Back to Journals » International Medical Case Reports Journal » Volume 17
Pneumonia Caused by Chlamydia psittaci and SARS-CoV-2 Coinfection Diagnosed Using Metagenomic Next-Generation Sequencing: A Case Report
Authors Zhang A, Liang J, Lao X, Xia X, Liang J
Received 17 January 2024
Accepted for publication 18 March 2024
Published 21 March 2024 Volume 2024:17 Pages 187—194
DOI https://doi.org/10.2147/IMCRJ.S458131
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Vinay Kumar
Anbing Zhang,1,* Jinguang Liang,2,* Xiaoli Lao,1,3 Xiuqiong Xia,1 Jianping Liang1,3
1Department of Respiratory and Critical Care Medicine, Zhongshan People’s Hospital, Zhongshan, Guangdong Province, People’s Republic of China; 2Department of Respiratory and Critical Care Medicine, Zhongshan Huangpu People’s Hospital, Zhongshan, Guangdong Province, People’s Republic of China; 3Graduate School, Guangdong Medical University, Zhanjiang, Guangdong Province, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jianping Liang, Department of Respiratory and Critical Care Medicine, Zhongshan People’s Hospital, 2 Sunwen East Road, Zhongshan, 528400, People’s Republic of China, Tel +86-159-8910-7884, Fax +86-760-8988-0256, Email [email protected]
Abstract: We report a case of pneumonia caused by coinfection with Chlamydia psittaci and the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Omicron XBB.1 variant, confirmed using metagenomic next-generation sequencing (mNGS) and quantitative polymerase chain reaction (qPCR). C. psittaci and SARS-CoV-2 were detected in bronchoalveolar lavage fluid using mNGS. Additionally, mNGS detected C. psittaci in blood and nasopharyngeal specimens and was more sensitive than qPCR. The patient recovered after treatment with moxifloxacin. This report highlights the use of coinfections of C. psittaci and SARS-CoV-2, as mNGS has already been recognized to be a diagnostic tool for identifying coinfections.
Keywords: coinfection, quantitative reverse-transcription polymerase chain reaction, bronchoalveolar lavage fluid, zoonoses
Introduction
Psittacosis, also known as Chlamydia psittaci (C. psittaci) pneumonia, is a zoonotic infection transmitted to humans from birds contaminated with C. psittaci through the inhalation route. C. psittaci accounts for approximately 1% of cases of community-acquired pneumonia.1 It is underdiagnosed and, hence, insufficiently understood by clinicians and the public. Previously, psittacosis was diagnosed primarily using serology and polymerase chain reaction (PCR) testing.2 However, in recent years, metagenomic next-generation sequencing (mNGS) has been used extensively in hospitals at all levels in China to diagnose psittacosis.3,4
Familial clustering of psittacosis was reported during the coronavirus disease (COVID-19) pandemic,5 including clustering in a COVID-19 testing area.6 Han et al7 have previously reported a case of psittacosis complicated by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection; however, the authors did not report the SARS-CoV-2 variant and did not compare the results of pharyngeal swabs, blood, and bronchoalveolar lavage specimens for detecting C. psittaci or SARS-CoV-2, nor the respective advantages of mNGS and quantitative PCR (qPCR) testing. Here, we report a case of psittacosis complicated by SARS-CoV-2 infection. This report highlights the use of mNGS as an essential diagnostic tool in identifying coinfections of C. psittaci and SARS-CoV-2.
Case Description
In May 2023, a 75-year-old woman with a history of hypertension and no past SARS-CoV-2 infection was admitted to our hospital for chills, pyrexia (maximum temperature, 38°C), cough, headache, asthenia, myalgia, and lumbago. The patient had received two doses of Sinopharm/BIBP COVID-19 vaccine. Inquiring about her exposure to birds revealed that she had worked in a slaughterhouse and slaughtered chickens before the disease onset.
