Back to Journals » OncoTargets and Therapy » Volume 13
Phillygenin, a MELK Inhibitor, Inhibits Cell Survival and Epithelial–Mesenchymal Transition in Pancreatic Cancer Cells
Authors Li H, Chen M, Yang Z, Wang Q, Wang J, Jin D, Yang X, Chen F, Zhou X, Luo K
Received 17 November 2019
Accepted for publication 24 March 2020
Published 3 April 2020 Volume 2020:13 Pages 2833—2842
DOI https://doi.org/10.2147/OTT.S238958
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Hongchun Li,1 Miao Chen,2 Zhuying Yang,3 Qinxian Wang,1 Jiesheng Wang,1 Dong Jin,1 Xiuyun Yang,1 Fuxing Chen,1 Xiumin Zhou,4 Kexue Luo1
1Department of Cadre Health, Tongde Hospital of Zhejiang Province, Hangzhou 310012, Zhejiang, People’s Republic of China; 2Department of Oncology, Tongde Hospital of Zhejiang Province, Hangzhou 310012, Zhejiang, People’s Republic of China; 3Department of Gastroenterology, Tongde Hospital of Zhejiang Province, Hangzhou 310012, Zhejiang, People’s Republic of China; 4Department of Pharmacy, Tongde Hospital of Zhejiang Province, Hangzhou 310012, Zhejiang, People’s Republic of China
Correspondence: Kexue Luo
Department of Cadre Health, Tongde Hospital of Zhejiang Province, 234 Gucui Road, Hangzhou 310012, Zhejiang, People’s Republic of China
Tel +86 571 89972437
Email [email protected]
Introduction: Pancreatic cancer (PC) is one of the leading causes of cancer, with the lowest 5-year survival rate of all cancer types. Given the fast metastasis of PC and its resistance to surgery, radiotherapy, chemotherapy, and combinations thereof, it is imperative to develop more effective anti-PC drugs. Phillygenin (PHI) has been reported to exert anti-cancer, anti-bacterial, and anti‐inflammatory properties. However, the mechanism of PHI in the development of PC is still unclear.
Methods: The cytotoxicity of PHI in pancreatic cancer cells was evaluated by MTT assay, and clonogenic assay was used to test the anti-proliferation of PHI. The pro-apoptotic effect of PHI was detected by flow cytometry analysis. The changes of epithelial–mesenchymal transition (EMT) in pancreatic cancer cells treated with PHI were determined by Western blot. Transwell assay was used to test the migration and invasion of PC cells after treatment with PHI. Molecular docking was used to predict the potential binding site of candidate target with PHI.
Results: PHI could inhibit the proliferation, migration, and EMT of PC cells (PANC-1 and SW1990) and induce its apoptosis. Analysis of the Cancer Genome Atlas database indicated that elevated MELK levels correlated with poor overall survival (OS) and disease-free survival (DFS) of PC patients. In addition, molecular modeling showed that PHI may potentially target the catalytic domain of maternal embryonic leucine zipper kinase (MELK). Overexpression of MELK muted the anti-PC effects of PHI.
Conclusion: PHI holds promise as a potent candidate drug for the treatment of PC via targeted MELK.
Keywords: pancreatic cancer, phillygenin, MELK, proliferation, EMT, apoptosis
Introduction
Pancreatic cancer (PC) is one of the most lethal malignancies. Its median survival time after diagnosis is less than 6 months and the five-year survival rate is less than 6%.1,2 Currently, the most effective treatment for PC patients is surgery. However, fewer than 20% of PC patients are eligible for surgical resection because the disease has already reached an advanced stage at the time of diagnosis. At present, chemotherapy is a key treatment to prevent recurrence and prolong the survival of patients with metastatic or advanced PC.3 Notwithstanding, chemotherapy is also known to have some serious side effects on patients, such as loss of appetite, constipation, vomiting, nausea, etc. In light of this situation, there is an urgent need to develop new therapies with high efficiency and low toxicity.
