 rs2231142 CC genotype (51.2%; adjusted OR =1.53; 95% CI =1.05–2.24, P=0.028) and rs2622621 CG/GG genotypes (59.8%; adjusted OR =0.67; 95% CI =0.45–0.97, P=0.036) (Figure 1A and B and Table S3).
In addition, no significant association was observed between the distribution frequency of all six SNPs and preoperative serological biomarkers of CRC patients including B-mg, AFP, CEA, CA12-5, CA15-3, and CA19-9 (Table S4).
Prognostic markers’s evaluation in CRC patients
To test the hypothesis that all the selected SNPs are independent prognostic factors in our population, we performed the log-rank test and multivariate Cox proportional hazard regression model analysis including all variables to possibly affect DFS or OS among patients receiving oxaliplatin-based chemotherapy (n=580), outlined in Figures 2–4 and Tables 1 and 2. For simplicity, only clinical parameters in stratification analysis that were correlated with at least one genetic variant analyzed are shown and no significant data have been omitted.
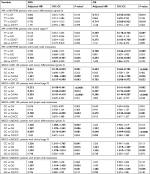
At log-rank test, XPA rs10817938 CT/TT genotypes retained its significant association with longer OS (62 months, 95% CI: 57–68 vs 55 months, 95% CI: 52–59; P=0.008; Figure 2B) in advanced CRC patients receiving oxaliplatin chemotherapy(n=580). This difference reflected into a better survival in the dominant model by using multivariate Cox analysis (adjusted HR: 0.730, 95% CI: 0.576–0.924; P=0.009) as shown in Table 1. However, XPA rs10817938 was found to withheld a tendency to DFS (P=0.055, Figure 2A). As to ABCG2 gene, a significant impact on the DFS and OS was found in rs2231142 polymorphism (log-rank test: P=0.001 and <0.0001, respectively; Figure 2A and B). The estimated mean DFS time and OS time were significantly prolonged among the rs2231142CA/AA carriers (DFS: 51 months, OS: 62 months) in comparison to those patients carrying CC genotype (DFS: 41 months, OS: 54 months). The multivariate Cox analysis also established rs2231142 as an independent prognostic factor (DFS: adjusted HR, 0.682; 95% CI: 0.540–0.861, P=0.001; OS: adjusted HR, 0.666; 95% CI: 0.527–0.843, P<0.0001), as illustrated in Table 1. Furthermore, the estimated median DFS time and OS time for patients who had rs2622621 CC genotype was 53 months (46–61) and 63 months (57–69) compared with the CG/GG genotype carriers (40 months [37–44] and 55 months [52–57]), respectively. The multivariate Cox analysis also identified that ABCG2 rs2622621 dominant model was a predictive of poor prognosis in DFS (adjusted HR: 1.372, 95% CI: 1.079–1.744; P=0.010) and OS (adjusted HR: 1.309, 95% CI: 1.027–1.669; P=0.030). However, in this cohort, there was no impact of the XPA rs2808668, XPC rs2607775, and WRN rs1346044 on DFS, the same applied on OS, data not shown.
Poor tumor differentiation status, clinical stage IV, and lymph node metastasis are biomarkers of severe outcome especially in the later stages of CRC development. Thus, we next evaluated whether correlations found between selected SNPs and subgroup of tumor differentiation grade 3 (n=523), clinical stage IV (n=73), and lymph node metastasis (n=557) as independent impacting factors for prognosis. When the SNPs results were considered in the stratification analysis, some SNPs such as XPA rs10817938 and ABCG2 rs2231142 and rs2622621 had significantly strengthened impact on clinical outcomes. These data showed evidence by the fact that XPA rs10817938 and ABCG2 rs2231142 dominant model and ABCG2 rs2622621 CC genotype indicating better prognosis in the subgroup patients with tumor differentiation grade 3 (n=523), clinical stage IV (n=73), and lymph node metastasis (n=557). Carrying XPA rs10817938 CT/CC genotype and ABCG2 rs2231142 CA/AA genotype and rs2622621 CC genotype were predictive of a prolonged OS time in the patients with tumor differentiation grade 3 (log-rank test: P=0.007, <0.0001, and 0.029, respectively; Figure 3A), or clinical stage IV (log-rank test: P=0.004, <0.0001, and 0.024, respectively; Figure 3B), or lymph node-positive status (log-rank test: P=0.002, 0.001, and 0.037, respectively; Figure 3C). Meanwhile, the multivariate Cox analysis further verified that XPA rs10817938 and ABCG2 rs2231142 and rs2622621 as independent better prognostic factors as illustrated in Table 2. As to DFS, note that the estimated median time for patients harboring the rs10817938 CT/CC genotype experience an increase in the subgroup of clinical stage IV (TT genotype: 37 months vs CT/TT genotype: 62 months; Figure 4B) and lymph node-positive status (TT genotype: 44 months vs CT/TT genotype: 49 months; Figure 4C). More importantly, ABCG2 rs2231142 CA/AA and rs2622621 CC genotypes also showed a significantly longer median DFS time in the patients with tumor differentiation grade 3 (log-rank test: P=0.001 and 0.007, respectively; Figure 4A), or clinical stage IV (log-rank test: P<0.0001 and 0.009, respectively; Figure 4B), or lymph node-positive status (log-rank test: P=0.002 and 0.018, respectively; Figure 4C), which was also verified to be independent prognostic markers in the multivariate Cox analysis, listed in Table 2.