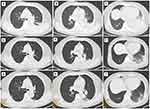
The patient was not tested for SARS-CoV-2 on admission to the hospital. Chest computed tomography (CT) on day 1 after admission revealed massive consolidation in the left lower lung and a left-sided pleural effusion (Figure 1), leading to a diagnosis of pneumonia. The laboratory test results revealed elevated white blood cell (WBC) and neutrophil counts, an elevated neutrophil percentage, and elevated levels of C-reactive protein (CRP), aspartate aminotransferase, fibrinogen, and D-dimer (Table 1).
 |
Table 1 Laboratory Test Results of the Patient with Chlamydia Psittaci Pneumonia and SARS-CoV-2 Coinfection |
Intravenous piperacillin-tazobactam (4 g/0.5 g) was administered every 8 hours to treat pneumonia. However, 3 days after admission, the patient developed recrudescent pyrexia (maximum temperature, 39.2°C). Fiberoptic bronchoscopy revealed tracheitis and bronchitis with bronchial mucosal hyperemia in the left lower lobe and various subsegments.
The patient underwent bronchoalveolar lavage of the left lower lobe, and bronchoalveolar lavage fluid (BALF), blood, and nasopharyngeal swab (NPS) samples were tested using mNGS to identify the pathogen. Clinical samples were collected according to the standard protocol of our hospital. DNA was extracted from BALF, blood, and other clinical samples using a TIANamp Micro DNA Kit (DP316, Tiangen Biotech Co., Beijing, China), followed by DNA/RNA library construction, high-quality sequencing, and classification analysis, as described previously.8 The mNGS analysis detected C. psittaci in the BALF (sequence number: 874), peripheral blood (sequence number: 2), and NPS (sequence number: 4) samples, as well as SARS-CoV-2 Omicron XBB.1 in the BALF (sequence number: 1,847,027) and NPS (sequence number: 224,671) samples (Figure 2).
The mNGS results were confirmed using qPCR tests for C. psittaci and SARS-CoV-2. The primer sets targeted the OmpA and ORF/N genes of C. psittaci and SARS-CoV-2, respectively (Table 2). The qPCR detected C. psittaci nucleic acid in the BALF only (Ct: 24.59) and SARS-CoV-2 nucleic acid in both the BALF and NPS specimens (Ct: 38.57 and 31.07, respectively; Table 3).
 |
Table 2 C. Psittaci and COVID-19 PCR Primers Used in This Study |
 |
Table 3 mNGS and Confirmative qPCR Results |
Serum samples were tested for SARS-CoV-2 immunoglobulin (Ig)M/IgG using chemiluminescence immunoassays, performed according to the instructions of Guangzhou Huayin Health Medical Group Co., Ltd. The patient tested negative for SARS-CoV-2 IgM (cutoff index, 0.87) and positive for SARS-CoV-2 IgG (cutoff index, 64.58).
Based on these findings, the patient was diagnosed with psittacosis and SARS-CoV-2 coinfection. The antibiotic therapy was switched to intravenous moxifloxacin (MFX) (400 mg daily), and 2 days later, the fever resolved (Figure 3). The patient did not require specific treatment for SARS-CoV-2 infection. Repeat chest CT 1 week after starting MFX showed a reduction in the size of the lung lesions (Figure 1). As she was clinically stable, she was discharged with a prescription for oral MFX (400 mg daily). At a follow-up visit 1 week after discharge, she had almost fully recovered clinically. Follow-up chest CT showed a marked reduction in the size of the lung lesions (Figure 1), and SARS-CoV-2 nucleic acid testing of an NPS was negative.