Many studies have shown that maternal embryonic leucine zipper kinase (MELK) plays a critical role in the proliferation and development of cancer cells.4 As a result, MELK has become a potential molecular target for cancer therapy, including liver cancer and breast cancer.5,6 A well-known MELK inhibitor, OTS167, has been reported to exhibit effective growth suppression in mice xenograft models for a number of cancer types, including lung cancer, kidney cancer, and acute myeloid leukemia.7–11 Currently, some clinical studies, such as NCT01910545 and NCT02926690, are being performed to evaluate the therapeutic potential of OTS167.12,13 For example, Chung and his colleagues have shown that an orally administrative MELK-targeting inhibitor could suppress the growth of tumor-initiating cells and has the potential to be widely used for the treatment of a variety of cancer types, including PC.14 As the role of MELK in PC remains largely unclarified, their study opens up the possibility for further studies to explore this issue.
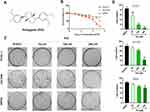
Phillygenin (PHI, Figure 1A), a lignan extracted from Osmanthus fragrans flower, has been reported to have many biological functions, such as anti-inflammation and anti-tumor effects.15,16 PHI could induce mitochondrial-mediated apoptosis and ROS production, and inhibit the nuclear factor kappa B (NF-κB) signaling pathway.16 In addition, in vitro experiments have shown that PHI could also inhibit NF-κB signaling-mediated macrophage inflammation.15 However, there is little research on the role of PHI in tumors or its effect on PC cells. In this study, we examined the effect of PHI on PC using cultured PC cells; meanwhile, the target of PHI in PC was validated using bioinformatics analysis, molecular modeling, and experimental assays.
Methods and Materials
Compound, Antibodies, Plasmids, and siRNA
PHI was obtained from the National Institutes for Food and Drug Control (China, Figure 1A). PHI was dissolved in 0.1% DMSO with 0.1% DMSO as vehicle control. Antibodies against MELK, Vimentin, E-cadherin, and GAPDH were procured from Cell Signaling Technology (Danvers, MA). p-CMV6-MELK and p-CMV6-control plasmids were purchased from OriGene (USA). MELK siRNA was purchased from GenePharma (China).
Cell Lines
Human PC cell lines (PANC-1 and SW1990) and normal pancreatic cells (HPNE) were purchased from the Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (China). Cells were cultured in RPMI-1640 medium (Gibco, Eggenstein, Germany) supplemented with 10% heat-inactivated fetal bovine serum (Gibco), 100 U/mL penicillin, and 100 μg/mL streptomycin. Cells were treated with different concentrations of PHI for different time periods as indicated.
MTT Assay
To measure cell viability after exposure to PHI, cells were seeded in 96-well plates at a density of 3000 per well and allowed to attach overnight. Cells were then exposed to different concentrations of PHI for 24 h. Following the treatment, 10 μL MTT reagent was added through fresh media and cells were cultured for an additional 4 h. 150 μL DMSO was used to dissolve the formazan product and absorbance was measured at 450 nm using SpectraMax M5 microplate reader (Molecular Devices, USA). The percent inhibition was calculated as [1 − (treated/control)] × 100.
Clonogenic Growth Assay
Cells were seeded at 500 cells per well in 6-well plates and treated with PHI for 12 h. Cells were grown in normal growth media for 10 days and stained with crystal violet solution (0.5 in 25% methanol) to assess colony growth. A colony is defined as a cluster of at least 50 cells that can often be determined only through the microscope.
Flow Cytometric Analysis of Apoptosis
The expression of phosphatidylserine on the cell membrane (an indicator of apoptotic cells) was detected using the Annexin V-FITC apoptosis detection kit. The cells were briefly harvested and washed twice with PBS, gently resuspended in Annexin V binding buffer, and incubated with Annexin V-FITC in the dark for 10 min. They were then analyzed by flow cytometry using Cell Quest software (BD Biosciences, USA).
Transwell Migration and Invasion Assay
Transwell chamber: 24-well, 8.0-μm pore membranes (Corning USA) were used according to the manufacturer’s protocol. 1 × 105 cells per well were seeded in the upper chamber in 100 μL of serum-free medium, while 600 μL of complete medium was added simultaneously as a chemoattractant to the lower chamber. After 24 h incubation at 37°C, the remaining cells at the upper surface of the membrane were removed with cotton swabs, and the cells on the lower surface of the membrane were the migrated cells. After staining with 4% paraformaldehyde and 0.1% crystal violet solution, the cells passing through the filter were photographed with an inverted fluorescence microscope. The transwell invasion assay was performed as described above, except that 100 μL of 1:8 DMEM-diluted Matrigel (BD, USA) was added to each well at 37°C for 6 h. Then, the cells were seeded onto the membrane and incubated for 48 h.