High-order interactions with CRC prognosis by MDR analysis
To further evaluate the existence of possible gene, environmental and clinicopathological parameters’ interaction in association with the clinical outcomes, high-order interactions assessed by using the MDR analysis were conducted with inclusion of the six selected SNPs (ie, rs10817938, rs2808668, rs2607775, rs1346044, rs2231142, and rs2622621) and five known risk factors (ie, gender, age at diagnosis, body weight, first-degree family history of cancer, and smoking). In the MDR analysis, the combination of five risk factors was the best model with the highest cross-validation consistency (CVC) and the lowest prediction error in comparison to the one-factor model among all five risk factors. Moreover, the 11-factor model had a maximum CVC and a minimum prediction error, with the prediction error being statistically significant (Table 3) both in DFS and OS. Taken together, the 11-factor model showed a better prediction prognosis than the five-factor model and represented the best model to predict CRC prognosis for this study population.
Discussion
Oxaliplatin-based chemotherapy has been the mainstay of primary therapy for CRC; however, some patients will develop oxaliplatin resistance and then recurrence. Increasingly, studies provided evidence that the genetic variation in the DNA repair genes especially in “damage incision” step was significantly associated with cancer risk and had interactions with environmental factors.17,37 Thus, searching potential genetic markers has always been a central topic in the field of oncology as to the clinical application with the perspective of individualized treatment.
In the present study, these six candidate SNPs in the NER pathway and MDR-associated genes were chosen; since previous studies mainly focused on the cancer risk, interaction with clinicopathological parameters and prognostic influence on the CRC of those SNPs needs to be further elucidated. First, a statistically significant distribution frequency difference was found between lymph node metastases status and XPC rs2607775 and ABCG2 rs2622621, and harboring rs2607775 CG/GG or rs2622621CG/GG genotypes showed significantly higher distribution frequency than those without lymph node metastases. To data, only two studies investigated the interaction between XPC rs2607775 and gastric cancer and gastrointestinal stromal cancer involved in environmental factors, but not CRC. Liu et al17 conducted a case (n=898)–control (n=937) study and found that XPC rs2607775 had significant interaction with smoking gastric cancer patients. In another gastrointestinal stromal cancer study, the DRC levels of UV induced-DNA damage were measured and a significantly difference in mean DRC per genotype for XPC rs2607775 was observed.38 Importantly, in our study, a significantly increased distribution frequency was observed in carrying rs2231142 CC genotype patients who had the first-degree family history of cancer, which has not been reported yet. These data indicated that above SNPs could serve as potential biomarkers for evaluating the interaction of clinicopathological parameters and advanced CRC-associated polymorphisms.
