Back to Journals » Infection and Drug Resistance » Volume 16
Plasmodium falciparum Genetic Diversity and Resistance Genotype Profile in Infected Placental Samples Collected After Delivery in Ouagadougou
Authors Sawadogo H, Soulama I , Zida A, Zongo C , Sawadogo PM, Guiguemde KT , Nikiema S, Badoum SE, Sawadogo S, Tou A, Sombié S, Tchekounou C , Sermé SS, Ouedraogo-Traoré R, Guiguemdé TR, Savadogo A
Received 5 July 2023
Accepted for publication 4 October 2023
Published 12 October 2023 Volume 2023:16 Pages 6673—6680
DOI https://doi.org/10.2147/IDR.S420004
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Héctor Mora-Montes
Haffsatou Sawadogo,1,2 Issiaka Soulama,3,4 Adama Zida,2,5 Cheikna Zongo,1 Patindoilba Marcel Sawadogo,2,5 Kiswendsida Thierry Guiguemde,5,6 Seni Nikiema,7 Salimata Emilie Badoum,1,8 Salam Sawadogo,7 Aïcha Tou,4 Salif Sombié,4 Chanolle Tchekounou,1,9 Sindié Samuel Sermé,1,8 Rasmata Ouedraogo-Traoré,9 Tinga Robert Guiguemdé,10 Aly Savadogo1
1Laboratory of Applied Biochemistry and Immunology (LABIA), Joseph KI - ZERBO University, Ouagadougou, Burkina Faso; 2Parasitology-Mycology Department, Centre Hospitalier Universitaire Yalgado Ouédraogo (CHU-YO), Ouagadougou, Burkina Faso; 3Health Science Research Institute (IRSS), Ouagadougou, Burkina Faso; 4National Malaria Research and Training Center (CNRFP), Ouagadougou, Burkina Faso; 5Health Sciences Training and Research Unit (UFR/SDS), Joseph KI - ZERBO University, Ouagadougou, Burkina Faso; 6Centre Hospitalier Universitaire Pédiatrique – Charles de Gaulle (CHU-CDG), Ouagadougou, Burkina Faso; 7Molecular Biology and Genetics Laboratory (LABIOGENE), Joseph KI - ZERBO University, Ouagadougou, Burkina Faso; 8Health Action Research Group (GRAS), Ouagadougou, Burkina Faso; 9International Institute of Science and Technology (Iistech), Ouagadougou, Burkina Faso; 10Parasitology-Mycology Laboratory, National Institute of Health Sciences (INSP), Nazi Boni University, Bobo-Dioulasso, Burkina Faso
Correspondence: Cheikna Zongo, Laboratory of Applied Biochemistry and Immunology (LABIA), Joseph KI - ZERBO University, Burkina Faso 03 BP 7021 Ouagadougou 03, Ouagadougou, Burkina Faso, Tel +226 76579605, Fax +226 25307242, Email [email protected]
Purpose: Intermittent preventive treatment with sulfadoxine-pyrimethamine is widely used for the prevention of malaria in pregnant women in Africa. Known resistance cases of sulfadoxine-pyrimethamine during pregnancy need to be follow up to support IPTp implementation in Burkina Faso. However, data on the development and spread of resistance to this molecule are lacking. This study aimed to investigating the genetic diversity of P. falciparum and the mutation prevalence in the dhfr and dhps genes infected from postpartum infected placentas.
Patients and Methods: This was a prospective and cross-sectional study conducted between April 2019 and March 2020 in four health districts of Ouagadougou capital city. From the placentas collected after delivery, P. falciparum detection and mps1 and msp2 polymorphism analysis were performed by nested PCR. The resistance profile was checked after analyzing the mutation point on dhfr and dhps genes.
Results: PCR-positive samples were estimated at 96% for msp1 and 98% for msp2. The polymorphism analysis showed that the RO33 and 3D7 allelic families were the most widespread with 62.5% and 91.83%, respectively. Multiple infections by msp1 and msp2 were frequent with 12.50% and 92.92%, respectively. The prevalence of individual dhfr mutation point, 51I, 108A, and 59R, was 1.96, 15.68, and 7.84, respectively, and the dhps mutation point, 437G, was 3.92. There is no detected mutation at the point 164L and 540E. The triple (51I+108A+59R) in dhfr and quadruple (51I+108A+59R+ 437G) mutation were not found.