Western Immunoblot Analysis
A total of 30–50 μg of cell and tissue lysates were separated by 10% SDS-PAGE and electrotransferred to a PVDF membrane. Membranes were blocked for 1.5 h in Tris-buffered saline containing 0.05% Tween20 and 5% non-fat milk. The PVDF membrane was then incubated with specific primary antibodies. Immunoreactive bands were detected by incubating with a secondary antibody conjugated with horseradish peroxidase and enhanced chemiluminescence reagent (Bio-Rad). The amount of the proteins was analyzed using Image J analysis software version 1.38e and normalized to their respective control.
Molecular Modeling of MELK Bound with PHI
The crystal coordinates for human MELK (PDB code: 5K00) were retrieved from the RCSB Brookhaven Protein Data Bank (PDB) database.17 The missing hydrogen atoms, loop segments, and residues were constructed using Chimera software.18 Then, the LeDock software was used to estimate the binding mode between PHI and MELK.19 During docking calculations, 200 conformations were generated for molecular dynamics (MD) simulations using the optimal binding energy conformation. Prior to MD simulation, the partial atomic charges of PHI were estimated by the restrained electrostatic potential (RESP) method on the basis of HF/6-13G* basis set. Afterwards, the ff14SB force field and General Amber Force Field (GAFF) were assigned to MELK and PHI, respectively.20,21 The complex was then immersed into a water box with TIP3P water model extending 12 Å from any solute atom. Finally, counter ions were placed into the water box to neutralize the system.
Two-stage minimizations were performed to relax the system. First, 5000 steps of steepest descent (SD) followed by 5000 steps of conjugate gradient (CG) were utilized to remove the bad contacts between solvent and solute. Then, the bad contacts of the whole system were removed using10,000 steps of SD, followed by 10,000 steps of CG. The system was then heated up from 0 to 310 K in 400 ps and equilibrates with 1000 ps in the isothermal isobaric (NPT) ensemble. Finally, 300 ns MD simulation was performed at a constant temperature and pressure of 310 K and 1 atm to produce trajectory. Coordinates were recorded every 10 ps for the subsequent numerical analysis using CPPTRAJ module in AmberTools 18 package.22 The last 100 ns MD simulation trajectory with 1000 snapshots were employed for binding free energy calculations and per-residue decomposition using molecular mechanics/generalized Born surface area (MM/GBSA) method.23,24
Enzyme-Linked Immunosorbent Assay for MELK Antigen Detection
MELK activity was detected using the MELK Human ELISA detection kit (No. SEH665Hu, Cloud-Clone Corp., China), according to manufacturer’s protocol.
Cell Transfections for Gene Silencing and Overexpression
MELK siRNA duplexes used for the silencing study were purchased from Genepharma (China) and sequenced as follows: 5ʹ-GACAUCCUAUCUAGCUGCA-3ʹ. Negative Control (NC) was used as the control. p-CMV6-MELK and p-CMV6-control plasmids were used for the overexpression study. PANC-1 cells (2×105/well) were seeded in 6-well plates and cultured for 24 h, and were then transfected with first group: siRNA duplexes against human MELK or control siRNA, and second group: p-CMV6-MELK and p-CMV6-control plasmids by lipofectamine 3000 (Invitrogen) according to manufacturer’s protocol.
Statistical Analysis
The results are presented as means ± SEMs. In GraphPad Prism 7.0, different groups were compared using one-way analysis of variance (ANOVA), with the differences statistically significant at P<0.05.