Previous studies demonstrated the role of enhanced NER activity and ABC transporters in oxaliplatin resistance and diminished NER activity and ABC transporters in oxaliplatin sensitivity,3,5,39 therefore, identifying the prognostic markers of oxaliplatin resistance that can be targeted effectively to increase sensitivity and improve the treatment efficacy of CRC. Thereafter, we assessed the association of six SNPs with PFS and OS in advanced CRC patients receiving oxaliplatin-based treatment. For XPA gene, a significant gene-clinical variables’ interaction between OS or DFS and CT/CC genotype of rs10817938, but not rs2808668, was observed. The log-rank test and multivariate Cox data showed that rs10817938 CT/CC genotype carriers had better OS in the advanced CRC patients with oxaliplatin treatment, tumor differentiation grade 3, clinical stage IV, and lymph node-positive status. As to DFS, note that the estimated median time for patients harboring the rs10817938 CT/CC genotype experiences an increase in the patients with clinical stage IV (n=73) and lymph node-positive status (n=557). Although reports on the SNP in the XPA gene have been published previously, these did not share a relation with the significant XPA SNPs that was identified in our current study. Most of those previous studies mainly focused on identifying the relationships between the SNPs and cancer risk in different populations.17,38,40–49 Only Gao et al50 found a prognostic importance related to theXPArs10817938 C allele, which indicated that it is a potential functional SNP and a biomarker for prognosis in oral squamous cell carcinoma patients, similarly with our results, no significant association of rs2808668 polymorphism with OSCC prognosis observed. However, in another single tag SNP analysis study, XPA rs2808668 was found significantly associated with lung cancer risk in 265 lung cancer patients and 301 healthy controls.40 It should be noted that all the studies did not include CRC patients. Our data suggest that rs10817938 may represent novel markers of CRC response to oxaliplatin-based chemotherapy, especially in the advanced CRC patients with tumor differentiation grade 3, clinical stage IV, and lymph node-positive status.
In the present study, our results for the first time demonstrated that SNPs rs2231142 CA/AA genotype and rs2622621 CC of ABCG2 gene showed significant association with DFS and OS in advanced CRC patients treated with oxaliplatin regimens and multivariate analysis further confirmed the significant prognostic value of the SNPs as independent of standard prognostic markers. MDR analysis consistently revealed a gene–gene interaction among those SNPs associated with CRC prognosis. More importantly, results from stratification analysis with clinical variables between the SNPs and prognosis and found that harboring rs2231142 CA/AA and rs2622621 CC genotypes showed better prognosis in the subgroup patients with tumor differentiation (grade III), clinical stage IV, or lymph node metastases. The rs2231142 AA genotype was related to a remarkably better response to the anthracycline treatment of breast cancer in our previous study.34 In a recent study including series of metastatic renal cell carcinoma (mRCC) patients, polymorphism rs2231142 in the efflux pump ABCG2gene was associated with hypothyroidism in mRCC patients treated with sunitinib.51 These suggest that ABCG2SNPs are useful SNPs and potential biomarkers for predicting the individualized response variability to anticancer agents in CRC patients.
Therefore, a systematic evaluation of the relationship between above SNPs and interindividual variability in oxaliplatin response and the clinical outcome are urgently required. Through the multiple candidate gene approach, we extensively evaluated for the first time the significant associations between polymorphisms in DNA repair genes and ABC transporter gene, clinical pathological features, and clinical outcome in CRC patients. Our results provided evidence that XPA and ABCG2 polymorphisms are associated with oxaliplatin-based chemotherapy in a large and well-characterized cohort of advanced CRC patients, especially in the subgroup of tumor differentiation (grade 3), clinical stage IV, or lymph node-positive status. Therefore, the variation of NER pathway or ABC transporter polymorphisms such as XPA and ABCG2 genes might be applied in the prediction of advanced CRC prognosis and personalized treatment in future.
Conclusion
- In gene–environmental factor interactions, the distribution frequency of harboring XPC rs2607775 CG/GG or ABCG2 rs2622621CG/GG genotypes was increased in lymph node metastases patients. Meanwhile, rs2231142 CC genotype was higher in patients who had the first-degree family history of cancer.
- A significantly better OS was observed between CT/CC genotype of XPA rs10817938 and carriers with oxaliplatin treatment, tumor differentiation grade 3, clinical stage IV, or lymph node-positive status.
- It is notable that a remarkably better prognosis for SNPs rs2231142 CA/AA genotype, rs2622621 CC of ABCG2 gene in advanced CRC patients with oxaliplatin-based chemotherapy, tumor differentiation grade 3, clinical stage IV, or lymph node-positive status.
- Future in vitro and in vivo studies in more numerous cancers of patients are needed to confirm these preliminary results.