Conclusion: The results showed that Plasmodium falciparum has a high genetic diversity of msp1 and msp2. This suggests that dhfr and dhps mutant genotypes are potential early warning factors in the increase in the sulfadoxine-pyrimethamine resistance.
Keywords: sulfadoxine-pyrimethamine, Plasmodium falciparum, msp1, msp2, dhfr, dhps, placenta
Introduction
The number of malaria cases continues to increase worldwide with nearly 247 million cases in 2021, compared to 245 in 2020. Most of this increase was reported in Africa. In 33 countries in the WHO African Region, in 2020, approximately 11.6 million (34%) pregnant women were exposed to malarial infection during pregnancy out of all the 33.8 million pregnancies recorded in the world.1 The malarial occurrence during pregnancy could lead to consequences such as worsening of anaemia in pregnant women, prematurity, perinatal mortality, foetal growth retardation, or low birth weight.2
To reduce the burden of malaria, the World Health Organization (WHO) recommends chemoprevention in pregnant women by administering sulfadoxine-pyrimethamine (SP) during antenatal visits. This intermittent preventive treatment (IPTp) is widely used in areas with moderate and high malarial transmission. Plasmodium falciparum is the most dangerous parasite species due to its strong involvement in the mortality attributed to malaria and its ability to develop drug resistance. Genetically, P. falciparum presents a high polymorphism. There exists a great diversity of Plasmodium strains that infect people living in malarial-endemic areas.3 Parasitic diversity and parasitic antigenic variation are the two main factors responsible for the slow acquisition (several years) of protection against malaria. The study of this genetic diversity takes certain factors into account, in particular the transmission intensity and the host’s immunity. The msp1 and msp2 genes are located on chromosome 9 and chromosome 2, respectively. Variations in the repeat sequences of block 2 (msp1) and the polymorphism of the central block (msp2) allow the parasitic populations to be subdivided into three allelic families K1, MAD20, and RO33 for msp1, and into two allelic families 3D7 and FC273 for msp2.3 The development of malaria vaccine candidates based on the surface proteins MSP1 and MSP2 contributes to further extensively analyze the distinct subpopulations of the parasite during clinical trial.4 At the same time, the resistance of P. falciparum to drugs constitutes a real obstacle to the fight against malaria in endemic countries. Nowadays, the estimation of the level of resistance of P. falciparum to antimalarial drugs on a national scale is done using molecular markers. Data obtained by measuring these markers indicate gene mutations for two enzymes (dihydropteroate synthase (dhps) and dihydrofolate reductase (dhfr)) in Burkina Faso.5 These enzymes are thought to be associated with sulfadoxine resistance and pyrimethamine resistance, respectively.6 However, whether the use of intermittent preventive treatment can greatly affect the spread and distribution of the genotypic resistance profile in pregnant women remains debatable.
This work aimed to study the genetic polymorphism of P. falciparum and the prevalence of the mutation in the dhfr and dhps genes in the placenta after delivery of women living in Ouagadougou, Burkina Faso.
Materials and Methods
Study Site and Period
This is a prospective and cross-sectional study conducted between April 2019 and March 2020 in four health districts of the city of Ouagadougou. The health facilities concerned were Noongr-Massom District Hospital, Schiphra District Hospital, Boulmiougou District Hospital, and Paul VI District Hospital as previously described.7 The climate in the study site is tropical savannah with rainy seasons between June and October.
Study Population
In 2020, the population of the city of Ouagadougou was approximately 2,684,052 inhabitants. The study population was all pregnant women admitted to the health facilities having given their informed consent to participate in the study (n = 630). Those who were infected with HIV were not included in the study. All pregnant women received sulfadoxine-pyrimethamine combination during the antenatal care visits. All doses were administrated at the hospital.
Sample Collection and Laboratory Analysis
The women’s placenta obtained after delivery made it possible to make samples of dried blood (DBS) on Whatman paper and thin smear slides for PCR and microscopy detection, respectively.