Results
PHI Effectively Suppressed Cell Growth and Induced Apoptosis in Pancreatic Cells
First, we determined the effect of PHI on cell viability of PANC-1, SW1990 and HPNE. Using the MTT assay, PHI treatment significantly decreased the cell viability of PANC-1, SW1990, and HPNE in 24 h (Figure 1B). Then, we confirmed this finding by assessing the colony-forming ability of PC cells after PHI treatment. Indeed, our results showed that PHI caused a dose-dependent decrease in colony formation of PANC-1, SW1990, and HPNE cells (Figure 1C and D). Flow cytometry assay further indicated that PHI could significantly induce apoptosis in PANC-1 and SW1990 cells (Figure 2A and B). However, HPNE cells were not sensitive to PHI treatment. The above results suggest that PHI exhibits potent anti-PC activity in a dose-dependent manner by inducing apoptosis of PC cells. In addition, 100 μm PHI and PANC-1 cells were selected for subsequent experiments due to the toxicity and sensitivity of PHI.
PHI Effectively Suppressed PANC-1 Cells Migration and Invasion
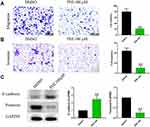
Cancer cells are highly migratory and invasive, especially in PCs, where cancer cells are further induced to metastasize.25 Thus, we investigated the effect of PHI on PANC-1 cells. Using transwell assay, the results showed that PHI could inhibit PANC-1 migration and invasion (Figure 3A and B). Epithelial–mesenchymal transition (EMT) is a serious consequence of excessive migration and invasion of cancer cells. To investigate whether PHI affects the expression of genes that regulate an EMT, we investigated the expression of the epithelial (E‐cadherin) and mesenchymal markers (Vimentin) in PC cells. As shown in Figure 3C, compared to the DMSO group, PANC-1 cells treated with PHI significantly reduced Vimentin and increased E-cadherin expressions. These results confirm that PHI has a protective effect on cell migration, invasion, and EMT.
MELK Is Highly Expressed in Human PC and Involved in Survival Curve
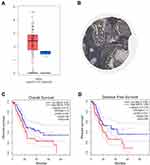
Previous studies have shown that MELK is a candidate drug target that may be related to the development of PC.26 In this study, we analyzed MELK expression in PC using the TCGA database. As shown in Figure 4A, MELK expression was higher in the tumor group than in the normal group. Then, the immunohistochemistry (IHC) of PC slide also showed strong expression of MELK (Figure 4B). The survival curves further revealed that MELK was negatively correlated with both overall survival (OS) and disease-free survival (DFS) in PC patients (Figure 4C and D). These results suggest that MELK may be a potential target for PC therapy.
PHI Might Target the Catalytic Domain of MELK
In this study, we first applied molecular docking to the construction of the initial binding pose between MELK and PHI. As shown in Figure 5A, the docking results indicated that PHI could bind efficiently to the catalytic domain of MELK. In order to explore the stability and dynamic behavior of molecular docking, the binding mode of molecular docking prediction was submitted to 300 ns molecular dynamics (MD) simulations. Firstly, the root-mean-square deviations (RMSDs) of the protein backbone atoms (Cα) of MELK, and the heavy atoms of PHI were monitored to investigate the stability of the simulated complex. As shown in Figure 5B, both the RMSDs of Cα of MELK and PHI have relatively small fluctuations after 100 ns MD simulation, indicating that we obtained a series of stable conformations via MD simulations. To highlight the key residues during the binding of PHI to MELK, the binding-free energies for each residue were decomposed based on the molecular mechanics/generalized Born surface area (MM/GBSA) method. As shown in Figure 5C, the most contributed residues were Ile-17, Phe-151, Val-25, Leu-139, Leu-27, Ala-38, Ile-139, Tyr-88, Leu-86, and Gly-92. Structural analysis showed that the predominant residues were hydrophobic amino acid around the PHI (Figure 5D). Next, using ELISA assay, PHI was shown to have a strong inhibitory effect on MELK in a dose-dependent manner (Figure 5E).