Ethical approval
All procedures performed in the present study involving human participants were in accordance with the ethical standards of the institutional and national research committee and the 1964 Declaration of Helsinki.
Informed consent
Written informed consent was obtained from all individual participants included in the study.
Acknowledgments
This study was supported by the First Hospital and the Shengjing Hospital of China Medical University. The authors thank all the doctors and nurses for their great help in collecting tissue samples. This work was supported by grants from the National Natural Science Foundation of China (nos 31828005, 81872905, 81673475, 81501346, 81603149, and 81601370), National Natural Science Foundation of China and Liaoning joint fund key program (no U1608281), Shenyang S&T Projects (17-123-9-00 and Z18-4-020), the Key Laboratory Foundation from Shenyang S&T Projects (F16-094-1-00), and Key Laboratory Foundation from Liaoning Province (no LS201617).
Author contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. | ||
Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. | ||
Ge L, Wang YF, Tian JH, et al. Network meta-analysis of Chinese herb injections combined with FOLFOX chemotherapy in the treatment of advanced colorectal cancer. J Clin Pharm Ther. 2016;41(4):383–391. | ||
El Khoury F, Corcos L, Durand S, Simon B, Le Jossic-Corcos C. Acquisition of anticancer drug resistance is partially associated with cancer stemness in human colon cancer cells. Int J Oncol. 2016;49(6):2558–2568. | ||
Perez M, Lucena-Cacace A, Marín-Gómez LM, et al. Dasatinib, a Src inhibitor, sensitizes liver metastatic colorectal carcinoma to oxaliplatin in tumors with high levels of phospho-Src. Oncotarget. 2016;7(22):33111–33124. | ||
Kilic L, Ordu C, Yildiz I, et al. Current adjuvant treatment modalities for gastric cancer: from history to the future. World J Gastrointest Oncol. 2016;8(5):439–449. | ||
Guo Y, Xiong BH, Zhang T, Cheng Y, Ma L. XELOX vs FOLFOX in metastatic colorectal cancer: an updated meta-analysis. Cancer Invest. 2016;34(2):94–104. | ||
Di Francia R, Siesto RS, Valente D, et al. Current strategies to minimize toxicity of oxaliplatin: selection of pharmacogenomic panel tests. Anticancer Drugs. 2013;24(10):1069–1078. | ||
Cvitkovic E, Bekradda M. Oxaliplatin: a new therapeutic option in colorectal cancer. Semin Oncol. 1999;26(6):647–662. | ||
Misset JL, Bleiberg H, Sutherland W, Bekradda M, Cvitkovic E. Oxaliplatin clinical activity: a review. Crit Rev Oncol Hematol. 2000;35(2):75–93. | ||
Dilruba S, Kalayda GV. Platinum-based drugs: past, present and future. Cancer Chemother Pharmacol. 2016;77(6):1103–1124. | ||
Quiroga D, Lyerly HK, Morse MA. Deficient mismatch repair and the role of immunotherapy in metastatic colorectal cancer. Curr Treat Options Oncol. 2016;17(8):41. | ||
Da Silva FC, Wernhoff P, Dominguez-Barrera C, Dominguez-Valentin M. Update on hereditary colorectal cancer. Anticancer Res. 2016;36(9):4399–4406. | ||
Kap EJ, Seibold P, Richter S, et al. Genetic variants in DNA repair genes as potential predictive markers for oxaliplatin chemotherapy in colorectal cancer. Pharmacogenomics J. 2015;15(6):505–512. | ||
Mccullough LE, Santella RM, Cleveland RJ, et al. Polymorphisms in DNA repair genes, recreational physical activity and breast cancer risk. Int J Cancer. 2014;134(3):654–663. | ||
Corral R, Lewinger JP, van den Berg D, et al. Comprehensive analyses of DNA repair pathways, smoking and bladder cancer risk in Los Angeles and Shanghai. Int J Cancer. 2014;135(2):335–347. | ||