The microscopy detection was applied according to the WHO reference method for malaria diagnostic. Thick and thin blood films were prepared on clean and grease-free glass microscope slides with the maternal face blood of the placenta and examined by two microscopists after staining with 5% Giemsa solution for 30 minutes as described previously.7
The QIAamp kit (QIAGEN) was used for parasite DNA extraction on the DBS according to the manufacturer’s instructions. Microscopic examinations were performed on the prepared thin smear slides. The extracted Plasmodium falciparum DNA was either used immediately for the amplification reactions or kept at a temperature of –20 ℃ before being used. The nested polymerase chain reaction (nested PCR) was the DNA amplification technique used based on small subunit ribosomal RNA (ssrRNA) genes.8 The primer sequences for the first (nested) amplification were: rPLU5 5-CCT GTT GTT GCC TTA AAC TTC-3 (forward) rPLU6 5-TTA AAA TTG TTG CAG TTA AAA CG-3 (reverse). The primer sequences for the second (nested) amplification were used rFAL1 5-TTA ACC TGG TTT GGG AAA ACC AAA TAT ATT-3 (forward) rFAL2 5-ACA CAA TGA ACT CAA TCA TGA CTA CCC GTC-3 (reverse).
Applied Biosystem 2720 Thermal cycler tool was used for amplification. A volume of 20 µL of reaction containing 1 µL of genomic DNA, 2 µL of polymerase chain reaction (PCR) buffer 10X, 0.5 µL of 10 µM of each primer, 0.8 µL of 50 mM MgCl2, 1.25 µL of mM of dNTP and 0.1 of 5UI Taq polymerase was used for amplification. The initial denaturation at 95°C was programmed at 5 min, the denaturation at 94°C at 1 min followed by 24 cycles. Each cycle corresponds to a denaturation at 94°C for 1 minute, an annealed primer for 2 min at 58°C, and an extension for 2 minutes at 72°C. The extension was completed by adding an additional 5 minutes of 72°C incubation at the end of the cycle. The 30-cycle condition was set for the second amplification. The msp1 and msp2 typing, extracted DNA was amplified by using sequence-specific primers according to a modified protocol. After the primary amplifications, secondary PCR reactions were performed using allelic primers for Mad20, K1, RO33 (for msp1), FC27 and 3D7 (for msp2).
Then, the amplification of the dhfr and dhps genes was performed by PCR restriction fragment length polymorphism. PCR- positive samples for P. falciparum were analyzed at dhfr (codons 51, 59, 108, and 164) and dhps (codons 436 and 540). The amplicons (DNA fragment amplified by PCR obtained) were analyzed by electrophoresis on agarose gel or stored at +4°C.
Data Collection and Processing
Data on the demographic and clinical information of pregnant women were collected through a structured questionnaire designed for the study. The information covered age, level of education, occupation, marital status, current and previous pregnancies, as well as environmental and living conditions. Information such as the use of intermittent preventive treatment (IPT) and the use of insecticide-treated bed nets (ITN) was also collected.
Statistical Analysis
R software was used to analyze the data. The chi-square was used to compare the proportions, and the student’s test and ANOVA were used for normally distributed continuous data.
Ethical Considerations
The investigations during this study were carried out following the rules of the Helsinki’s Declaration. The protocol of this study was submitted for ethical approval by the National Ethics Committee for Health Research in Burkina Faso (deliberation N° 2019-4-056). The study received the authorization of the chief medical officers of the various health districts, the mayors of the various communes, and the administrative officials of each health structure. Each pregnant woman’s informed and signed consent was obtained. This made it possible for them to participate in the study.
Results
A total of 630 pregnant women participated in the study, and all available samples were systematically considered, including 51 for genotyping. Of the 51 Plasmodium falciparum-positive samples by species nested PCR, 48 were positive for msp1 and 49 were positive for msp2. A flowchart was made to explain the results obtained (Figure 1).
Frequency of msp1 and msp2 Genes Family
The allelic distribution of the msp1 gene (K1, MAD20, and RO33) revealed that there was no statistically significant difference between the frequencies of the allelic families K1 52.08%, RO33 62.50% and MAD20 41.66% (P = 0.1418). However, a predominance of RO33 alleles was noted followed by K1 alleles. Allelic analysis of the msp2 gene (3D7 and FC27) revealed a statistically high frequency of the 3D7 allelic family at 91.83% (P = 0.04039) (Table 1).
 |
Table 1 Prevalence of Allelic Families of msp1 and msp2 Genes |
Frequency of msp1 and msp2 Individual Alleles
A total of 42 alleles were found and classified according to their size in base pairs. Eleven different alleles of the msp1 gene had been obtained with five alleles for K1 (150–200Pb), two alleles for RO33 (150–200Pb), and four for MAD20 (190–400Pb). For the msp2 gene, 27 alleles were found with 15 alleles for 3D7 (100–1000Pb) and 12 alleles for FC27 (240–700Pb).