Overexpression of MELK Reduced PHI-Induced Cytotoxicity
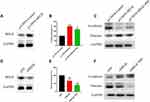
Furthermore, we confirmed that MELK is involved in PHI-mediated PC cell cytotoxicity. To this end, we modulated MELK expression through overexpression of plasmid and siRNA, and then exposed the cells to PHI. First, we ensured successful transfer of the p-CMV6-MELK plasmid to PANC-1 cells (Figure 6A). Overexpression of MELK in PANC-1 cells decreased PHI-induced cytotoxicity (Figure 6B). Meanwhile, PHI-inhitied EMT effects were strongly blocked after overexpression of MELK (Figure 6C). Then, we determined the effects of siRNA-mediated depletion of MELK on human PC cells. Knockdown of MELK by siRNA markedly attenuated MELK expression in PANC-1 cells (Figure 6D). The silencing of MELK in PANC-1 cells increased the cytotoxicity of PHI (Figure 6E), thereby increasing PHI-induced EMT levels (Figure 6F). These results suggest that PHI may be a candidate inhibitor of MELK in PC.
Discussion and Conclusion
As one of the most common and lethal forms of cancer in the world, PC remains a huge global health concern. Among the existing therapies, systemic chemotherapy is still the main treatment for PC patients.27,28 Over the years, the use of chemotherapeutic drugs, such as administration schedules and frequency of toxicity monitoring, has led to steady improvements in patient management.29 However, there remains an urgent need to discover and develop novel, effective, and non-toxic agents for patient care. Although gemcitabine-based chemotherapy has improved survival in PC patients, the improvements are still limited.1,27 A number of factors could account for the limited performance of chemotherapy, including the extrinsic or intrinsic resistance to conventional therapies.30 Therefore, increasing the sensitivity of cancer cells to chemotherapeutic agents and finding new therapeutic targets can improve the survival of patients. One strategy toward this end may be traditional Chinese medicine (TCM), which has shown great promise in PC-related research and clinical studies. The existing literature has already documented good anti-tumor effects with more than 50 kinds of TCM in the treatment of PC, like oblongifolin C, oridonin, and Wumei.31–33
PHI is a phenylethanoid glycoside isolated from the Chinese medicinal Osmanthus fragrans flower, which has been proven to have various pharmacological properties, such as anti-oxidation, hypolipidemic inhibition, and inhibition of low-density lipoprotein oxidation.34,35 However, the mechanism of PHI-mediated anti-tumor action has not been fully elucidated. In this study, we found that PHI dose-dependently decreased PANC-1 and SW1990 cells viability by MTT assay and induced PANC-1 and SW1990 cells apoptosis by flow cytometric analysis. In addition, PHI significantly inhibited the migration, invasion and EMT progression of PANC-1 cells.
Another interesting finding is that we first discovered that PHI may directly bind to MELK in PC cells, leading to cell proliferation and migration. MELK is expressed in stem/progenitor cells in some adult tissues and is involved in a variety of biological processes, including the control of cell proliferation. Chung et al found that the silencing of MELK significantly decreased the development of PC.26 Some research shows that MELK promotes CDK1 involvement in cell cycle and cell progression in cancers.36 Through molecular modeling, we predicted that PHI might bind to the catalytic binding site of MELK and inhibit the development and progression of EMT PC cells. However, the mechanisms underlying these processes remain to be further clarified and should become the focus of future studies.
In summary, we first investigated and reported the anti-PC effects of PHI and its underlying mechanisms. We found that PHI reduced the growth of PC cells through inhibition of MELK. These changes led to the arrest of PC cells and induction of apoptosis. These results indicate that TCM-PHI holds great promise as a potential candidate agent for the treatment of PC. In addition, our results also suggest that MELK activation could be a potential target for the development of new anti-cancer agents.
Acknowledgment
The authors gratefully acknowledge financial support from Zhejiang Province Bureau of Health (2019RC029, 2019KY350) and Zhejiang Province Bureau of Traditional Chinese Medicine (2017ZA029, 2020ZA024).
Disclosure
The authors disclose no potential conflicts of interest in this work.