Discussion
Psittacosis in humans primarily presents as a respiratory tract infection; however, it may also lead to systemic infections.9 The lungs are particularly susceptible to C. psittaci infection,10 and C. psittaci can be detected in both the upper and lower respiratory tracts of patients infected with C. psittaci. In a recent large-scale retrospective study, qPCR detected C. psittaci in samples from the upper and lower respiratory tracts and fecal samples. The detection rate of C. psittaci in the lower respiratory tract was significantly higher than that in the upper respiratory tract; however, specimens were more readily collected from the upper respiratory tract than from the lower respiratory tract.11 In the current case, C. psittaci was detected in the upper and lower respiratory tracts and the blood specimen using mNGS. The C. psittaci sequence number was highest in the BALF specimen and was low in the NPS and blood specimens. Hence, BALF should be the preferred specimen for detecting C. psittaci using mNGS.12
The accurate diagnosis of C. psittaci infection poses a challenge. Commonly employed methods include serological testing, in vitro culture, qPCR, and mNGS. Serological tests are prone to inaccuracies and may produce false-positive results,13 whereas in vitro cultures are challenging due to stringent criteria. Furthermore, although qPCR is specific, it is limited to suspected cases and cannot diagnose coinfections. Contrastingly, mNGS, which is superior to PCR, aids in diagnosing psittacosis and coinfections;14,15 it can detect C. psittaci in blood, NPS, BALF, and cerebrospinal fluid specimens. Because C. psittaci does not colonize the human body, C. psittaci infection should be diagnosed if C. psittaci sequences are detected in upper or lower respiratory tract specimens or other non-respiratory tract specimens. As mNGS can accurately and rapidly identify pathogens, it is useful for diagnosing C. psittaci infection, and promptly adjusting the antibiotic regimen following diagnosis can contribute to a favorable prognosis. In this case, the patient experienced recurrent pyrexia 3 days after treatment with piperacillin combined with tazobactam; therefore, mNGS was used to confirm the diagnosis. After mNGS detected C. psittaci in the BALF specimen, the antibiotic was changed to MFX, which is a more effective treatment. The patient’s temperature normalized, preventing progression to critical illness.16
Viral infections can compromise host immunity, facilitating the progression of viral and bacterial coinfections.17,18 C. psittaci, Legionella pneumophila, Streptococcus pneumoniae, and Mycoplasma pneumoniae have been identified as bacterial pathogens with SARS-CoV-2 coinfection.19 Studies indicate two mechanisms of action in viral and bacterial coinfections: (i) direct interactions, where viruses exploit bacterial components, and (ii) indirect interactions, where bacteria exploit viral infections. These interactions involve complex regulatory mechanisms, such as cell receptor upregulation, epithelial layer damage, symbiotic bacterial turnover, and immune system suppression, leading to further damage to the host.20 Elevated erythrocyte sedimentation rate and CRP levels are common laboratory parameters for diagnosing psittacosis.21 In this case, the increased WBC and neutrophil counts and elevated CRP levels indicated that the patient may have been infected with C. psittaci on admission. However, it is unknown which infection occurred first in this case.
Both SARS-CoV-2 and C. psittaci cause pneumonia. Distinguishing SARS-CoV-2 infection from C. psittaci infection is challenging.22,23 In this case, Omicron XBB.1 and C. psittaci were detected in BALF and NPS specimens. Chest CT revealed massive consolidation in the left lower lung field and left-sided pleural effusion. Patients with psittacosis typically show varying degrees of effusion and consolidation, including patchy shadows.
Patients with severe psittacosis may present with lobar shadows, extensive double pneumonia, and sometimes, pleural effusion, and chest lesions usually resolve within 2–4 weeks of initiating pharmacotherapy.24–26 In this case, SARS-CoV-2 was detected in BALF, blood, and NPS specimens using mNGS. Therefore, the patient was diagnosed with C. psittaci and SARS-CoV-2 coinfection. C. psittaci and SARS-CoV-2 (Omicron XBB.1) were detected in the BALF specimen from the left lower lung. The NPS specimen showed the same variant as that in the lung.
Common chest imaging findings for SARS-CoV-2 include ground-glass opacity and multifocal consolidation, with pleural effusion (6%) being uncommon.27 The imaging findings in this patient were atypical. The Omicron XBB.1 variant causes less severe disease than other SARS-CoV-2 variants do.28,29
In this case, piperacillin combined with tazobactam was initially administered; however, C. psittaci is not sensitive to these antibiotics. After detecting C. psittaci in the BALF, the antibiotic therapy was changed to MFX, leading to recovery. Quinolones are first-line antibiotics for community-acquired pneumonia, covering atypical pathogens such as C. psittaci and Mycoplasma pneumoniae, and MFX should be administered empirically in cases of suspected psittacosis.4 The recommended first-line treatment for psittacosis is doxycycline rather than quinolones30 because quinolones have lower intracellular activity against C. psittaci than tetracycline does. In this case, the patient was treated with MFX instead of tetracycline because our hospital did not have doxycycline.
This case report compared mNGS with qPCR for testing BALF, blood, and NPS specimens in a patient with C. psittaci and SARS-CoV-2 coinfection and highlights the utility of mNGS in patients with pneumonia of unknown cause that is unresponsive to antibiotics and is diagnosing coinfections with more than one pathogen.