Polyclonality of infection was assessed for each gene. It corresponds to the proportion of pregnant women with more than one clone (allele) of Plasmodium falciparum. The estimated polyclonality was 12.50% for msp1 and 95.92% for msp2. The study of the polymorphism of the msp1 gene indicated that the number of parasite clones per person in the study population ranged from 1 to 5 clones and the number of parasite clones per person in the msp-2 gene ranged from 1 to 8 clones. As presented in Figure 2, the monoclonal infection was estimated at 87.50% for msp1 and 4.8% for msp2 (Figure 2).
 |
Figure 2 Polyclonal and monoclonal infection. |
Prevalence of dhfr and dhps Gene Mutations
Table 2 represents the proportion of different dhfr and dhps points of mutation. The prevalence of individual dhfr mutation, 51I, 108A, and 59R, was 1.96%, 15.68%, and 7.84%, respectively, and that of dhps, 437G, was 3.92. There was no mutant parasite with 164L or 540E point of mutation (Table 2).
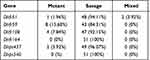 |
Table 2 Proportion of the Different dhfr and dhps Points of Mutations |
Out of the samples that showed mutations, 25.49% (13/51) contained at least one dhfr gene mutation. The dhfr’s gene mutation at codon 59 was the most represented one with a proportion of 15.68% followed by dhfr 108 mutation (7.84%). The double mutation dhfr (51 + 59) and (59+108) were found with a proportion of 3.92% and 1.96%, respectively (Table 3). A level of the dhps gene, only the mutation at point 437 was found with a proportion of 3.92%. We did not find any mutation of dhfr I164L and dhps K540E mutation.
 |
Table 3 Prevalence of Different Mutations |
Discussion
The genetic variation of P. falciparum strains and their complexity in pregnant women using intermittent preventive treatment with sulfadoxine-pyrimethamine were described in our study. Our study showed that the frequency of successfully analyzed samples was higher with msp2 than msp1. All polymorphic profiles of msp1 (K1, MAD20, and RO33) and msp2 (3D7 and FC27) genes were encountered in our study. The RO33 and 3D7 allelic families were the most dominant one followed by the FC27 allelic family, K1 and MAD20. The dominance of RO33 is reported by similar studies conducted in Burkina Faso by Nébié in 2002,9 and by Hamid in Sudan in 2013.10 In contrast, other studies conducted in Burkina Faso by Soulama in 2009 and Somé in 2018,4,11 and Issoufou in 2001 in Bénin12 found a predominance of the K1 allelic family. As regards the dominance of the 3D7 allele of msp-2, there was a consensus with previous studies in Burkina4 in 2018, in Côte d’Ivoire, and in Gabon in 2016.13 On the other hand, they are different from previous studies in Africa. Indeed, the Soulama’s studies in 20099 and Badoum14 in 2019 in Burkina Faso and Hounto’s in Benin in 201315 reported that the K1 and FC27 allelic families were the most predominant one for msp-1 for msp-2 respectively. The predominance of RO33 and 3D7 alleles in our study could be explained by the effect of IPT/SP in favour of an increase in the “biological fitness” of these parasitic clones. In addition, IPT/SP seems to have an effect on allelic variability leading to a predominance of RO33 and 3D7. Finally, it could be explained by the fact that our study population consisted of asymptomatic individuals. Indeed, previous studies have already shown the association of the RO33 and 3D7 alleles in the case of asymptomatic malaria.16 For mixed infections, RO33+MAD20 has the highest frequency followed by K1+RO33. The two genes msp1 and msp2 are highly polymorphic, as there are 11 alleles of the msp1 gene and 27 alleles of the msp2 gene. Similar results are observed in Senegal17 and Benin where 23 msp1 and 12 msp2 alleles are reported.18