References
1. van Huijgevoort NCM, Del Chiaro M, Wolfgang CL, van Hooft JE, Besselink MG. Diagnosis and management of pancreatic cystic neoplasms: current evidence and guidelines. Nat Rev Gastroenterol Hepatol. 2019;16(11):676–689. doi:10.1038/s41575-019-0195-x
2. Wei MY, Shi S, Liang C, et al. The microbiota and microbiome in pancreatic cancer: more influential than expected. Mol Cancer. 2019;18(1):97. doi:10.1186/s12943-019-1008-0
3. Luo W, Yang G, Qiu J, et al. Novel discoveries targeting gemcitabine-based chemoresistance and new therapies in pancreatic cancer: how far are we from the destination? Cancer Med. 2019;8(14):6403–6413. doi:10.1002/cam4.2384
4. Pitner MK, Taliaferro JM, Dalby KN, Bartholomeusz C. MELK: a potential novel therapeutic target for TNBC and other aggressive malignancies. Expert Opin Ther Targets. 2017;21(9):849–859. doi:10.1080/14728222.2017.1363183
5. Xia H, Kong SN, Chen J, et al. MELK is an oncogenic kinase essential for early hepatocellular carcinoma recurrence. Cancer Lett. 2016;383(1):85–93. doi:10.1016/j.canlet.2016.09.017
6. Giuliano CJ, Lin A, Smith JC, Palladino AC, Sheltzer JM. MELK expression correlates with tumor mitotic activity but is not required for cancer growth. eLife. 2018;7:e32838.
7. Stefka AT, Park JH, Matsuo Y, et al. Anti-myeloma activity of MELK inhibitor OTS167: effects on drug-resistant myeloma cells and putative myeloma stem cell replenishment of malignant plasma cells. Blood Cancer J. 2016;6(8):e460. doi:10.1038/bcj.2016.71
8. Chung S, Kijima K, Kudo A, et al. Preclinical evaluation of biomarkers associated with antitumor activity of MELK inhibitor. Oncotarget. 2016;7(14):18171–18182. doi:10.18632/oncotarget.7685
9. Kato T, Inoue H, Imoto S, et al. Oncogenic roles of TOPK and MELK, and effective growth suppression by small molecular inhibitors in kidney cancer cells. Oncotarget. 2016;7(14):17652–17664. doi:10.18632/oncotarget.7755
10. Alachkar H, Mutonga MB, Metzeler KH, et al. Preclinical efficacy of maternal embryonic leucine-zipper kinase (MELK) inhibition in acute myeloid leukemia. Oncotarget. 2014;5(23):12371–12382. doi:10.18632/oncotarget.2642
11. Inoue H, Kato T, Olugbile S, et al. Effective growth-suppressive activity of maternal embryonic leucine-zipper kinase (MELK) inhibitor against small cell lung cancer. Oncotarget. 2016;7(12):13621–13633. doi:10.18632/oncotarget.7297
12. ClinicalTrials.gov. Phase 1 Study of OTS167 in Patients with Solid Tumors. Available from: https://clinicaltrials.gov/ct2/show/NCT01910545.
13. ClinicalTrials.gov. Safety study of MELK inhibitor to treat patients with advanced breast cancer and triple negative breast cancer. Available from: https://clinicaltrials.gov/ct2/show/NCT02926690.
14. Chung S, Suzuki H, Miyamoto T, et al. Development of an orally-administrative MELK-targeting inhibitor that suppresses the growth of various types of human cancer. Oncotarget. 2012;3(12):1629–1640. doi:10.18632/oncotarget.790
15. Hu N, Wang C, Dai X, et al. Phillygenin inhibits LPS-induced activation and inflammation of LX2 cells by TLR4/MyD88/NF-kappaB signaling pathway. J Ethnopharmacol. 2019;248:112361.