Liu J, Sun L, Xu Q, et al. Association of nucleotide excision repair pathway gene polymorphisms with gastric cancer and atrophic gastritis risks. Oncotarget. 2016;7(6):6972–6983. | ||
Liu J, Zhang Z, Cao XL, et al. XPA A23G polymorphism and susceptibility to cancer: a meta-analysis. Mol Biol Rep. 2012;39(6):6791–6799. | ||
Zou JH, An L, Chen S, Ren LQ. XPA A23G polymorphism and lung cancer risk: a meta-analysis. Mol Biol Rep. 2012;39(2):1435–1440. | ||
Wang G, Sun H, Liu Z, et al. Lack of associations between XPC polymorphisms and colorectal cancer: a meta-analysis. J Buon. 2015;20(3):770–774. | ||
Sankhwar M, Sankhwar SN, Bansal SK, Gupta G, Rajender S. Polymorphisms in the XPC gene affect urinary bladder cancer risk: a case-control study, meta-analyses and trial sequential analyses. Sci Rep. 2016;6:27018. | ||
Shi YH, Wang B, Xu BP, et al. The association of six non-synonymous variants in three DNA repair genes with hepatocellular carcinoma risk: a meta-analysis. J Cell Mol Med. 2016;20(11):2056–2063. | ||
Loghin A, Bănescu C, Nechifor-Boila A, et al. XRCC3 Thr241Met and XPD Lys751Gln gene polymorphisms and risk of clear cell renal cell carcinoma. Cancer Biomark. 2016;16(2):211–217. | ||
Pichierri P, Ammazzalorso F, Bignami M, Franchitto A. The Werner syndrome protein: linking the replication checkpoint response to genome stability. Aging. 2011;3(3):311–318. | ||
Zins K, Frech B, Taubenschuss E, Schneeberger C, Abraham D, Schreiber M. Association of the rs1346044 polymorphism of the Werner syndrome gene RECQL2 with increased risk and premature onset of breast cancer. Int J Mol Sci. 2015;16(12):29643–29653. | ||
Jiang S, Hu N, Zhou J, et al. Polymorphisms of the WRN gene and DNA damage of peripheral lymphocytes in age-related cataract in a Han Chinese population. Age. 2013;35(6):2435–2444. | ||
Ehrenberg M, Dratviman-Storobinsky O, Avraham-Lubin BR, Goldenberg-Cohen N. Lack of association of the WRN C1367T polymorphism with senile cataract in the Israeli population. Mol Vis. 2010;16:1771–1775. | ||
Frank B, Hoffmeister M, Klopp N, Illig T, Chang-Claude J, Brenner H. Colorectal cancer and polymorphisms in DNA repair genes WRN, RMI1 and BLM. Carcinogenesis. 2010;31(3):442–445. | ||
Sugimoto Y, Tsukahara S, Ishikawa E, Mitsuhashi J. Breast cancer resistance protein: molecular target for anticancer drug resistance and pharmacokinetics/pharmacodynamics. Cancer Sci. 2005;96(8):457–465. | ||
Pollex EK, Anger G, Hutson J, Koren G, Piquette-Miller M. Breast cancer resistance protein (BCRP)-mediated glyburide transport: effect of the C421A/Q141K BCRP single-nucleotide polymorphism. Drug Metab Dispos. 2010;38(5):740–744. | ||
Noguchi K, Katayama K, Mitsuhashi J, Sugimoto Y. Functions of the breast cancer resistance protein (BCRP/ABCG2) in chemotherapy. Adv Drug Deliv Rev. 2009;61(1):26–33. | ||
Ohtsuka K, Ohnishi H, Morii T, et al. Downregulated ABCG2 enhances sensitivity to topoisomerase I inhibitor in epidermal growth factor receptor tyrosine kinase inhibitor-resistant non-small cell lung cancer. J Thorac Oncol. 2010;5(11):1726–1733. | ||
Wen CC, Yee SW, Liang X, et al. Genome-wide association study identifies ABCG2 (BCRP) as an allopurinol transporter and a determinant of drug response. Clin Pharmacol Ther. 2015;97(5):518–525. | ||
Wu H, Liu Y, Kang H, et al. Genetic variations in ABCG2 gene predict breast carcinoma susceptibility and clinical outcomes after treatment with anthracycline-based chemotherapy. Biomed Res Int. 2015;2015:279109-12. | ||
Kobayashi D, Ieiri I, Hirota T, et al. Functional assessment of ABCG2 (BCRP) gene polymorphisms to protein expression in human placenta. Drug Metab Dispos. 2005;33(1):94–101. | ||