As regards mutation associated with resistance to SP, only dhfr59 mutation prevalence was high (61.54%). The high prevalence of dhfr59 mutation was found at 99.9% in Mali19 in 2014. Thus, the study states that this mutation is no longer relevant for monitoring sulfadoxine-pyrimethamine resistance. The other dhfr (51, 108, 164) and dhps (437,540) mutations were poorly represented, negligible, or absent in our study. dhps 437 mutation was 15.38%, whereas a study conducted in Mali by Coulibaly found 21.4%. In Cameroon, Chauvin published a high prevalence of the dhps 437 gene mutation (76.5%).20
In this study, we did not record dhfr 164L and dhps 540E mutations, suggesting that they are still absent in Burkina Faso. This is consistent with previous reports in the country. This result is similar to data reported in other studies in Burkina Faso in pregnant women21,22 on the one hand and in children and in the general population on the other hand.2,23
Our samples did not carry any triple and quadruple mutations. A study by Nikiema in 202224 shows little to no prevalence of triple and quadruple mutation. The quintuple mutation is strongly associated with the clinical failure of SP treatment.25
In Mali, Coulibaly also reported lower rates of triple mutation (4.7%).26 In Nigeria, Iwalokun reported a higher prevalence of triple mutation of 35%.27 In Central Africa in Gabon, Bouyou-Akotet in a survey reported triple mutation and quadruple mutation at high proportions of 80% and 53%, respectively.28 In Cameroon, Chauvin published a high prevalence of the triple mutation at 96%.20
This different reported prevalence is higher than those found in our study area and could be related to drug pressure being higher in these different areas of Africa than in Burkina Faso.29–31 The difference could be related to the higher level of endemicity in these regions than in our study area. Indeed, according to some authors, it is likely that in hyper-endemic areas, resistance spread more rapidly in the population. This is especially true for resistance induced by mutations in a single gene for a given molecule. Even with a multiplicity of clones, there is competition between strains, which will facilitate the recolonization of the organism by mutant strains after the elimination of susceptible strains by chemoprevention.
Conclusion
This study using msp1 and msp2 genes in the population of low parasitemia strains from placentas obtained after delivery confirmed a high polymorphism of these two genes with statistically high frequency of the 3D7 allelic family. The study also showed a high polyclonality using msp2 gene. Also, it is to be noted that there is a contribution to the reduction of the prevalence of Plasmodium falciparum infection by sulfadoxine-pyrimethamine.
Mutations in the Pfdhfr and Pfdhps genes associated with SP resistance were relatively common among pregnant women in Ouagadougou. Nevertheless, the absence of dhfr triple mutation shows the efficacy of intermittent preventive treatment with SP, and molecular markers linked to SP resistance should continue to be monitored.
Acknowledgments
The authors would like to thank pregnant women for their participation and collaboration. The authors would like to thank all the health personnel, particularly the midwives and the administrative staff of the various health structures for their collaboration, and support, in particular Drs OUEDRAOGO Sambo, SANOU Guillaume and SANNI Sekossounon. This study was funded by the West African Research Association.
Author Contributions
All the authors made a significant contribution to the work reported, whether it was in the conception, study design, execution, acquisition of data, analysis, and interpretation, or in all these areas; took part in drafting, revising, or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. World Health Organisation. World malaria report 2021; 2021.