This study has one limitation. The clinical characteristics features of this case of C. psittaci and SARS-CoV-2 coinfection are derived from this specific patient; therefore, the findings may not be generalizable.
Data Sharing Statement
All relevant data are provided within the manuscript. All information in the article has been anonymized.
Ethics Approval and Informed Consent
The Ethics Committees of Zhongshan People’s Hospital (K2021-137) approved this study. Informed consents were obtained from the patient. Zhongshan People’s Hospital granted approval to publish the case details.
Consent for Publication
The patient has provided informed consent for the publication of her case details and the associated images.
Funding
This work was supported by the Science and Technology Plan Project of Zhongshan under grant number 2020B1113.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Hogerwerf L, Gier DE, Baan B, et al. Chlamydia psittaci (psittacosis) as a cause of community-acquired pneumonia: a systematic review and meta-analysis. Epidemiol Infect. 2017;145(15):3096–3105. doi:10.1017/S0950268817002060
2. Nieuwenhuizen AA, Dijkstra F, Notermans DW, et al. Laboratory methods for case finding in human psittacosis outbreaks: a systematic review. BMC Infect Dis. 2018;18(1):442. doi:10.1186/s12879-018-3317-0
3. Zhang A, Xia X, Yuan X, et al. Severe Chlamydia psittaci pneumonia complicated by rhabdomyolysis: a case series. Infect Drug Resist. 2022;15:873–881. doi:10.2147/IDR.S355024
4. Zhang A, Xia X, Yuan X, et al. Clinical characteristics of 14 cases of severe Chlamydia psittaci pneumonia diagnosed by metagenomic next-generation sequencing: a case series. Med Baltim. 2022;101(24):e29238. doi:10.1097/MD.0000000000029238
5. Li N, Li S, Tan W, et al. Metagenomic next-generation sequencing in the family outbreak of psittacosis: the first reported family outbreak of psittacosis in China under COVID-19. Emerg Microbes Infect. 2021;10(1):1418–1428. doi:10.1080/22221751.2021.1948358
6. Lei JH, Xu Y, Jiang YF, et al. Clustering cases of Chlamydia psittaci pneumonia in coronavirus disease 2019 screening ward staff. Clin Infect Dis. 2021;73(9):e3261–e3265. doi:10.1093/cid/ciaa1681
7. Han L, Sun LX, Chen YL, et al. Clinical characteristics of a case of severe pneumonia caused by coinfection of COVID-19 and Chlamydia psittaci. Zhonghua Jie He He Hu Xi Za Zhi. 2023;46(11):1118–1120. Chinese. doi:10.3760/cma.j.cn112147-20230906-00143
8. Liang Y, Dong T, Li M, et al. Clinical diagnosis and etiology of patients with Chlamydia psittaci pneumonia based on metagenomic next-generation sequencing. Front Cell Infect Microbiol. 2022;12:1006117. doi:10.3389/fcimb.2022.1006117
9. Čechová L, Halánová M, Babinská I, et al. Chlamydiosis in farmed chickens in Slovakia and zoonotic risk for humans. Ann Agric Environ Med. 2018;25(2):320–325. doi:10.26444/aaem/82948
10. Gu L, Liu W, Ru M, et al. The application of metagenomic next-generation sequencing in diagnosing Chlamydia psittaci pneumonia: a report of five cases. BMC Pulm Med. 2020;20(1):65. doi:10.1186/s12890-020-1098-x
11. McGovern OL, Kobayashi M, Shaw KA, et al. Use of real-time PCR for Chlamydia psittaci detection in human specimens during an outbreak of psittacosis – Georgia and Virginia, 2018. MMWR Morb Mortal Wkly Rep. 2021;70(14):505–509. doi:10.15585/mmwr.mm7014a1
12. Wang K, Liu X, Liu H, et al. Metagenomic diagnosis of severe psittacosis using multiple sequencing platforms. BMC Genomics. 2021;22(1):406. doi:10.1186/s12864-021-07725-9
13. Stewardson AJ, Grayson ML. Psittacosis. Infect Dis Clin North Am. 2010;24(1):7–25. doi:10.1016/j.idc.2009.10.003