Bram EE, Ifergan I, Grimberg M, Lemke K, Skladanowski A, Assaraf YG. C421 allele-specific ABCG2 gene amplification confers resistance to the antitumor triazoloacridone C-1305 in human lung cancer cells. Biochem Pharmacol. 2007;74(1):41–53. | ||
Wu H, Kang H, Liu Y, et al. Association of ABCB1 genetic polymorphisms with susceptibility to colorectal cancer and therapeutic prognosis. Pharmacogenomics. 2013;14(8):897–911. | ||
Perez-Mayoral J, Pacheco-Torres AL, Morales L, Acosta-Rodriguez H, Matta JL, Dutil J. Genetic polymorphisms in RAD23B and XPC modulate DNA repair capacity and breast cancer risk in Puerto Rican women. Mol Carcinogen. 2013;52(Suppl):E127–E138. | ||
Zhao J, Li W, Zhu D, et al. Association of single nucleotide polymorphisms in MTHFR and ABCG2 with the different efficacy of first-line chemotherapy in metastatic colorectal cancer. Med Oncol. 2014;31(1):802. | ||
Mei C, Hou M, Guo S, et al. Polymorphisms in DNA repair genes of XRCC1, XPA, XPC, XPD and associations with lung cancer risk in Chinese people. Thorac Cancer. 2014;5(3):232–242. | ||
Ravegnini G, Nannini M, Simeon V, et al. Polymorphisms in DNA repair genes in gastrointestinal stromal tumours: susceptibility and correlation with tumour characteristics and clinical outcome. Tumour Biol. 2016;37(10):13413–13423. | ||
Wang B, Xu Q, Yang HW, Sun LP, Yuan Y. The association of six polymorphisms of five genes involved in three steps of nucleotide excision repair pathways with hepatocellular cancer risk. Oncotarget. 2016;7(15):20357–20367. | ||
Sun Y, Tan L, Li H, Qin X, Liu J. Association of NER pathway gene polymorphisms with susceptibility to laryngeal cancer in a Chinese population. Int J Clin Exp Pathol. 2015;8(9):11615–11621. | ||
Zhao F, Shang Y, Zeng C, Gao D, Li K. Association of single nucleotide polymorphisms of DNA repair genes in NER pathway and susceptibility to pancreatic cancer. Int J Clin Exp Pathol. 2015;8(9):11579–11586. | ||
Choi HY, Bae KS, Cho SH, et al. Impact of CYP2D6, CYP3A5, CYP2C19, CYP2A6, SLCO1B1, ABCB1, and ABCG2 gene polymorphisms on the pharmacokinetics of simvastatin and simvastatin acid. Pharmacogenet Genomics. 2015;25(12):595–608. | ||
Campa D, Pardini B, Naccarati A, et al. A gene-wide investigation on polymorphisms in the ABCG2/BRCP transporter and susceptibility to colorectal cancer. Mutat Res. 2008;645(1–2):56–60. | ||
Oberstadt MC, Bien-Möller S, Weitmann K, et al. Epigenetic modulation of the drug resistance genes MGMT, ABCB1 and ABCG2 in glioblastoma multiforme. BMC Cancer. 2013;13:617. | ||
Han W, Kim KY, Yang SJ, Noh DY, Kang D, Kwack K. SNP-SNP interactions between DNA repair genes were associated with breast cancer risk in a Korean population. Cancer. 2012;118(3):594–602. | ||
Tian C, Ambrosone CB, Darcy KM, et al. Common variants in ABCB1, ABCC2 and ABCG2 genes and clinical outcomes among women with advanced stage ovarian cancer treated with platinum and taxane-based chemotherapy: a Gynecologic Oncology Group study. Gynecol Oncol. 2012;124(3):575–581. | ||
Gao C, Wang J, Li C, Zhang W, Liu G. A Functional polymorphism (rs10817938) in the XPA promoter region is associated with poor prognosis of oral squamous cell carcinoma in a Chinese Han population. PLoS One. 2016;11(9):e0160801. | ||
Werbrouck E, Bastin J, Lambrechts D, et al. ABCG2 Polymorphism rs2231142 and hypothyroidism in metastatic renal cell carcinoma patients treated with sunitinib. Acta Clin Belg. Epub 2018 May 23. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.