2. Geiger C, Compoaré G, Coulibaly B, et al. Substantial increase in mutations in the genes pfdhfr and pfdhps puts sulfadoxine-pyrimethamine-based intermittent preventive treatment for malaria at risk in Burkina Faso. Trop Med Int Health. 2014;19(6):690–697. doi:10.1111/tmi.12305
3. Sondo P, Bihoun B, Kabore B, et al. Polymorphisme de Plasmodium falciparum et mutations des gènes de résistances Pfcrt et Pfmdr1 dans la zone de Nanoro, Burkina Faso [Polymorphisms in Plasmodium falciparum parasites and mutations in the resistance genes Pfcrt and Pfmdr1 in Nanoro area, Burkina Faso]. Pan Afr Med J. 2021;39(118). doi:10.11604/pamj.2021.39.118.26959
4. Somé AF, Bazié T, Zongo I, et al. Plasmodium falciparum msp1 and msp2 genetic diversity allele frequencies in parasites isolates from symptomatic malaria patients in Bobo-Dioulasso, Burkina Faso. Parasit Vectors. 2018;11:323. doi:10.1186/s13071-2895
5. Cissé M, Awandare GA, Somé FA, Hayette MP, Guiguemdé RT. High concordance of Pfdhfr and Pfdhps genotypes between matched peripheral and placental isolates of delivered women in Bobo-Dioulasso, Burkina Faso. Ann Parasitol. 2017;63(2):111–116. PMID: 28802281. doi:10.17420/ap6302.93
6. Amimo F, Lambert B, Magit A, Sacarlal J, Hashizume M, Shibuya K. Plasmodium falciparum resistance to Sulfadoxine-pyrimethamine in Africa: a systematic analysis of national trends. BMJ Global Health. 2020;e003217. doi:10.1136/bmjgh-2020-003217
7. Haffsatou S, Zida A, Zongo C, et al. Prevalence of placental infection with Plasmodium falciparum detected by a polymerase chain reaction and associated risk factors in women after delivered in Ouagadougou (Burkina Faso). Int J Adv Res. 2021;9(9):132–141. doi:10.21474/IJAR01/13386
8. Snounou G, Viriyakosol S, Zhu XP, et al. High sensitivity of detection of human malaria parasites by the use of nested polymerase chain reaction. Mol Biochem Parasitol. 1993;61(2):315–320. PMID: 8264734. doi:10.1016/0166-6851(93)90077-b
9. Nébié I. Influence à long terme des rideaux imprégnés d’insecticide sur l’immunité anti-palustre chez des enfants vivant en zone rurale au Burkina Faso [Long-term influence of insecticide-impregnated curtains on malaria immunity in children living in rural areas in Burkina Faso] [dissertation]. Burkina Faso: Université de Ouagadougou; 2002.
10. Abdel Hamid MM, Mohammed SB, El hassan IM. Diversité génétique des isolats de terrain de Plasmodium falciparum au Soudan central déduite par génotypage PCR desprotéines de surface de mérozoïte 1 et 2 [Genetic diversity of Plasmodium falciparum field isolates in central Sudan inferred by PCR genotyping of Merozoite Surface Protein 1 and 2]. N Am J Med Sci. 2013;5:95–101.
11. Soulama I, Nébié I, Ouédraogo A, et al. Plasmodium falciparum genotypes diversity in symptomatic malaria of children living in an urban and a rural setting in Burkina Faso. Malar J. 2009;8:135–142. doi:10.1186/1475-2875-8-135
12. Issifou S, Djikou S, Sanni A, Lekoulou F, Ntoumi F. No influence of season of the transmission nor age of patients on the complexity and genetic diversity of Plasmodium falciparum infection in Cotonou, Benin. Bull Soc Pathol Exot. 2001;94(2 Pt 2):195.
13. Yavo W, Konaté A, Mawili-Mboumba DP, et al. Genetic polymorphism of msp1 and msp2 in Plasmodium falciparum Isolates from Côte d’Ivoire versus Gabon. J Parasitol Res. 2016;7. doi:10.1155/2016/3074803
14. Badoum SE, Bougouma E, Sombie S, et al. Relationship between human genetic factors and Plasmodium falciparum genetic diversity of msp1, msp2 and glurp in a malaria endemic area of Burkina Faso. Biomed Genet Genomics. 2019;4(2). doi:10.15761/BGG.1000144
15. Hounto AO, Gazard DK, Ndam N, et al. Genetic polymorphism of merozoite surface protein-1 and merozoite surface protein-2 in Plasmodium falciparum isolates from children in South of Benin. Parasite. 2013;20:37. doi:10.1051/parasite/2013039
16. Al-Yaman F, Genton B, Reeder JC, Anders RF, Smith T, Alpers MP. Reduced risk of clinical malaria in children infected with multiple clones of Plasmodium falciparum in a highly endemic area: a prospective community study. Trans R Soc Trop Med Hyg. 1997;91(5):602. doi:10.1016/S0035-9203(97)90046-8