14. Duan Z, Gao Y, Liu B, et al. The application value of metagenomic and whole-genome capture next-generation sequencing in the diagnosis and epidemiological analysis of psittacosis. Front Cell Infect Microbiol. 2022;12:872899. doi:10.3389/fcimb.2022.872899
15. Shi Y, Chen J, Shi X, et al. A case of Chlamydia psittaci caused severe pneumonia and meningitis diagnosed by metagenome next-generation sequencing and clinical analysis: a case report and literature review. BMC Infect Dis. 2021;21(1):621. doi:10.1186/s12879-021-06205-5
16. Raeven VM, Spoorenberg SM, Boersma WG, et al. Atypical aetiology in patients hospitalised with community-acquired pneumonia is associated with age, gender and season; a data-analysis on four Dutch cohorts. BMC Infect Dis. 2016;16:299. doi:10.1186/s12879-016-1641-9
17. Morris DE, Cleary DW, Clarke SC. Secondary bacterial infections associated with influenza pandemics. Front Microbiol. 2017;8:1041. doi:10.3389/fmicb.2017.01041
18. Public Health Agency of Canada. A tool for the potential fall 2009 wave of pandemic H1N1 to guide public health decision-making: an overview of the public health agency of Canada’s planning considerations, September 2009. Can Commun Dis Rep. 2010;36(S3):1–20. doi:10.14745/ccdr.v36i00as3
19. Li H, Chen K, Liu M, et al. The profile of peripheral blood lymphocyte subsets and serum cytokines in children with 2019 novel coronavirus pneumonia. J Infect. 2020;81(1):115–120. doi:10.1016/j.jinf.2020.04.001
20. Bosch AA, Biesbroek G, Trzcinski K, et al. Viral and bacterial interactions in the upper respiratory tract. PLOS Pathog. 2013;9(1):e1003057. doi:10.1371/journal.ppat.1003057
21. Khadka S, Timilsina B, Pangeni RP, et al. Importance of clinical history in the diagnosis of psittacosis: a case report. Ann Med Surg Lond. 2022;82:104695. doi:10.1016/j.amsu.2022.104695
22. Frutos MC, Origlia J, Gallo Vaulet ML, et al. SARS-CoV-2 and Chlamydia pneumoniae co-infection: a review of the literature. Rev Argent Microbiol. 2022;54(3):247–257. doi:10.1016/j.ram.2022.05.009
23. Laine C, Cotton D. Current clinical challenges in the prevention and management of COVID-19. Ann Intern Med. 2022;175(10):1475. doi:10.7326/M22-2684
24. Branley JM, Weston KM, England J, et al. Clinical features of endemic community-acquired psittacosis. New Microbes New Infect. 2014;2(1):7–12. doi:10.1002/2052-2975.29
25. Balsamo G, Maxted AM, Midla JW, et al. Compendium of measures to control Chlamydia psittaci infection among humans (psittacosis) and pet birds (avian chlamydiosis), 2017. J Avian Med Surg. 2017;31(3):262–282. doi:10.1647/217-265
26. Rybarczyk J, Versteele C, Lernout T, et al. Human psittacosis: a review with emphasis on surveillance in Belgium. Acta Clin Belg. 2020;75(1):42–48. doi:10.1080/17843286.2019.1590889
27. Jeong YJ, Wi YM, Park H, et al. Current and emerging knowledge in COVID-19. Radiology. 2023;306(2):e222462. doi:10.1148/radiol.222462
28. Lewnard JA, Hong VX, Patel MM, et al. Clinical outcomes associated with SARS-CoV-2 Omicron (B.1.1.529) variant and BA.1/BA.1.1 or BA.2 subvariant infection in Southern California. Nat Med. 2022;28(9):1933–1943. doi:10.1038/s41591-022-01887-z
29. Wolter N, Jassat W, von Gottberg A, et al.; DATCOV-Gen author group. Clinical severity of omicron lineage BA.2 infection compared with BA.1 infection in South Africa. Lancet. 2022;400(10346):93–96. doi:10.1016/S0140-6736(22)00981-3
30. Mandell LA, Wunderink RG, Anzueto A, et al. Infectious Diseases Society of America/American Thoracic Society consensus guidelines on the management of community-acquired pneumonia in adults. Clin Infect Dis. 2007;44(suppl 2):S27–S72. doi:10.1086/511159
 © 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2024 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.