16. He J, Wei W, Yang Q, Wang Y. Phillygenin exerts in vitro and in vivo antitumor effects in drug-resistant human esophageal cancer cells by inducing mitochondrial-mediated apoptosis, ROS generation, and inhibition of the nuclear factor kappa B NF-κB signalling pathway. Med Sci Monit: Intl Med J Exp Clin Res. 2019;25:739–745. doi:10.12659/MSM.913138
17. Chen X, Giraldes J, Sprague ER, et al. “Addition” and “subtraction”: selectivity design for type II maternal embryonic leucine zipper kinase inhibitors. J Med Chem. 2017;60(5):2155–2161. doi:10.1021/acs.jmedchem.7b00033
18. Yang Z, Lasker K, Schneidman-Duhovny D, et al. UCSF Chimera, MODELLER, and IMP: an integrated modeling system. J Struct Biol. 2012;179(3):269–278. doi:10.1016/j.jsb.2011.09.006
19. Zhao H, Caflisch A. Discovery of ZAP70 inhibitors by high-throughput docking into a conformation of its kinase domain generated by molecular dynamics. Bioorg Med Chem Lett. 2013;23(20):5721–5726. doi:10.1016/j.bmcl.2013.08.009
20. Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11(8):3696–3713. doi:10.1021/acs.jctc.5b00255
21. Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi:10.1002/jcc.20035
22. Roe DR, Cheatham TE. PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data. J Chem Theory Comput. 2013;9(7):3084–3095. doi:10.1021/ct400341p
23. Genheden S, Ryde U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin Drug Discov. 2015;10(5):449–461. doi:10.1517/17460441.2015.1032936
24. Wang E, Sun H, Wang J, et al. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem Rev. 2019;119(16):9478–9508. doi:10.1021/acs.chemrev.9b00055
25. Stuelten CH, Parent CA, Montell DJ. Cell motility in cancer invasion and metastasis: insights from simple model organisms. Nat Rev Cancer. 2018;18(5):296–312. doi:10.1038/nrc.2018.15
26. Chung CH, Miller A, Panopoulos A, et al. Maternal embryonic leucine zipper kinase regulates pancreatic ductal, but not β-cell, regeneration. Phys Rep. 2014;2(9):e12131. doi:10.14814/phy2.12131
27. Neoptolemos JP, Kleeff J, Michl P, Costello E, Greenhalf W, Palmer DH. Therapeutic developments in pancreatic cancer: current and future perspectives. Nat Rev Gastroenterol Hepatol. 2018;15(6):333–348. doi:10.1038/s41575-018-0005-x
28. Halbrook CJ, Lyssiotis CA. Employing metabolism to improve the diagnosis and treatment of pancreatic cancer. Cancer Cell. 2017;31(1):5–19. doi:10.1016/j.ccell.2016.12.006
29. Mulkerin DL, Bergsbaken JJ, Fischer JA, Mulkerin MJ, Bohler AM, Mably MS. Multidisciplinary optimization of oral chemotherapy delivery at the University of Wisconsin Carbone Cancer Center. J Oncol Pract. 2016;12(10):e912–e923. doi:10.1200/JOP.2016.013748
30. Gebregiworgis T, Bhinderwala F, Purohit V, Chaika NV, Singh PK, Powers R. Insights into gemcitabine resistance and the potential for therapeutic monitoring. Metabolomics. 2018;14(12):156. doi:10.1007/s11306-018-1452-7
31. Li Y, Xi Z, Chen X, et al. Natural compound Oblongifolin C confers gemcitabine resistance in pancreatic cancer by downregulating Src/MAPK/ERK pathways. Cell Death Dis. 2018;9(5):538. doi:10.1038/s41419-018-0574-1
32. Wang B, Shen C, Li Y, et al. Oridonin overcomes the gemcitabine resistant PANC-1/Gem cells by regulating GST pi and LRP/1 ERK/JNK signalling. Onco Targets Ther. 2019;12:5751–5765. doi:10.2147/OTT.S208924
33. Wan Y, Xu L, Liu Z, et al. Utilising network pharmacology to explore the underlying mechanism of Wumei Pill in treating pancreatic neoplasms. BMC Complement Altern Med. 2019;19(1):158. doi:10.1186/s12906-019-2580-y
34. Song W, Ao MZ, Shi Y, Yuan LF, Yuan XX, Yu LJ. Interaction between phillygenin and human serum albumin based on spectroscopic and molecular docking. Spectrochim Acta A Mol Biomol Spectrosc. 2012;85(1):120–126. doi:10.1016/j.saa.2011.09.044
35. Song W, Wu J, Yu L, Peng Z. Evaluation of the pharmacokinetics and hepatoprotective effects of phillygenin in mouse. Biomed Res Int. 2018;2018:7964318. doi:10.1155/2018/7964318
36. Ganguly R, Mohyeldin A, Thiel J, Kornblum HI, Beullens M, Nakano I. MELK-a conserved kinase: functions, signaling, cancer, and controversy. Clin Transl Med. 2015;4(1):11. doi:10.1186/s40169-014-0045-y
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.