17. Mercereau-Puijalon O, Ménard D. Plasmodium vivax and the Duffy antigen: a paradigm revisited. Transfus Clin Biol. 2010;17(3):176–183. doi:10.1016/j.tracli.2010.06.005
18. Gunawardena S, Karunaweera ND. Advances in genetics and genomics: use and limitations in achieving malaria elimination goals. Pathog Glob Health. 2015;109(3):123–141. doi:10.1179/2047773215Y.0000000015
19. Mahamar A, Sumner KM, Levitt B, et al. Effect of three years’ seasonal malaria chemoprevention on molecular markers of resistance of Plasmodium falciparum to sulfadoxine-pyrimethamine and amodiaquine in Ouelessebougou, Mali. Malar J. 2022;21:39. doi:10.1186/s12936-022-04059-z
20. Coulibaly SO, Kayentao K, Taylor S, et al. Parasite clearance following treatment with sulphadoxine-pyrimethamine for intermittent preventive treatment in Burkina-Faso and Mali: 42-day in vivo follow-up study. Malar J. 2014;13:41. doi:10.4269/ajtmh.2011.10-0507
21. Chauvin P, Sandie M, Iriart X, et al. Prevalence of Plasmodium falciparum parasites resistant to sulfadoxine/pyrimethamine in pregnant women in Yaoundé, Cameroon: emergence of highly resistant pfdhfr/pfdhps alleles. J Antimicrob Chemother. 2015;70(5). doi:10.1093/jac/dkv160
22. Tahita MC, Tinto H, Erhart A, et al. Prevalence of the dhfr and dhps mutations among pregnant women in rural Burkina Faso five years after the introduction of intermittent preventive treatment with sulfadoxine-pyrimethamine. PLoS One. 2015;10(9):e0137440. doi:10.1371/journal.pone.0137440
23. Tinto H, Ouédraogo JB, Zongo I, et al. Sulfadoxine-pyrimethamine efficacy and selection of Plasmodium falciparum DHFR mutations in Burkina Faso before its introduction as intermittent preventive treatment for pregnant women. Am J Trop Med Hyg. 2007;76(4):608. doi:10.4269/ajtmh.2007.76.608
24. Nikiema S. Etude de l’influence de la chimioprévention du paludisme saisonnier sur la prévalence de l’infection palustre, la diversité génétique du parasite et la prévalence des marqueurs de résistance de Plasmodium falciparum chez les enfants vivant en zone rurale au Burkina Faso [Study of the influence of seasonal malaria chemoprevention on the prevalence of malaria infection, parasite genetic diversity and the prevalence of Plasmodium falciparum resistance markers in children living in rural areas of Burkina Faso]. [Dissertation] [Thèse PhD]. Université Joseph KI-ZERBO Ouagadougou Burkina Faso; 2022.
25. Baba E, Hamade P, Kivumbi H, et al. Effectiveness of seasonal malaria chemoprevention at scale in west and Central Africa: an observational study. Lancet. 2020;396:1829–1840. doi:10.1016/S0140-6736(20)32227-3
26. Iwalokun BA, Iwalokun SO, Adebodun V, Balogun M. Carriage of mutant dihydrofolate reductase and dihydropteroate synthase genes among plasmodium falciparum isolates recovered from pregnant women with asymptomatic infection in Lagos, Nigeria. Med Princ Pract. 2015;24(5):436. doi:10.1159/000430987
27. Bouyou-Akotet MK, Mawili-Mboumba DP, Tchantchou TD, Kombila M. High prevalence of sulfadoxine/pyrimethamine-resistant alleles of Plasmodium falciparum isolates in pregnant women when introducing intermittent preventive treatment with sulfadoxine/pyrimethamine in Gabon. J Antimicrob Chemother. 2010;65(3):
28. Talisuna AO, Bloland P, Alessandro D, Alessandro UD. History, dynamics, and public health importance of malaria parasite resistance. Clin Microbiol Rev. 2004;17(1):
29. Hastings I. Modelling parasite drug resistance: lessons for management and control strategies. Trop Med Int Health. 2001;6:883–890. doi:10.1046/j.1365-3156.2001.00800.x
30. Hastings I, D’Alessandro U. Modelling a predictable disaster: the rise and spread of drug-resistant malaria. Parasitol Today. 2000;16:340. doi:10.1016/S0169-4758(00)01707-5
31. Cisse M, Awandare GA, Soulama A, Tinto H, Hayette MP, Guiguemdé RT. Recent uptake of intermittent preventive treatment during pregnancy with sulfadoxine-pyrimethamine is associated with increased prevalence of Pfdhfr mutations in Bobo-Dioulasso, Burkina Faso. Malar J. 2017;16(1):38. PMID: 28114990; PMCID: PMC5259838. doi:10.1186/s12936-017-1695-1
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

